Abstract
Objective: Elite Controllers or Suppressors (ES) are patients who control HIV replication without antiretroviral therapy. In this study, we compared baseline and inducible HIV-1 mRNA levels in CD4+ T cells from ES and chronic progressors (CPs) receiving suppressive antiretroviral therapy. Methods: We quantified basal levels of cell associated HIV-1 mRNA in CD4+ T cells isolated from CPs and ES. Additionally, we measured the fold upregulation of intracellular HIV-mRNA after stimulation of CD4+ T cells with phorbol 12-myristate 13-acetate (PMA) and ionomycin, and quantified the amount of HIV-mRNA levels released into culture supernatant. Results: ES have significantly less cell associated HIV-mRNA per 5x106 cells (p = 0.003); 8 of 10 CPs had quantifiable HIV-1 mRNA at baseline, whereas this was present in only 2 of 10 ES. Upon stimulation with PMA and ionomycin, 4 of 5 CPs and 7 of 9 ES showed increased cell associated HIV-mRNA. Interestingly, released HIV-1 mRNA could be detected in supernatants of CD4+ T cells stimulated with PMA/ionomycin from 5 of 8 ES. Conclusion: Our results demonstrate that while the baseline levels of cell associated HIV-1 mRNA are significantly lower in ES compared to CPs, stimulation of CD4+ T cells results in a comparable relative upregulation of viral transcription.
Keywords: Elite suppressors, Elite controllers, reservoirs, mRNA transcription
Introduction
Elite controllers or suppressors (ES) are individuals infected with HIV that maintain undetectable viral loads without receiving antiretroviral therapy (ART). Prior studies have shown that replication-competent virus can be cultured from CD4+ T cells from some ES [1-7] and full genome sequence analysis of replication competent virus has not revealed the presence of mutations associated with attenuation [1,2]. Furthermore, a recent study demonstrated that HIV-1 isolates from ES replicate vigorously and cause CD4+ T cell depletion in humanized mice [2]. Low levels of HIV-1 RNA can be detected in the plasma of most ES with ultrasensitive PCR assays [8-11], and sequence analysis has revealed evolution of plasma virus present in these patients [12-16]. These studies suggest that there is ongoing low-level replication in these patients, and yet in spite of this, ES maintain very small latent reservoirs as measured by total [5,17,18] and integrated DNA [19] as well as by the quantitative viral outgrowth assay [1] and the mouse viral outgrowth assay [20]. Despite the small reservoirs present in these patients, we hypothesized that ES would have relatively high levels of cell associated mRNA as a result of ongoing viral replication.
Hatano et al. have measured cell-associated in PBMCs isolated from ES [11] and have compared cell associated RNA present in CD4+ T cells of ES and CPs isolated from gut-associated lymphoid tissue [21]. These studies, however, did not specifically quantify HIV-1 mRNA which may be a more direct indicator of HIV-1 transcription [22]. In the present study, we measure baseline and inducible levels of cell associated HIV-mRNA in peripheral CD4+ T cells of ES and CPs using a previously validated primer probe set [23]. These results further our understanding of the HIV-1 latent reservoir in ES.
Materials and Methods
Study Participants
All studies were approved by the Johns Hopkins Institutional Review Board. All patients provided written informed consent before participation in this study. CPs are patients with undetectable viral loads on suppressive ART regimens for at least one year. ES have maintained undetectable viral loads by standard commercial assays without ART.
Isolation of HIV-1 mRNA and DNA
PBMCs were isolated from fresh blood samples by Ficoll gradient centrifugation. CD4+ T Cells were isolated using Miltenyi CD4+ T Cell Isolation Kit and cultured in RPMI1640 supplemented with 1% Pen/Strep and 10% FBS. 5x106 cells in 2 mL of complete media were either cultured in the presence of DMSO (0.2%) or PMA (50 ng/mL) and ionomycin (2 µM), or microbeads coated with antibodies against CD3 and CD28 (Dynal) for 24 hours. Stimulation with either PMA and ionomycin or the antibody coated microbeads resulted in greater than 90 percent of T cell activation as determined by CD69 expression. Supernatants were mixed with Trizol LS and cells were lysed using Trizol. Total RNA was isolated as described previously [22]. Human genomic DNA was isolated using Puregene kit (Qiagen) as per manufacturer’s instructions.
Quantification of HIV-1 mRNA and DNA
Total intracellular RNA was reverse transcribed using qScript from Quanta Biosciences as per manufacturer’s instructions. Reverse transcribed HIV-1 mRNA was quantified as described previously [22] using a primer and probe set that anneals to a highly-conserved region of the 3’ end of HIV-1 mRNA [23]. The limit of quantification was set as the dilution point at which the Ct of the plasmid molecular standard replicates had an s.d. > 0.5. We determined that the limit of quantification for all intracellular HIV-mRNA transcripts was 10 copies [22]. HIV-1 proviral DNA levels were quantified by qPCR using ToughMix (Quanta Biosciences). Previously published primers that target a small region of Gag [24] were used to detect HIV-1 proviral DNA, and cellular input was quantified measuring RNaseP levels (ThermoFisher) of a human genome standard (Roche).
Results
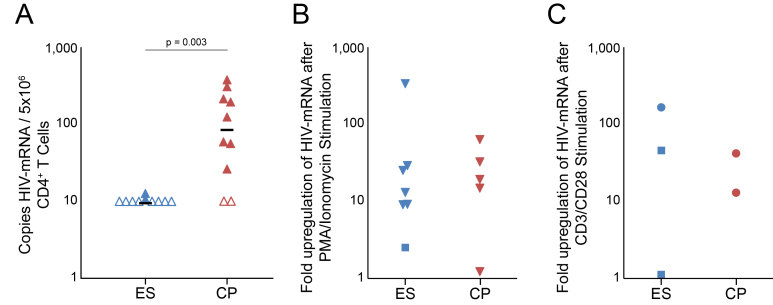
Cell associated HIV-1 mRNA from unstimulated CD4+ T cells ex vivo was quantifiable in only 2 of 10 ES (median < 10 copies/5M cells) as opposed to 8 of 10 CPs tested (median = 92 copies/5M cells; p = 0.03) (Figure 1a). Proviral HIV DNA was measured in 10 ES (median of 52 copies per 106 CD4+ T cells) and 10 CPs (median of 309 copies per 106 CD4+ T cells) to verify that our patients represent previously seen ES and CP cohorts (data not shown).
Figure 1.
Intracellular HIV-mRNA in CD4+ T cells before and after PMA and ionomycin stimulation. (a) HIV-mRNA isolated from CD4+ T cells of ES (blue triangles) and CPs (red triangles) was quantified by qPCR. Samples below the limit of detection are represented by open triangles. The black horizontal bar represents the median value of HIV-mRNA for each population. HIV-mRNA levels observed were higher in CD4+ T cells of CPs than of ES (p = 0.003). (b,c) CD4+ T cells of ES (blue symbols) and CPs (red symbols) were stimulated with either DMSO or PMA and ionomycin (b) or microbeads coated with antibodies against CD3 and CD28 (c) for 24 hours and HIV-mRNA was isolated and quantified by qPCR. Fold upregulation upon stimulation is plotted. Squares are used to represent subjects where either the DMSO control value and/or the value from stimulated cells were below the limit of detection. In these subjects the limit of detection was arbitrarily used as the DMSO control value. No significance difference in fold upregulation was detected between the two populations after stimulation with PMA and ionomycin (p = 0.27 with all symbols, 0.24 without squares).
To determine whether transcription was induced with T cell activation, we compared cell associated HIV-1 mRNA at baseline and after maximal T cell activation. Treatment of CD4+ T cells with the T cell activation control of PMA plus ionomycin (PMA/I) increased cell associated HIV-1 mRNA levels in 7 of 9 ES and 4 of 5 CPs (Figure 1b). The measured cell associated HIV-1 mRNA per 106 CD4+ T cells remained statistically higher for CPs (median = 33,676 per 106 cells) than for ES (median = 873 per 106 cells) (p = 0.047). However, there was no statistical difference between fold increase of cell associated HIV-mRNA between ES (13.1-fold, interquartile range = 12.1) and CPs (19.2-fold, interquartile range = 13.4, p = 0.22) per 106 stimulated cells. Interestingly, cell associated HIV-1 mRNA levels remained below the limit of detection in 2 ES after stimulation with PMA and ionomycin stimulation. In order to determine whether more physiological T cell stimulation would result in similar upregulation of transcription, CD4+ cells from a subset of the patients were activated with microbeads that were coated with antibodies to CD3 and CD28. There was a robust increase in intracellular mRNA in 2 of 3 ES whereas mRNA was undetectable at baseline and after T cell activation in the third subject. The level of mRNA upregulation seen in the 2 ES was comparable to the level seen in 2 CPs (Figure 1C).
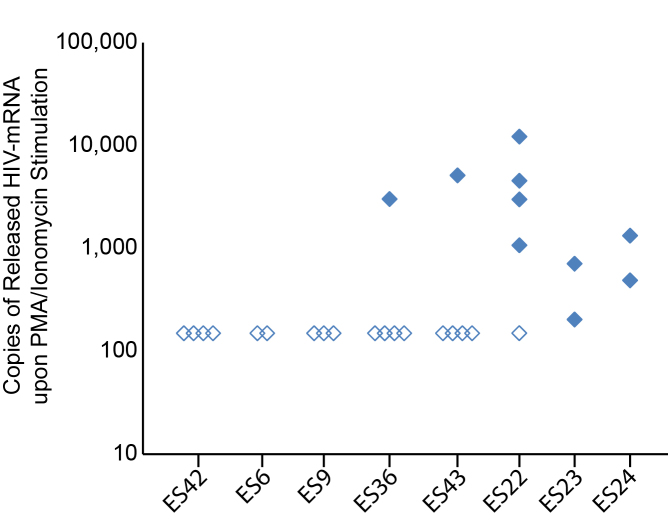
We also assessed the effect of T cell activation on viral release into culture supernatant. Upon stimulation of replicates of 5x106 CD4+ T cells from ES with PMA and ionomycin, HIV-1 mRNA was detected in the culture supernatant from 5 of 8 ES (Figure 2). Three ES (ES6, ES9, and ES42) had undetectable HIV-1 mRNA measurements in culture supernatant even when a total of 10-20x106 CD4+ T cells were stimulated. In contrast, in ES22, ES23, and ES24 we detected HIV-1 mRNA in the culture supernatant from the majority of stimulated CD4+ T cell replicates. Interestingly in ES36 and ES43, HIV-1 mRNA was detected in the culture supernatant in only 1 of 5 stimulated replicates, with 3,012 and 5,116 copies of HIV-1 mRNA present in the positive wells. The frequency of detected released HIV-1 RNA suggests an 89 percent probability by Poisson distribution that the signal observed reflects a single cell releasing virus, and our HIV-1 mRNA measurements fit with our recent estimates of the burst size of an infected CD4+ T cell in vitro [25]. Thus, of the 25x106 cells stimulated, it appears that only one cell harbored an inducible virus in both of these patients. Interestingly, ES36 and ES43 also had undetectable cell associated HIV-1 mRNA in 5x106 CD4+ T cells after stimulation with PMA and ionomycin. Taken together, these data suggest that there is significant heterogeneity in the inducible HIV-1 reservoir seen in ES which is consistent with results obtained from the viral outgrowth assay [1,2].
Figure 2.
HIV-mRNA release from CD4+ T cells upon PMA and ionomycin stimulation. Replicates of 5x106 CD4+ T cells from ES were stimulated with PMA and ionomycin for 24 hours, and supernatant was collected for measurement of released HIV-mRNA. Open diamonds indicate replicates with supernatant values below the limit of detection. Single replicates from 2 ES (ES 36 and ES43) were positive, suggesting release of HIV-mRNA from individual cells.
Discussion
Control of HIV-1 without ART by ES can be considered a model of a functional cure of infection. We have previously shown that these patients harbor much lower frequencies of latently infected CD4+ T cells [1] than CPs on suppressive ART regimens [26], and in this study we further analyze the inducible reservoir in ES. We demonstrate that cell associated HIV-1 mRNA levels observed in ES CD4+ T cells at baseline were significantly lower than those observed in CPs. These results further characterize the nature the latent reservoir in ES patients. Recent studies have found that ES had low levels of HIV-1 RNA in peripheral blood and in the GALT [11,21]. However, despite the low baseline levels of HIV-1 mRNA, there was no significant difference in the fold increase of cell associated HIV-1 mRNA levels upon PMA and ionomycin stimulation in CD4+ T cells isolated from ES and CPs, suggesting a similar induction of transcription in response to PMA and ionomycin treatment in both patient populations. This is consistent with results from a recent study that found a correlation between copies of proviral DNA and the inducibility of the HIV reservoir in ES and CPs [27]. Another recent study has shown that some defective proviruses present in CP CD4+ T cells can be transcribed [28]. It is not known whether this phenomenon also occurs in ES and so while we cannot determine whether the observed increase in HIV-1 mRNA was due to transcription of defective or intact proviral sequences, our data are consistent with our ability to culture replication-competent virus from some ES.
There was significant heterogeneity in the amount of cell associated and released virus seen in CD4+ T cells from different ES. Interestingly, ES23 and ES24 had detectable HIV-1 mRNA at baseline and both patients had a relatively high frequency of CD4+ T cells that released virus into culture supernatant following T cell stimulation. In contrast, the frequency of CD4+ T cells that released virus into culture supernatant was much lower in other ES. However, we were able to culture and characterize replication-competent virus from two such patients, ES9 [1] and ES36 [12] in prior studies. Further studies will be needed to determine whether virus that is releases into culture supernatant following CD4+ T cell activation is in fact replication-competent.
Studies have shown that virus in the plasma of ES evolves over time suggesting that there is ongoing viral replication [12-16]. In spite of that we still found very low levels of HIV-1 mRNA in peripheral CD4+ T cells in some of these patients. It is possible that the robust CTL (cytotoxic T lymphocytes) response in these patients [29] is able to eliminate transcriptionally active CD4+ T cells in peripheral blood that may be producing viral proteins, while CD4+ T cells in anatomical compartments that exclude CTL are the true source of ongoing viral replication. Lymph nodes represent one such compartment [30,31] and results from a recent study that found that CD8+ T cells are excluded from B cell follicles in ES monkeys may be consistent with this hypothesis [32]. Furthermore, sequence analysis from controllers with low level viremia suggest that there is ongoing viral replication in lymphoid tissue [33]. It will be interesting to look at mRNA expression in ES CD4+ T cells in B cell follicles and in other potential reservoirs such as myeloid cells. It will also be interesting to determine whether there is a correlation between the HIV-1 mRNA levels present in CD4+ T cells and the immune phenotype of ES and CPs.
In summary, we characterize baseline CD4+ T cell associated HIV-1 mRNA and show a robust upregulation following T cell stimulation in some ES. The results will have implications for patients who are subjected to curative procedures but still have residual low level HIV-1 reservoirs.
Acknowledgments
This work was funded by NIH grants R56AI080328-05A1 and 1R01AI120024-01 to J.N.B. and by the Johns Hopkins University Center for AIDS Research, an NIH-funded program (1P30AI094189).
Glossary
- ES
elite suppressors
- CPs
chronic progressors
- PMA
phorbol 12-myristate 13-acetate
- ART
antiretroviral therapy
- CTL
cytotoxic T lymphocytes
Author Contributions
CPW, CKB, ARM, and GML contributed equally to this study. CPW, CKB, GML, and ARM generated and analyzed the data and wrote the paper. SUC and VEKW generated data. JNB analyzed the data and wrote the paper.
References
- Blankson JN, Bailey JR, Thayil S. et al. Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors. J Virol. 2007;81:2508–2518. doi: 10.1128/JVI.02165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado M, Swanson MD, Pohlmeyer CW. et al. HLA-B*57 Elite Suppressor and Chronic Progressor HIV-1 Isolates Replicate Vigorously and Cause CD4+ T Cell Depletion in Humanized BLT Mice. J Virol. 2014;88:3340–3352. doi: 10.1128/JVI.03380-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamine A, Caumont-sarcos A, Saez-Cirion A. et al. Replication-competent HIV strains infect HIV controllers despite undetectable viremia (ANRS EP36 study). AIDS. 2007;21:1043–1058. doi: 10.1097/QAD.0b013e3280d5a7ac. [DOI] [PubMed] [Google Scholar]
- Buckheit III RW, Allen TG, Alme A. et al. Host factors dictate control of viral replication in two HIV-1 controller/chronic progressor transmission pairs. Nat Commun. 2012;3:716. doi: 10.1038/ncomms1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julg B, Buzon MJ, Piechocka-Trocha A. et al. Infrequent Recovery of HIV from but Robust Exogenous Infection of Activated CD4 + T Cells in HIV Elite Controllers. Clin Infect Dis. 2010;51:233–238. doi: 10.1086/653677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JR, O’Connell K, Yang H-C. et al. Transmission of human immunodeficiency virus type 1 from a patient who developed AIDS to an elite suppressor. J Virol. 2008;82:7395–7410. doi: 10.1128/JVI.00800-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard S, Dinoso JB, Marsh JA. et al. Sustained elite suppression of replication competent HIV-1 in a patient treated with rituximab based chemotherapy. J Clin Virol. 2011;51:195–198. doi: 10.1016/j.jcv.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinoso JB, Kim SY, Siliciano RF. et al. A Comparison of Viral Loads between HIV‐1–Infected Elite Suppressors and Individuals Who Receive Suppressive Highly Active Antiretroviral Therapy. Clin Infect Dis. 2008;47:102–104. doi: 10.1086/588791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migueles SA, Osborne CM, Royce C. et al. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity. 2008;29:1009–1021. doi: 10.1016/j.immuni.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereyra F, Palmer S, Miura T. et al. Persistent Low-Level Viremia in HIV-1 Elite Controllers and Relationship to Immunologic Parameters. J Infect Dis. 2009;200:984–990. doi: 10.1086/605446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano H, Delwart EL, Norris PJ. et al. Evidence for persistent low-level viremia in individuals who control human immunodeficiency virus in the absence of antiretroviral therapy. J Virol. 2009;83:329–335. doi: 10.1128/JVI.01763-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JR, Williams TM, Siliciano RF. et al. Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J Exp Med. 2006;203:1357–1369. doi: 10.1084/jem.20052319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell KA, Brennan TP, Bailey JR. et al. Control of HIV-1 in elite suppressors despite ongoing replication and evolution in plasma virus. J Virol. 2010;84:7018–7028. doi: 10.1128/JVI.00548-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado M, Brennan TP, O’Connell KA. et al. Evolution of the HIV-1 nef gene in HLA-B*57 Positive Elite Suppressors. Retrovirology. 2010:7. doi: 10.1186/1742-4690-7-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mens H, Kearney M, Wiegand A. et al. HIV-1 Continues To Replicate and Evolve in Patients with Natural Control of HIV Infection. J Virol. 2010;84:12971–12981. doi: 10.1128/JVI.00387-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado M, Gandhi SK, Buckheit RW. et al. Evolution of an Attenuated HIV-1 Isolate in an Elite Suppressor. AIDS Res Hum Retroviruses. 2014;30:284–288. doi: 10.1089/aid.2013.0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulck E Van, Bracke L, Heyndrickx L. et al. Immune and Viral Correlates of “Secondary Viral Control” after Treatment Interruption in Chronically HIV- 1 Infected Patients. PLoS One. 2012;7:e37792. doi: 10.1371/journal.pone.0037792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambotte O, Boufassa F, Madec Y. et al. HIV Controllers: A Homogeneous Group of HIV-1–Infected Patients with Spontaneous Control of Viral Replication. Clin Infect Dis. 2005;41:1053–1056. doi: 10.1086/433188. [DOI] [PubMed] [Google Scholar]
- Graf EH, Mexas AM, Yu JJ. et al. Elite suppressors harbor low levels of integrated HIV DNA and high levels of 2-LTR circular HIV DNA compared to HIV+ patients on and off HAART. PLoS Pathog. 2011;7:e1001300. doi: 10.1371/journal.ppat.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf Pate K a, Pohlmeyer CW, Walker-Sperling VE. et al. A murine viral outgrowth assay to detect residual HIV-1 in patients with undetectable viral loads. J Infect Dis. 2015:1–10. doi: 10.1093/infdis/jiv230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano H, Somsouk M, Sinclair E. et al. Comparison of HIV DNA and RNA in Gut-Associated Lymphoid Tissue of HIV-Infected Controllers and Non-controllers. AIDS. 2013;27:2255–2260. doi: 10.1097/QAD.0b013e328362692f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen CK, Laird GM, Durand CM. et al. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat Med. 2014;20:425–429. doi: 10.1038/nm.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, Rabi SA, Laird GM. et al. A Novel PCR Assay for Quantification of HIV-1 RNA. J Virol. 2013;87:6521–6525. doi: 10.1128/JVI.00006-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S, Wiegand AP, Maldarelli F. et al. New Real-Time Reverse Transcriptase-Initiated PCR Assay with Single-Copy Sensitivity for Human Immunodeficiency Virus Type 1 RNA in Plasma. J Clin Microbiol. 2003;41:4531–4536. doi: 10.1128/JCM.41.10.4531-4536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell K a, Rabi SA, Siliciano RF. et al. CD4+ T cells from elite suppressors are more susceptible to HIV-1 but produce fewer virions than cells from chronic progressors. Proc Natl Acad Sci U S A. 2011;108:E689–E698. doi: 10.1073/pnas.1108866108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano JD, Siliciano RF. Enhanced Culture Assay for Detection and Quantitation of Latently Infected, Resting CD4+ T-Cells Carrying Replication-Competent Virus in HIV-1-Infected Individuals. Methods Mol Biol. 2005;304(3):15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- Noel N, Peña R, David A. et al. Long-Term Spontaneous Control of HIV-1 Is Related to Low Frequency of Infected Cells and Inefficient Viral Reactivation. J Virol. 2016;90(13):6148–6158. doi: 10.1128/JVI.00419-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Pollack R, Yong P. et al. Defective HIV-1 Proviruses Can Be Transcribed Upon Activation [Abstract]. CROI. 2015:392. [Google Scholar]
- Migueles SA, Connors M. Success and failure of the cellular immune response against HIV-1. Nat Immunol. 2015;16:563–570. doi: 10.1038/ni.3161. [DOI] [PubMed] [Google Scholar]
- Folkvord JM, Armon C, Connick E. Lymphoid follicles are sites of heightened human immunodeficiency virus type 1 (HIV-1) replication and reduced antiretroviral effector mechanisms. AIDS Res Hum Retroviruses. 2005;21(5):363–370. doi: 10.1089/aid.2005.21.363. [DOI] [PubMed] [Google Scholar]
- Connick E, Mattila T, Folkvord JM. et al. CTL fail to accumulate at sites of HIV-1 replication in lymphoid tissue. J Immunol. 2007;178:6975–6983. doi: 10.4049/jimmunol.178.11.6975. [DOI] [PubMed] [Google Scholar]
- Fukazawa Y, Lum R, Okoye AA. et al. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med. 2015;21:132–139. doi: 10.1038/nm.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boritz EA, Darko S, Swaszek L. et al. Multiple Origins of Virus Persistence during Natural Control of HIV Infection. Cell. 2016;166(4):1004–1015. doi: 10.1016/j.cell.2016.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]