Abstract
Endothelial abnormalities play a critical role in the pathogenesis of malaria caused by the human pathogen, Plasmodium falciparum. In serious infections and especially in cerebral malaria, red blood cells infected with the parasite are sequestered in small venules in various organs, resulting in endothelial activation and vascular occlusion, which are believed to be largely responsible for the morbidity and mortality caused by this infection, especially in children. We demonstrate that after incubation with infected red blood cells (iRBCs), cultured human umbilical vein endothelial cells (HUVECs) contain parasite protein, genomic DNA, and RNA, as well as intracellular vacuoles with apparent parasite-derived material, but not engulfed or adherent iRBCs. The association of this material with the HUVECs is observed over 96 hours after removal of iRBCs. This phenomenon may occur in endothelial cells in vivo by the process of trogocytosis, in which transfer of material between cells depends on direct cell contact. This process may contribute to the endothelial activation and disruption involved in the pathogenesis of cerebral malaria.
Keywords: Malaria, Plasmodium falciparum, endothelial cells, trogocytosis
Introduction
Plasmodium falciparum is the most lethal malarial pathogen, with a majority of deaths occurring in infected children in Africa. Although P. falciparum can infect a high proportion of red blood cells, the level of parasitemia correlates poorly with the severity of disease caused in children, which may be uncomplicated or may present as severe anemia, severe malaria, or the most lethal presentation of cerebral malaria. The interaction between endothelial cells and parasite-infected erythrocytes plays a major role in the pathogenesis of human malaria caused by P. falciparum. Following infection of the erythrocyte, the parasite produces adhesion molecules including P. falciparum erythrocyte membrane protein 1 (PfEMP1) located in “knobs” on the surface of infected red blood cells (iRBCs) that allow the iRBCs to adhere to the vascular wall through interaction with a series of endothelial cell surface proteins. This adhesion results in peripheral sequestration of parasites and vascular occlusion accompanied by endothelial cell activation [1-5]. In experiments using human umbilical vein endothelial cells (HUVECs), this activation occurs rapidly, and requires direct contact between the iRBCs and HUVECS; thus, activation is not due to soluble factors from iRBCs acting on the HUVECs without cell-cell contact [6,7]. Sequestered vascular wall-bound iRBCs are protected, as they are not cleared from circulation by the spleen. There are multiple var forms of PfEMP1 in the P. falciparum genome (which vary among parasite isolates), each of which interact preferentially with different endothelial cell surface receptors; some of these endothelial cell receptors are preferentially expressed in different organs (reviewed in [8]). The PfEMP1 receptors on endothelial cells include intercellular adhesion molecule 1 (ICAM-1), endothelial protein C receptor (EPCR) and CD36. Intercellular chondroitin sulfate A (CSA) in the placenta appears to be the receptor for the PfEMP1 molecules in strains causing malaria of pregnancy [9]. Tissue binding of iRBCs appears to be critical to the pathogenesis of cerebral malaria in children, where the main endothelial receptors in the brain vasculature are ICAM-1 and EPCR [10]. Analysis of PfEMP1 expression in children with malaria has shown that var alleles in which the CIDRα1 domain binds to EPCR are the main and consistent driver for development of severe disease, including the severe malaria, severe anemia, and cerebral malaria syndromes [11]. Adhesion of iRBCs leads to increased expression of pro-inflammatory cytokines by the endothelial cells [12] and transfer of iRBC membrane components and cytoplasmic malaria antigens, aldolase, and histidine rich protein 2 (HRP-2), into the endothelial cells [13]. These processes may be an important mechanism by which malaria induces the dysfunction of the endothelial barrier and activation of endothelial cells [14-16].
Mammalian endothelial cells are non-professional phagocytes and antigen presenting cells which can incorporate a variety of pathogens. Various fungi, including Aspergillus fumigatus [17,18] as well as zymosan particles [19] can be taken up into HUVECs, as can bacterial pathogens such as Staphylococcus aureus [20] along with many others. The result of this uptake varies with the pathogen; endothelial cell uptake of viable pathogens can shield them from antimicrobial agents or host immune responses, and can alter gene expression or function of the endothelial cells. It has been suggested that this process plays a role in pathogen persistence [18] or in disease pathogenesis and dissemination [21].
Cell-cell interactions in the immune system can occur by a contact-mediated transfer of membrane components between cells during a brief contact in a process termed trogocytosis [22,23]. This process involves the transfer of membrane fragments and associated molecules from one cell to another. Endothelial cells can apparently engage in trogocytosis with other cells. In addition to the role of this process in immune cell interactions, trogocytosis can also spread bacterial pathogens between cells in a manner which shields them from the immune system; this has been observed for the transfer of Francisella tularensis and Salmonella enterica between infected and uninfected macrophages [24]. Using brain endothelial cell cultures, it has been shown that P. falciparum antigenic material and membrane lipids can be transferred from iRBCs to endothelial cells in a trogocytosis-like process, which occurs relatively rapidly and requires cell-cell contact [14]. This process may be responsible for the changes which occur in the endothelium in response to sequestration of iRBCs in malaria. Much work has been done on the interaction with iRBCs with endothelial cells in vivo and in culture systems. However, relatively little is known about the mechanism by which parasite-derived materials are transferred to the endothelial cells in this process. In a study of the interaction of iRBCs infected with various strains of P. falciparum with different endothelial cell lines, photomicrographs show iRBCs bound to endothelial cells [25]. Although not explicitly commented upon, the micrographs also show many apparent darkly staining bodies which are not within RBCs overlying the endothelial cells; it cannot be determined if this material is adherent to the endothelial cell surface or internal, but this raises the possibility that intracellular and/or membrane associated parasite-derived particulate material might be transferred from iRBCs to endothelial cells.
In this study, we investigated the ability of P. falciparum to interact with human endothelial cells in culture. Association of parasite-derived material with HUVECs was observed by immunofluorescence, electron microscopy, and quantitative PCR (qPCR) after culture with iRBC. This work suggests that parasites or subcellular fragments derived from them can associate with and persist in endothelial cells, apparently as free particles not obviously within iRBCs.
Materials and Methods
Cell Culture
Pooled HUVECs were obtained from Lonza (Walkersville, MD). HUVECs were grown in endothelial basal medium 2 (EBM-2) supplemented with EGM-2 SingleQuots (Lonza). Cells were maintained at 37°C and 5 percent CO2. HUVECs were used before passage six. P. falciparum 3D7 cells expressing green fluorescent protein (3D7HT-GFP) [26] were obtained from the Malaria Research and Reference Reagent Resource Center (MR4, http://www.mr4.org/). Cells were grown in P. falciparum culture medium, containing RPMI 1640 with 25 mM HEPES (Sigma-Aldrich, Saint Louis, MO) supplemented with 0.5 percent Albumax (GIBCO, Carlsbad, CA), sodium bicarbonate (2 g/l), 0.1 mM hypoxanthine, and gentamycin (50 μg/l). Albumax is an inexpensive substitute for human serum in P. falciparum culture medium, which has less batch-to-batch variation than does serum [27], although it may not be suitable for drug sensitivity studies [27,28]. Albumax has been widely used for culture of P. falciparum. Human erythrocytes were obtained from Innovative Research (Novi, MI). Purified human RBCs were added to P. falciparum cultures at 2 percent hematocrit. Culture flasks were gassed with 90 percent N2, 5 percent O2, and 5 percent CO2 and incubated at 37°C.
HUVEC Parasite Association Assay
HUVEC cell cultures were grown to 80 percent confluence in EBM-2 medium (37°C, 5 percent CO2). 3D7-GFP P. falciparum-iRBCs, at ~10 percent parasitemia level were added to HUVEC cultures at 0.25 percent hematocrit in 50 percent EBM-2 media and 50 percent P. falciparum culture medium. As a control, uninfected RBCs were added to HUVEC cultures. Flasks were gassed with 90 percent N2, 5 percent O2, 5 percent CO2 and cell cultures were incubated at 37°C. For the 0 hour exposure, iRBC (or uninfected RBCs in controls) were added to HUVECs and the cells were immediately washed three times in an equal volume of RPMI to remove iRBCs and RBCs. For the 4 hour and 24 hour time points, iRBCs and RBCs were removed from adherent HUVEC cultures after 4 or 24 hours, respectively, and HUVECs were washed. For the 48 hour and 96 hour time points, iRBCs and RBCs were removed after 24 hours of co-incubation and HUVEC cultures were washed to thoroughly remove iRBCs. Fresh medium consisting of 50 percent EBM-2 medium and 50 percent P. falciparum culture medium was added to the cultures. Flasks were gassed with 90 percent N2, 5 percent O2, 5 percent CO2 and cell cultures were incubated at 37°C for an additional 24 or 72 hours, respectively.
Giemsa Staining
HUVECs were fixed in 100 percent methanol for 1 minute. Fixed cells were air dried, then stained with Giemsa stain for 30 minutes. Stained cells were visualized using a 1000X oil immersion lens.
Fluorescence Detection of Parasites
Adherent HUVEC cells in culture flasks were washed three times with RPMI. Cells were fixed in culture flasks with 2 percent formaldehyde in RPMI for 2 hours at room temperature, then in 2 percent formaldehyde, 0.0075 percent glutaraldehyde in 1x PBS overnight at 4°C. Fixed cells were washed three times with PBS and blocked with 1X PBS, 3 percent BSA, 0.3 percent Triton X-100 for 1 hour at room temperature. Parasite-derived proteins were detected with either P. falciparum-specific anti-MSP1 (ATCC/MR4) or anti-aldolase (Abcam, Cambridge, UK) primary antibody. Cells were incubated with primary antibody diluted in dilution buffer (1X PBS, 1 percent BSA, 0.3 percent Triton X-100, 0.01 percent sodium azide) for 1 hour at room temperature, and then washed three times with PBS. Alexa Fluor 546 anti-rabbit IgG (Invitrogen, Grand Island, NY) secondary antibody was diluted 1:1000 in dilution buffer, added to cells, followed by incubation in the dark for 1 hour at room temperature. Cells were washed three times with PBS. Prolong Gold Antifade Reagent with DAPI (Invitrogen) was added to cells. Cells were visualized by fluorescence microscopy using 600X magnification.
Electron Microscopy
RBCs and iRBCs were removed from culture flasks and HUVECs were washed with RPMI then removed from the flasks with TypLE Express (GIBCO). HUVECs and/or RBCs (1X106) were fixed in 2.5 percent gluteraldehyde/4 percent paraformaldehyde for 30 min at room temperature. Cells were pelleted, then dehydrated in graded series alcohol, and embedded in epon resin. Thin sections (90 nm) were collected on copper grids and stained with uranyl acetate and lead citrate. Grids were visualized at 80 kV on a JEOL (Peabody, MA) 1200 electron microscope.
Nucleic Acid Purification and qPCR
Following the HUVEC invasion assay, adherent HUVEC cells in T-75 culture flasks were washed three times with RPMI. Total RNA and genomic DNA (gDNA) were purified from HUVEC cells using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. For RNA purification, total RNA was treated with DNase I (NEB) at 37°C for 10 min, then DNase I was inactivated at 75°C for 10 min in the presence of 5 mM EDTA. cDNA was prepared from 2 μg of DNase-treated RNA using the Clontech (Mountain View, CA) RNA to cDNA EcoDry Premix (Double Primed). qPCR experiments were carried out using RT2 SYBR Green/ROX qPCR Master Mix (SA Biosciences, Frederick, MD). Briefly, 250 ng of cDNA or total RNA + gDNA was added to a 25 μl reaction containing 1X Master Mix, and 400 nM forward and reverse primers. Reactions were run in a Stratagene Mx3005P system (Agilent Technologies, Santa Clara, CA). Cycling conditions were as follows: 95°C for 10 min for cDNA synthesis, 45 cycles 95°C for 10 sec, 55°C for 30 sec, and 72°C for 30 sec. Following the PCR program, melting curve analysis was run from 55°C to 99°C. PCR products were separated by 1.5 percent agarose gel electrophoresis to confirm the presence of only one amplified PCR product. cDNA and gDNA were amplified using primers specific for P. falciparum small subunit (SSU) rRNA [29] (Forward 5’-TCTAGGGGAACTATTTTAGCTT-3’; Reverse 5’-CACAGTAAATGCTTTAACTGTT-3’) and human GAPDH (Forward 5’-ATGGGGAAGGTGAAGGTCG-3’; Reverse 5’-GGGGTCATTGATGGCAACAATA-3’). All rRNA-related PCR products had the expected Tm, although the bands for the 48 hour and 96 hour samples were faint and diffuse. Primers for GAPDH were generated using PrimerBank [30]. Relative RNA levels were calculated using a standard curve. Relative P. falciparum gDNA and rRNA amounts were normalized to human GAPDH amounts for each sample.
Results
P. falciparum from Infected RBCs can Associate with HUVECs in Culture
HUVECs were incubated with iRBC for 0, 4, or 24 hours. Following incubation, iRBCs were removed and the cultures were washed as described in Materials and Methods. Following these washes, very few RBCs were observed in the cultures by light microscopy (Figure 1). For 48 hour and 96 hour time points, HUVECs were incubated with iRBCs for 24 hours, washed, and incubation was continued. When HUVECs were exposed to iRBC for 4 or 24 hours, Giemsa-positive bodies about the size of merozoites were seen over the cytoplasm of the HUVECs after 4 hours and remained present for up to 96 hours (Figure 2). Such bodies were not seen in HUVECs without treatment or in those exposed to uninfected RBCs or exposed to iRBCs for “0 hour” time (at least 100 cells examined under each of these control conditions).
Figure 1.
Removal of RBCs from HUVEC cells by washing. HUVEC cultures were incubated with P. falciparum-infected RBCs for 24 hours. After 24 hours, RBCs were removed and washed 3 times with RPMI. Images show HUVEC culture before removal of RBCs (0 washes) and HUVEC culture after 1, 2, or 3 washes.
Figure 2.
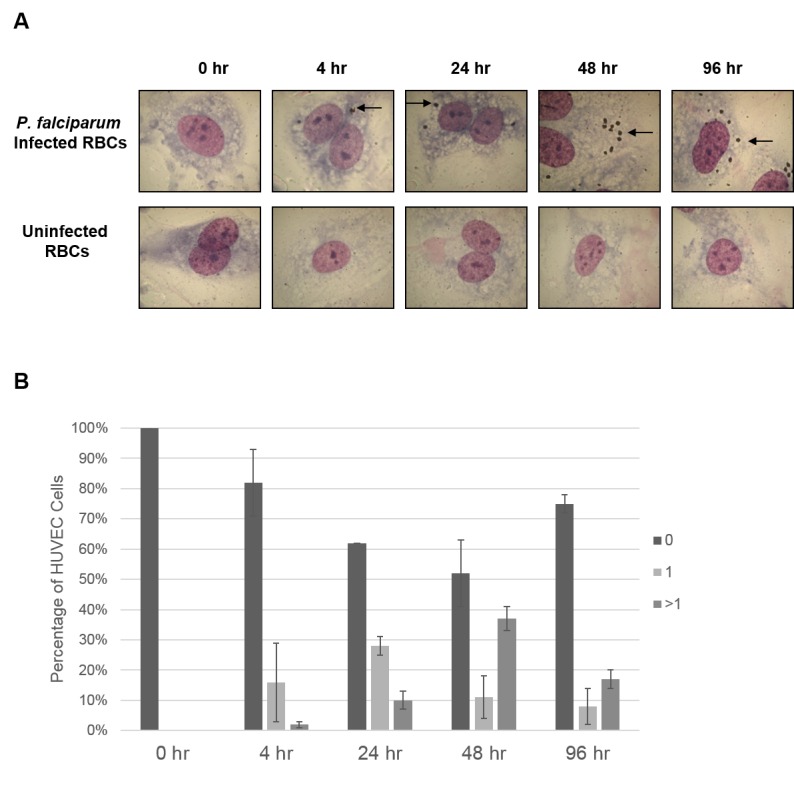
Detection of P. falciparum associated with HUVECs by Giemsa staining. P. falciparum iRBCs or uninfected RBCs were added to HUVEC cell cultures at the indicated times, as described in Materials and Methods. A. Following the HUVEC association assay, cells were stained with Giemsa. HUVECS incubated with P. falciparum iRBCS (top row) show multiple parasite-like bodies (arrow). No such bodies are seen in HUVECs incubated with uninfected RBCs (bottom row). B. The percentage of HUVEC cells containing 0, 1, or > 1 parasite-like bodies was calculated for each time point.
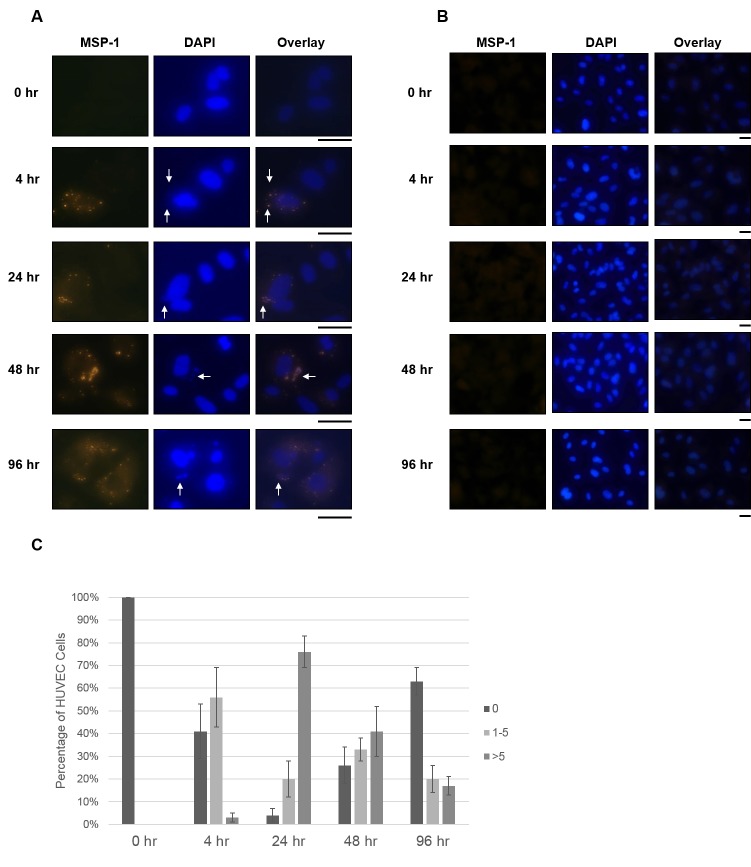
The HUVEC cultures were then stained with 4’, 6-diamidino-2-phenylindole (DAPI) and an antibody specific for the parasite protein merozoite surface protein 1 (MSP-1), a highly processed membrane-anchored surface protein abundant on merozoites of P. falciparum [31]. Fluorescence analysis of these images demonstrated parasites present over the HUVECs, but not elsewhere on the slides. DAPI staining showed DNA over the endothelial cell cytoplasm, which overlapped with MSP-1 expression, indicating the presence of parasite-derived particles (Figure 3A). This fluorescence was not seen with HUVECs incubated with uninfected RBCs (Figure 3B).
Figure 3.
Detection of P. falciparum associated with HUVECs by MSP-1 staining. P. falciparum iRBCs (A) or uninfected RBCs (B) were added to HUVEC cell cultures for the indicated times, as described in Materials and Methods. A. Following incubation with HUVECs, P. falciparum-derived particles were detected by immunofluorescence assay with α-MSP1 primary antibody (first column). DAPI staining shows nucleic acid of HUVECs and parasite-like bodies (indicated by arrows, second column). Overlays of MSP1 and DAPI staining shows overlap of MSP1 and DAPI fluorescence (arrows). B. HUVECs incubated with uninfected RBCs showed no spots of bright fluorescence from MSP1 staining (first column). DAPI staining shows HUVEC nucleic acid, but no smaller spots corresponding to parasite nucleic acid were observed (second column). The black bars below the micrographs indicated 25 μm. C. The percentage of HUVEC cells containing 0, 1 to 5, or > 5 P. falciparum-derived particles was calculated for each time point.
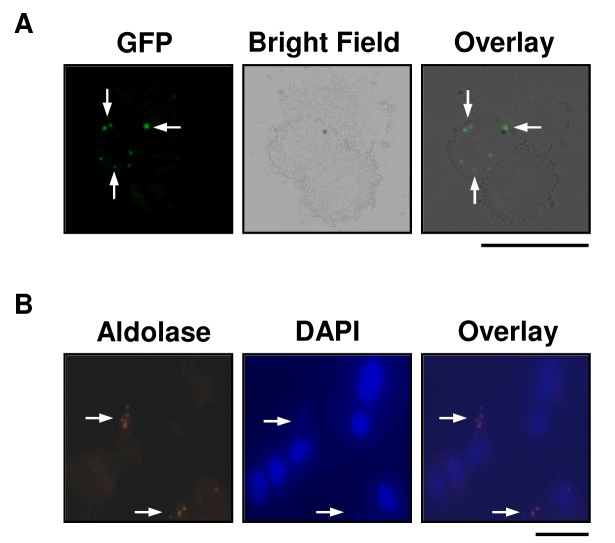
For HUVEC binding experiments, we used the P. falciparum strain, 3D7HT-GFP, which expresses green fluorescent protein (GFP). Parasite-GFP expression was observed in live HUVECs after 24 hours of exposure to iRBCs (Figure 4A). The cells were then fixed and stained with DAPI as well as an antibody recognizing P. falciparum aldolase (Figure 4B). Parasite-like particles expressing GFP were observed only over the cytoplasm of HUVECs and not elsewhere on the slides. Co-staining of intracellular particles with DAPI as well as for aldolase was observed. The HUVEC nuclei stained with DAPI but did not express GFP or malaria aldolase (Figure 4B). These results again indicate that the parasite-derived particles are localized over the cytoplasm of the HUVECs.
Figure 4.
Detection of P. falciparum-derived particles associated with HUVECs by GFP-expression and aldolase staining. A. HUVEC cultures were incubated with RBCs infected with GFP-expressing P. falciparum for 24 hours. After 24 hours, RBCs were removed, fresh media was added, and cultures were incubated an additional 24 hours. HUVECs were washed with RPMI and removed from flask with TypLE Express (GIBCO). Live cells were placed on a slide and coverslip and viewed for GFP-expression. Bright field and GFP overlay shows GFP fluorescence is localized over the HUVEC cytoplasm. B. P. falciparum infected RBCs were added to HUVEC cell cultures for 24 hours. Following this incubation, P. falciparum-derived protein was detected by immunofluorescence assay with α-aldolase primary antibody (first panel). DAPI staining shows nucleic acid of HUVECs and parasites (parasites indicated by arrows) (second panel). Overlays of aldolase and DAPI staining show overlap of aldolase and DAPI fluorescence (arrows). Black bars indicate 25 μm.
Electron Microscopy Shows Parasite-derived Forms within HUVECs
Electron microscopy was used to further define the morphology of the parasite-derived structures associated with the HUVECs. Although aberrant intracellular structures were noted within multiple HUVECs after exposure to iRBCs (Figure 5), no clear evidence of active infection of these cells was seen (compare with iRBCs, Figure 5, top row). Similar structures were not seen in HUVECs incubated with uninfected RBCs (Figure 5, bottom row). Of note, bound or internalized erythrocytes were not evident in endothelial cells, suggesting that the bound particles are not inside iRBCs bound to the endothelial cells, although we cannot determine whether binding of iRBCs to the HUVECs is required for association of these parasite-derived particles. However, these results are consistent with trogocytosis being responsible for the transfer of this parasite-derived material from iRBCs to HUVECs.
Figure 5.
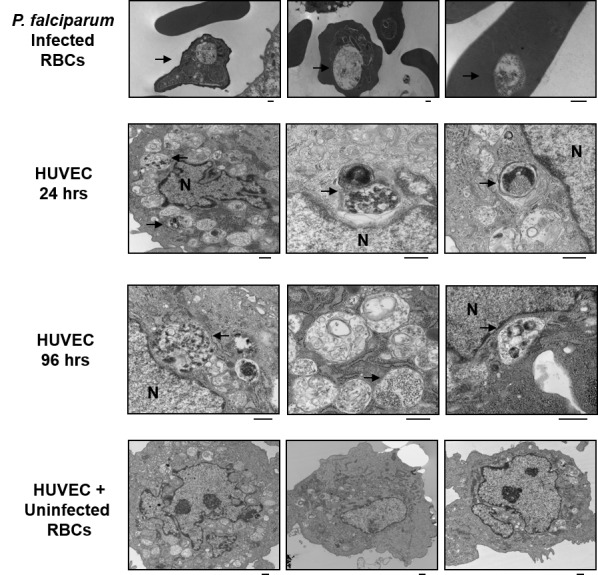
Visualization of HUVECs after incubation with iRBCs by electron microscopy. (Top row) P. falciparum cultures were grown in RBCs and fixed for electron microscopy. (Second row) HUVEC cultures were incubated with RBCs infected with GFP-expressing P. falciparum for 24 hours. (Third row) For 96 hr cultures, RBCs were removed after 24 hours, fresh medium was added, and cultures were incubated an additional 72 hours. (Bottom row) Aberrant structures seen only in HUVECs incubated with iRBCs are indicated by arrows. As a control, HUVECs were incubated with uninfected RBCs for 24 hours. No such aberrant structures were seen in these cells. HUVEC nucleus is indicated (N). Black scale bar represents 500 nm.
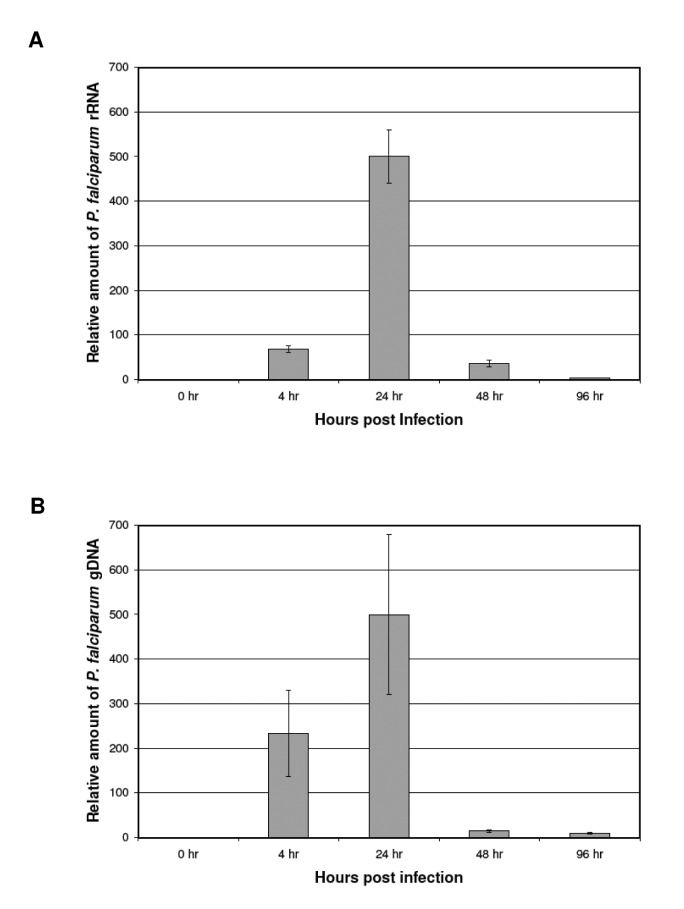
Levels of Parasite DNA and rRNA Increase during Culture of HUVECs Exposed to iRBCs
In order to determine whether the putative parasite-derived particles are metabolically active and replicating after association with HUVECs, we quantitated parasite genomic DNA and RNA by qPCR following exposure to iRBCs (Figure 6). To assess the presence of parasite DNA, the P. falciparum small subunit (SSU) ribosomal RNA (rRNA) gene was amplified from genomic DNA isolated from HUVEC cultures following exposure to iRBCs. Parasite SSU rRNA levels were measured by reverse-transcription qPCR using purified total RNA. The levels of parasite DNA and rRNA both increased up to 24 hours as HUVECs were incubated with iRBCs. After 24 hours, iRBCs were removed from the HUVEC cultures. The amount of plasmodium DNA and RNA present associated with the HUVECs dropped after iRBCs were removed, but was still detected after 96 hours in culture. This indicates that parasite-derived material persists in association with the endothelial cells, although there is no evidence of replication or metabolic activity. As a control, HUVECs were incubated with uninfected RBCs. No P. falciparum-specific gDNA or RNA was detected in these samples, however human GAPDH levels were normal (data not shown). In addition, no PCR products were detected in RNA samples that were not treated with reverse transcriptase, indicating the observed products were not due to contaminating gDNA.
Figure 6.
Measurement of P. falciparum rRNA and DNA associated with HUVECs by SYBR Green quantitative PCR. P. falciparum infected RBCs were added to HUVEC cell cultures for the indicated times, as described in Materials and Methods. Following these incubations, RNA and genomic DNA were purified from HUVECS. A. P. falciparum SSU rRNA amounts from HUVECs as measured by reverse-transcription qPCR. No PCR products were detected in RNA samples not treated with reverse-transcriptase (data not shown). Error bars represent standard deviation from three independent replicates. B. The SSU rRNA gene was amplified from P. falciparum genomic DNA (gDNA) from HUVECs and measured by qPCR. P. falciparum RNA and DNA amounts were normalized to human GAPDH RNA and DNA amounts, respectively. As a control, HUVECs were incubated with uninfected RBCs. No P. falciparum-specific PCR products were detected (data not shown). All experiments were done in triplicate.
Discussion
In this study, we demonstrate that P. falciparum-derived particles can bind to and persist in association with human endothelial cells after co-incubation with iRBCs. After incubation with iRBCs, these particles are evident in proximity to the endothelial cell cytoplasm within 4 hours and multiple parasite-derived bodies can be seen associated with an increasing number of cells over the next 24 hours during co-incubation. Levels of parasite DNA and rRNA present in these endothelial cell cultures increase until removal of iRBCs and decreased over the next 72 hours. These results are consistent with the transfer of cellular or subcellular parasite-derived material to the endothelial cells, and not just individual molecules of parasite products. In addition to the presence of plasmodium rRNA and gDNA, other products of the parasite such as recombinant GFP, aldolase and MSP-1 were observed associated with the HUVECs. Electron microscopy revealed aberrant morphology of the parasite-exposed endothelial cells which appear to contain necrotic material which could be parasite-derived, although the mechanism of entry and exact nature of this material cannot be determined unambiguously. The dependence of endothelial cell activation on direct iRBC-HUVEC cell contact [6,7] resembles the dependence on such contact of trogocytosis [22-24]. Future studies should determine whether the transfer of parasite-derived material to endothelial cells is, indeed, due to trogocytosis. We suggest that such transfer of parasite material from iRBCs to endothelial cells may be a process involved in endothelial cell activation, since both processes involve direct cellular contact. This hypothesis will require further testing.
The binding of iRBCs to endothelial and other cells in small blood vessels has long been known to be part of the pathogenesis of severe and cerebral malaria caused by P. falciparum. Previous work [25] has shown that cultured iRBCs containing several strains of P. falciparum, including strain 3D7 which was used in our experiments, bind poorly to endothelial cells in culture, including human brain endothelial cells (HBEC), human dermal endothelial cells (HDMEC) and human pulmonary microvascular endothelial cells (HPMEC). However, after four to seven rounds of selection by binding to and elution from endothelial cells, variants of three of the four P. falciparum strains tested showed many-fold enhanced binding to all three endothelial cell lines. The only malaria strain failing to show enhanced binding was strain Dd2, which is knobless and is known to have reduced cytoadherence [4,5]. Although it is noted that tumor necrosis factor (TNF) can activate endothelial cells, it was not required for their binding assay or selection [25]; similarly we did not utilize TNF in our studies. However, it would be of interest to test its effect on incorporation of parasite-derived material into endothelial cells. Based on our studies, it appears internalization or binding of parasite-derived bodies from iRBCs is constitutive in HUVECs, unlike the three endothelial cell lines previously tested. In our experiments, we saw parasite-derived bodies associated with the HUVECs, but did not see bound iRBCs. Although not commented on previously [25], in that work iRBCs appear visible on the HBECs, but parasite-like particles on HDMECs and HPMECs morphologically resemble our micrographs, in which iRBCs bound to the HUVECs are not visible.
It should be noted that our results were only obtained with experiments with one line of P. falciparum, 3D7, and with one type of endothelial cell, HUVEC. Other parasite and endothelial cell types should be tested to determine the generality of these results, although they are consistent with the literature cited. It should also be noted that our cell culture medium contained Albumax rather than human serum. It has been reported that cultivation of P. falciparum in Albumax results in fewer and smaller knobs on the iRBCs, and that those Albumax-cultured iRBCs show reduced adherence to endothelial receptors [32]. However, either growth medium resulted in iRBCs with knobs and capable of adhesion, and the effect of the medium was less on strain 3D7, used in our work, than on strain FCR3S1.2 of P. falciparum. We also look forward to future work correlating the morphological findings of this report with changes in gene expression in endothelial cells interacting with iRBCs.
Our data suggest that the parasite-derived particles derived from iRBCs can bind to and remain associated with endothelial cells. Interaction between infected RBCs and cerebral endothelium causes activation leading to iRBC sequestration [15] as well as up-regulation of pro-inflammatory pathways including NF-kB [11] and Tissue Factor [16]. However, it is unknown whether parasites or parasite-derived particles associated with endothelial cells might play a role in the changes in endothelial cell gene expression that are seen during human malaria. The internalization of parasite-derived particles into endothelial cells could thus contribute to the pathological changes seen in severe disease caused by P. falciparum.
Acknowledgments
This work was supported by a Grand Challenges Explorations Award (OPP 1008357) to MJL from the Bill and Melinda Gates Foundation. Partial support (AES) from NIH National Institute of General Medical Sciences, Research Centers in Minority Institutions Award 8G12 MD 007600, is also acknowledged. Debabarata Banerjee (Rutgers Cancer Institute of New Jersey) provided valuable discussions during this work. Raj Patel (Core Imaging Laboratory, Department of Pathology, Rutgers Robert Wood Johnson Medical School) provided superb technical assistance with electron microscopy. We are grateful to Manuel Llinas (Princeton University), Purnima Bhanot (Rutgers New Jersey Medical School), Ghislaine Mayer (Manhattan College), Shirley Luckhart (University of California-Davis), and Eduardo Alves (Sao Paulo University, Brazil) for their encouragement, advice, and technical assistance.
Glossary
- HUVEC
human umbilical vein endothelial cells
- DNA
deoxyribonucleic acid
- rRNA
ribosomal ribonucleic acid
- PCR
polymerase chain reaction
- RT
reverse transcriptase
- PfEMP1
Plasmodium falciparum erythrocyte membrane protein 1
- iRBC
infected red blood cell
- HRP-2
histidine-rich protein 2
- qPCR
quantitative PCR
- gDNA
genomic DNA
- cDNA
complementary DNA
- ICAM-1
intercellular adhesion molecule 1
- EPCR
endothelial protein C receptor
- CSA
chondroitin sulfate A
- DAPI
4’,6-diamidino-2-phenylindole
- MSP-1
merozoite surface protein 1
- HBEC
human brain endothelial cells
- HDED
human dermal endothelial cells
- HPMEC
human pulmonary microvascular endothelial cells
- TNF
tumor necrosis factor
Author's note:
At the time much of this work was done, the affiliations of the authors were as noted: CU, MJL: Department of Molecular Genetics, Microbiology & Immunology, Rutgers Robert Wood Johnson Medical School, Piscataway, NJ; CU, JWG, MJL: Rutgers Cancer Institute of New Jersey, New Brunswick, NJ; JWG: Department of Pharmacology and Department of Pediatrics, Rutgers Robert Wood Johnson Medical School, Piscataway, NJ.
References
- Cooke BM, Morris-Jones S, Greenwood BM. et al. Mechanisms of cytoadhesion of flowing, parasitized red blood cells from Gambian children with falciparum malaria. Am J Trop Med Hyg. 1995;53:29–35. [PubMed] [Google Scholar]
- Rowe JA, Claessens A, Corrigan RA. et al. Adhesion of Plasmodium falciparum-infected erythrocytes to human cells: molecular mechanisms and therapeutic implications. Expert Rev Mol Med. 2009;11:e16. doi: 10.1017/S1462399409001082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendt AR, Simmons DL, Tansey J. et al. Intercellular adhesion molecule-1 is an endothelial cell adhesion receptor for Plasmodium falciparum. Nature. 1989;341:57–59. doi: 10.1038/341057a0. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Hasler T, Morehead K. et al. Plasmodium falciparum –infected erythrocyte receptor(s) for CD36 and thrombospondin are restricted to knobs on the erythrocyte surface. J Histochem Cytochem. 1992;40:1419–1422. doi: 10.1177/40.9.1380530. [DOI] [PubMed] [Google Scholar]
- Herricks T, Meher A, Rathod PK. Deformability limits of Plasmodium falciparum-infected red blood cells. Cell Microbiol. 2009;11:1340–1353. doi: 10.1111/j.1462-5822.2009.01334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viebig NK, Wulbrand U, Förster R. et al. Direct activation of human endothelial cells by Plasmodium falciparum-infected erythrocytes. Infect Immun. 2005;73:3271–3277. doi: 10.1128/IAI.73.6.3271-3277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty SJ, Carret C, Nash GB. et al. Altered phenotype and gene transcription in endothelial cells, induced by Plasmodium falciparum-infected red blood cells: Pathogenic or protective? Int J Parasitol. 2007;37:975–987. doi: 10.1016/j.ijpara.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JD, Rowe JA, Higgins MK. et al. Malaria’s deadly grip: Cytoadhesion in Plasmodium falciparum infected erythrocytes. Cell Microbiol. 2013;15:1976–1983. doi: 10.1111/cmi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M, Duffy PE. Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science. 1996;272:1502–1504. doi: 10.1126/science.272.5267.1502. [DOI] [PubMed] [Google Scholar]
- Avril M, Bernabeu M, Benjamin M. et al. Interaction between endothelial protein C receptor and intercellular adhesion molecule 1 to mediate binding of Plasmodium falciparum-infected erythrocytes to endothelial cells. mBio. 2016;7:e000615–e000616. doi: 10.1128/mBio.00615-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen JS, Wang CW, Mkumbaye SI. et al. Plasmodium falciparum var genes expressed in children with severe malaria encode CIDRα1 domains. EMBO Molec Med. 2016;8:839–850. doi: 10.15252/emmm.201606188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi AK, Sha W, Shulaev V. et al. Plasmodium falciparum-infected erythrocytes induce NF-kappaB regulated inflammatory pathways in human cerebral endothelium. Blood. 2009;114:4243–4252. doi: 10.1182/blood-2009-06-226415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genrich GL, Guarner J, Paddock CD. et al. Fatal malaria infection in travelers: novel immunohistochemical assays for the detection of Plasmodium falciparum in tissues and implications for pathogenesis. Am J Trop Med Hyg. 2007;76:251–259. [PubMed] [Google Scholar]
- Jambou R, Combes V, Jambou MJ. et al. Plasmodium falciparum adhesion on human brain microvascular endothelial cells involves transmigration-like cup formation and induces opening of intercellular junctions. PLoS Pathog. 2010;6:e1001021. doi: 10.1371/journal.ppat.1001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges DJ, Bunn J, van Mourik JA. et al. Rapid activation of endothelial cells enables Plasmodium falciparum adhesion to platelet-decorated von Willebrand factor strings. Blood. 2010;115:1472–1747. doi: 10.1182/blood-2009-07-235150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francischetti IM, Seydel KB, Monteiro RQ. et al. Plasmodium falciparum-infected erythrocytes induce tissue factor expression in endothelial cells and support the assembly of multimolecular coagulation complexes. J Thromb Haemost. 2007;5:155–165. doi: 10.1111/j.1538-7836.2006.02232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris S, Boisvieux-Ulrich E, Crestani B. et al. Internalization of Aspergillus fumigatus conidia by epithelial and endothelial cells. Infect Immun. 1997;65:1510–1514. doi: 10.1128/iai.65.4.1510-1514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylnka JA, Moore MM. Uptake of Aspergillus fumigatus conidia by phagocytic and nonphagocytic cells in vitro: quantitation using strains expressing green fluorescent protein. Infect Immun. 2002;70:3156–3163. doi: 10.1128/IAI.70.6.3156-3163.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langeggen H, Namork E, Johnson E. et al. HUVEC take up opsonized zymosan particles and secrete cytokines IL-6 and IL-8 in vitro. FEMS Immunol Med Microbiol. 2003;36:55–61. doi: 10.1016/S0928-8244(03)00033-6. [DOI] [PubMed] [Google Scholar]
- Menzies BE, Kourteva I. Staphylococcus aureus alpha-toxin induces apoptosis in endothelial cells. FEMS Immunol Med Microbiol. 2000;29:39–45. doi: 10.1111/j.1574-695X.2000.tb01503.x. [DOI] [PubMed] [Google Scholar]
- Menzies BE, Kourteva I. Internalization of Staphylococcus aureus by endothelial cells induces apoptosis. Infect Immun. 1998;66:5994–5998. doi: 10.1128/iai.66.12.5994-5998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caumartin J, Favier B, Daouya M. et al. Trogocytosis-based generation of suppressive NK cells. EMBO J. 2007;26:1423–1433. doi: 10.1038/sj.emboj.7601570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner H, Frank C, Dechant C. et al. Intimate cell conjugate formation and exchange of membrane lipids precede apoptosis induction in target cells during antibody-dependent, granulocyte-mediated cytotoxicity. J Immunol. 2007;179:337–345. doi: 10.4049/jimmunol.179.1.337. [DOI] [PubMed] [Google Scholar]
- Steele S, Radlinski L, Taft-Benz S. et al. Trogocytosis-associated cell to cell spread of intracellular bacterial pathogens. eLife. 2016;5:e10625. doi: 10.7554/eLife.10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessens A, Rowe JA. Selection of Plasmodium falciparum parasites for cytoadhesion to human brain endothelial cells. J Vis Exp. 2012;59:e3122. doi: 10.3791/3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talman AM, Blagborough AM, Sinden RE. Plasmodium falciparum strain expressing GFP throughout the parasite’s life-cycle. PLoS One. 2010;5:e9156. doi: 10.1371/journal.pone.0009156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringwald P, Solange Meche F, Bickii J. et al. In vitro culture and drug sensitivity assay of Plasmodium falciparum with nonserum substitute and acute-phase serum. J Clin Microbiol. 1999;37:700–705. doi: 10.1128/jcm.37.3.700-705.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basco LK. Molecular epidemiology of malaria in Cameroon. XVII. Baseline monitoring of atovaquone-resistant Plasmodium falciparum by in vitro drug assays and cytochrome B gene sequence analysis. Am J Trop Med Hyg. 2003;69:179–183. [PubMed] [Google Scholar]
- Bell AS, Ranford-Cartwright LC. A real-time PCR assay for quantifying Plasmodium falciparum infections in the mosquito vector. Int J Parasitol. 2004;34:795–802. doi: 10.1016/j.ijpara.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Spandidos A, Wang X, Wang H. et al. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2010;38:D792–D799. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinski M, Dluzewski AR. A mechanistic approach to merozoite invasion of red blood cells: Merozoite biogenesis, rupture, and invasion or erythrocytes. In: Sherwin I, editor. Molecular Approaches to Malaria. Washington, DC: ASM Press; 2005. pp. 113–168. [Google Scholar]
- Tilly K-A, Thiede J, Metwally N. et al. Type of in vitro cultivation influences cytoadhesion, knob structure, protein localization and transcriptome profile of Plasmodium falciparum. Sci Rep. 2015;5:16766. doi: 10.1038/srep16766. [DOI] [PMC free article] [PubMed] [Google Scholar]