Abstract
Background
A possible relationship between mitochondrial haplogroups and psychiatric diseases (e.g. schizophrenia and bipolar disorder) has been postulated, but data regarding depression is still limited. We investigated whether any mitochondrial haplogroup carried a significant higher risk of depressive symptoms in a large prospective cohort of North American people included in the Osteoarthritis Initiative.
Methods
Cross sectional data was derived from the Osteoarthritis Initiative. The haplogroup was assigned through a combination of sequencing and PCR-RFLP techniques. All the mitochondrial haplogroups were named following this nomenclature: H, U, K, J, T, V, SuperHV, I, W, X or Others. Depression was ascertained through the 20-item Center for Epidemiologic Studies-Depression (CES-D), with ≥16 indicating depressive symptoms.
Results
Overall, 3601 Caucasian participants (55.9% women), mean age of 61.7 ± 9.3 years were included. No difference was observed in mitochondrial haplogroups frequency among those with depressive symptoms (n=285, =7.9% of the baseline population) compared to participants with no depressive symptoms (N=3316) (chi-square test=0.53). Using a logistic regression analysis, adjusted for eight potential confounders, with those having the haplogroup H as the reference group (the most common haplogroup), no significant mitochondrial haplogroup was associated with prevalent depressive symptoms. The same results were evident in secondary analysis in which we matched depressed and non-depressed participants for age and sex.
Limitations
Cross-sectional design; only CES-D for evaluating mood; participants not totally representative of general population.
Conclusions
We found no evidence of any relationship between specific mitochondrial haplogroups and depressive symptoms. Future longitudinal research is required to confirm/refute these findings.
Keywords: Mitochondrial haplogroups, Depression, Osteoarthritis initiative
1. Introduction
The human mitochondrial genome is a circular set of 16,569 base pairs encoding 37 genes and finally 13 proteins, involved in processes relevant to cellular function and survival. (Wang and Brinton, 2016) Similarly to nuclear DNA mitochondrial DNA (mtDNA) undergoes frequent mutations.(Copeland and Longley, 2014; Wang and Brinton, 2016) Mismatch, usually due to a recombination or repair, can lead to single nucleotide polymorphisms (SNPs) and clusters of these specific SNPs in the mitochondrial genome define the mitochondrial haplogroups. The evolution of mtDNA occurs at a much faster pace compared to the average nuclear DNA, and thus mutations have accumulated sequentially, even if only along radiating maternal lineages. (Wallace and Chalkia, 2013) Therefore, it is hardly surprising that the biology of mitochondrial DNA may partly explain the genetic predisposition to certain medical conditions, probably in a similar way to the germline mutations of nuclear DNA (Luchini et al., 2016; Mafficini et al., 2016). Recent studies have proposed that the different mitochondrial haplogroups might play a role in the development of several chronic disease states (Mishmar et al., 2003; Picard et al., 2016; Wallace and Chalkia, 2013). Each of the mitochondrial haplogroups, in fact, is probably involved not only in mitochondria physiology, but also in cell metabolism (Horan and Cooper, 2014; Wallace, 2015). Thus, organs at high consumption of energy (such as brain) are the main target of mitochondrial dysfunction (Mink et al., 1981).
From an epidemiological viewpoint, several psychiatric conditions, such as schizophrenia and mood disorders have a preferential transmission through the maternal line (Kirk et al., 1999). Moreover, more recent studies reported that mood disorders are more frequent in maternal relatives of children with mitochondrial diseases, compared with their paternal relatives, or compared with maternal relatives of the children with other metabolic diseases (Grigoroiu-Serbanescu et al., 1998; Kato, 2001; Linnane et al., 1989). In this argument, some authors reported an increased mtDNA- copy number in peripheral cells and plasma in depressed (Cai et al., 2015; Nicod et al., 2016) and suicidal (Lindqvist et al., 2016) subjects. Finally, several mitochondrial encephalomyopathies are characterized by a high prevalence of bipolar disorder and depression (Kato, 2001). Taken together, these findings suggest that mitochondrial dysfunction may be associated with mood disorders, including depression.
Even if several experimental and pre-clinical pieces of research report that mitochondrial dysfunction is plausible in the development of depression (e.g. depressed people use less ATP in some brain regions) (Gardner et al., 2003; Karabatsiakis et al., 2014), only two studies have investigated an association between mitochondrial haplotypes and the presence of depression in human being not affected by mitochondrial myopathies (Rollins et al., 2009; Zhang et al., 2016). Although these studies reported a significant association between mitochondrial haplotypes and mental illness, they were limited in sample size suggesting that further research is needed.
Given this background, we aimed to investigate whether any mitochondrial haplogroup carried a significant higher risk of depressive symptoms in a large cohort of North American people included in the Osteoarthritis Initiative.
2. Methods
2.1. Data source and subjects
All participants in this cross-sectional study were recruited as part of the ongoing, publicly and privately funded, multicenter Osteoarthritis Initiative (OAI study) (http://www.oai.ucsf.edu/). The specific dataset used was the OAI baseline data (November 2008) (V00). The OAI recruited participants with/at high risk of knee osteoarthritis (e.g. obese, with familiarity for OA) from four clinical sites in the US (Baltimore, MD; Pittsburgh, PA; Pawtucket, RI; and Columbus, OH) between February 2004 and May 2006. For this study, specific exclusion criteria are: pregnancy, presence of active rheumatoid arthritis, bilateral total knee replacement, unable to undergo knee magnetic resonance or to give a blood sample, any co-morbidity precluding the participation to the study and unwilling to sign the informed consent.
All participants provided written informed consent. The OAI study protocol was approved by the institutional review board of the OAI Coordinating Center, University of California at San Francisco.
2.2. Exposure
The haplogroup assignment was performed by a previously published method (Rego-Perez et al., 2008), which consisted in a combination of sequencing and PCR-RFLP techniques. The sequencing technique consisted in the multiplex assignment of the main 6 SNPs that contribute to the generation of the most prevalent Caucasian haplogroups (Torroni et al., 1996) (H, V, super HV, U, K, T, J) following the single base extension (SBE) assay. All the mitochondrial haplogroups have been named following this nomenclature in agreement with those suggested by the OAI (http://www.oai.ucsf.edu/): H, U, K, J, T, V, SuperHV, I, W, X or Others.
2.3. Outcome - depressive symptoms
The presence of depressive symptoms was derived from the 20-item Center for Epidemiologic Studies-Depression (CES-D) instrument. (Radloff, 1977) The range of possible values for this scores is 0–60, where higher scores indicate more depressive symptoms (Radloff, 1977). A cut-off of 16 is commonly used for the diagnosis of depressive symptoms (Radloff, 1977).
2.4. Covariates
A number of variables were identified from the OAI dataset to explore the relationship between mitochondrial haplogroups and depression. These included: (1) physical activity evaluated through the Physical Activity Scale for the Elderly (PASE) (Washburn et al., 1999). The PASE is validated in older populations, covers 12 different activities, such as walking, sports, and housework, and is scored from 0 to 400 and more; (2) race was defined as “white” vs. “others”; (3) smoking habits as “previous/current” vs. never; (4) educational level was categorized as “degree” vs. others; (5) yearly income as < vs. ≥ 50,000 $ or missing data; (6) co-morbidities assessed through the modified Charlson comorbidity score, with higher scores indicating an increased severity of conditions (Katz et al., 1996); (7) body mass index (BMI), as recorded by a trained nurse.
2.5. Statistical analyses
For continuous variables, normal distributions were tested using the Kolmogorov-Smirnov test. The data are shown as means and standard deviations (SD) for quantitative measures, and percentages for all discrete variables by presence of depression. For continuous variables, Student’s T test was used, whilst chi-square test was applied for discrete variables. Levene’s test was used to test the homoscedasticity of variances and, if its assumption was violated, then Welch’s ANOVA was used.
The strength of the association of mitochondrial haplogroups and depressive symptoms was assessed through a logistic regression analysis. Factors significantly associated with depression (taking a p-value < 0.05 as statistically significant) were included. Multi-collinearity among covariates was assessed through variance inflation factor, taking a cut-off of 2 as a reason of exclusion, but no variable was excluded due to this reason. The basic model was not adjusted for any confounders, while the fully adjusted model included baseline values of: age, BMI, PASE score, Charlson comorbidity index as continuous variables, and gender, education, smoking habits, yearly income as categorical variables. Data of logistic regression analysis were reported as odds ratios (ORs) with 95% confidence intervals (CIs).
In a secondary analysis, we used controls (matched for age and sex) to depressed participants in order to attenuate the presence of any selection bias (Pearce, 2016).
All analyses were performed using the SPSS 17.0 for Windows (SPSS Inc., Chicago, Illinois). All statistical tests were two-tailed and statistical significance was assumed for a p-value < 0.05.
3. Results
3.1. Study participants
At baseline, among 4796 potentially eligible individuals, 1047 subjects did not have a mitochondrial DNA assessment, 126 did not have this assessment for technical problems and 22 did not have information regarding CES-D. Thus, 3601 participants were enrolled in the current study. Compared to those included in this analysis, the 1195 people not enrolled were not significantly differences in terms of age and gender, but there was a higher presence of non-Caucasian ethnicities (chi-square test, p < 0.0001) (details not shown).
3.2. Baseline analyses
The 3601 participants included aged a mean of 61.7 ± 9.3 (range: 45–79) years, with a slight higher prevalence of women (=55.9%). All participants were Caucasians.
Participants were divided into those with depressive symptoms (n=285, 7.9% of the sample) and without (n=3316). The baseline characteristics of both groups are summarized in Table 1. Briefly, people with depressive symptoms were significantly older, more likely to be females, and obese compared to the non-depressed group. People with depressive symptoms were more frequently smokers, with a lower educational level, poorer and with more comorbidities than those with no depressive symptoms (Table 1).
Table 1.
Baseline characteristics by depression status.
| Variable | Depressive symptoms (n=285) | No depressive symptoms (n=3316) | p-value |
|---|---|---|---|
| Age (years) | 61.9 (9.3) | 59.3 (9.3) | < 0.0001 |
| Females (%) | 63.2 | 55.3 | 0.001 |
| BMI (kg/m2) | 29.1 (5.5) | 28.0 (4.5) | < 0.0001 |
| White race (%) | All whites | ||
| PASE (points) | 156.5 (81.1) | 164.1 (80.0) | 0.13 |
| Smoking (%) | 55.6 | 45.5 | 0.001 |
| Degree (%) | 24.6 | 34.3 | 0.001 |
| Yearly income (< 50,000 $) | 46.5 | 68.7 | < 0.0001 |
| Charlson co-morbidity score | 0.5 (0.8) | 0.3 (0.8) | < 0.0001 |
Notes:
Numbers are mean values (and standard deviations) or percentages, as appropriate.
Abbreviations: BMI: body mass index; PASE: physical activity scale for the elderly; CES-D: Center for Epidemiological Studies Depression.
3.3. Association between mitochondrial haplogroups and depression
As reported in Fig. 1, no significant differences emerged regarding in the prevalence of mitochondrial haplogroups (chi-square test=0.53). For example, the most frequent haplogroup (i.e. H) had the same prevalence among the depressed and no depressed (42.5% in both groups).
Fig. 1.
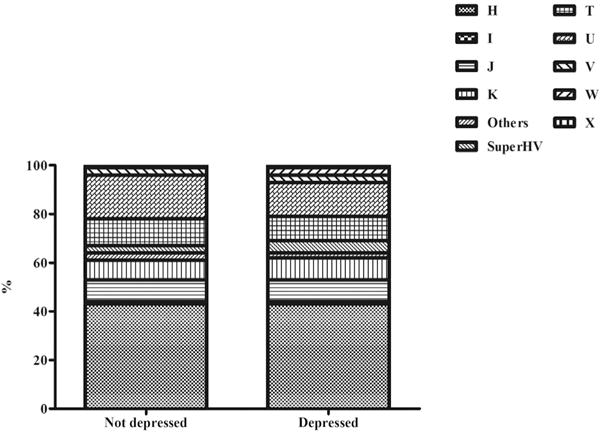
Prevalence of depressive symptoms by mitochondrial haplogroups. The prevalence of each haplogroup is represented as percentage in people with depressive symptoms or not.
Using a logistic regression analysis, adjusted for eight potential confounders, with those having the haplogroup H as the reference, no significant mitochondrial haplogroup was associated with a prevalent depressive symptoms. The OR ranged from 0.50 (95%CI: 0.18–1.43) of other haplogroup to 1.74 (95%CI: 0.75–4.07) of haplogroup W (Table 2).
Table 2.
Association between mitochondrial haplogroups and Depressive symptoms.
| All cohort
|
Matched (for age and sex)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Unadjusted OR (95%CI) |
P value | Fully-adjusted ORa (95%CI) |
P value | Unadjusted OR (95%CI) |
P value | Fully-adjusted ORa (95%CI) |
P value | |
| H | 1 [reference] | 1 [reference] | 1 [reference] | 1 [reference] | ||||
| I | 0.77 (0.23–2.50) | 0.66 | 0.57 (0.17–1.91) | 0.36 | 0.51 (0.15–1.74) | 0.29 | 0.37 (0.11–1.28) | 0.12 |
| J | 1.05 (0.67–1.64) | 0.83 | 0.99 (0.63–1.56) | 0.97 | 1.00 (0.62–1.62) | 0.99 | 0.93 (0.56–1.52) | 0.76 |
| K | 1.21 (0.77–1.91) | 0.41 | 1.32 (0.83–2.11) | 0.24 | 1.30 (0.79–2.13) | 0.31 | 1.36 (0.81–2.28) | 0.25 |
| Others | 0.61 (0.22–1.70) | 0.35 | 0.50 (0.18–1.43) | 0.20 | 0.72 (0.24–2.12) | 0.55 | 0.45 (0.14–1.43) | 0.18 |
| SuperHV | 1.39 (0.72–2.67) | 0.33 | 1.22 (0.63–2.38) | 0.56 | 1.08 (0.54–2.18) | 0.82 | 1.07 (0.52–2.22) | 0.85 |
| T | 0.99 (0.65–1.51) | 0.97 | 1.02 (0.66–1.57) | 0.94 | 0.95 (0.61–1.50) | 0.83 | 0.94 (0.58–1.50) | 0.78 |
| U | 0.76 (0.52–1.11) | 0.16 | 0.78 (0.53–1.14) | 0.20 | 0.84 (0.56–1.27) | 0.41 | 0.82 (0.54–1.50) | 0.37 |
| V | 0.89 (0.42–1.88) | 0.89 | 0.83 (0.39–1.79) | 0.64 | 0.66 (0.30–1.43) | 0.29 | 0.56 (0.25–1.27) | 0.17 |
| W | 1.91 (0.84–4.35) | 0.12 | 1.74 (0.75–4.07) | 0.20 | 2.12 (0.83–5.44) | 0.12 | 2.17 (0.80–5.91) | 0.13 |
| X | 1.15 (0.40–3.26) | 0.80 | 1.07 (0.36–3.15) | 0.91 | 1.05 (0.34–3.23) | 0.93 | 1.00 (0.32–3.21) | 0.99 |
Notes:
All the data are presented as odds ratios (ORs) with their 95% confidence intervals.
Abbreviations: CI: confidence intervals; OR: odds ratio.
Fully-adjusted model included as covariates: age (as continuous); sex; body mass index (as continuous); education (degree vs. others); smoking habits (current and previous vs. others); yearly income (categorized as ≥ or < 50,000$ and missing data); Physical Activity Scale for Elderly score (as continuous); Charlson co-morbidity index (as continuous).
In a secondary analysis, we explored whether using controls matched for age and sex, influenced our results, but the association between mitochondrial haplogroups and depressive symptoms remained not significant (Table 2).
4. Discussion
In this large cross-sectional paper, we found there was no significant association between mitochondrial haplotypes and depressive symptoms. Our findings remained unaltered after adjusting for potential confounders or using controls matched for sex and age. Thus, our data suggest that mitochondria haplogroups may not be related to depressive symptoms.
The interest of mitochondrial haplotypes in the development of psychiatric diseases is rapidly increasing. Several studies have reported that mitochondrial haplotypes are associated with the presence of some psychiatric conditions, particularly schizophrenia and bipolar disorder (Clay et al., 2011; Rajasekaran et al., 2015). The mechanisms linking mitochondrial haplogroups and related dysfunction and these conditions may be attributed to altered oxidative/nitrosative stress responses which could activate the immunoinflammatory pathways (Bano et al., 2017; Soysal et al.; Soysal et al., 2016) and subsequently lead to neuroprogressive changes, typical of bipolar disorder and schizophrenia (Clay et al., 2011; Rajasekaran et al., 2015). Since the same mechanisms are relevant for the development of depression, some researchers have tried to disentangle if any association exists between mitochondrial haplogroups and depressive mood. These studies, however, are very limited and included few participants. Initially Suomalainen et al. reported in a family affected by a chronic progressive ophthalmoplegia that the presence of depression was very common (Suomalainen et al., 1992). These authors interpreted this finding as a possible sign of mitochondrial encephalopathy. Similarly, Suzuki and collaborators investigated 15 people affected by a form of mitochondrial diabetes founding that four of them were affected by mood disorders (Suzuki et al., 1997). More recently, Rollins et al. showed that, in a peculiar subgroup of patients with schizophrenia, the rate of synonymous base pair substitutions in the coding regions of the mtDNA genome was significantly higher compared to controls (Rollins et al., 2009). However the same results were not confirmed in patients with depression compared with controls. These findings highlight the concept of a higher mtDNA instability in some mental disorders. Finally, in 2016, Zhang et al., reported in 744 Chinese subjects with major depression matched with 767 Chinese healthy controls that the T-C haplotype was significantly associated with depression (Zhang et al., 2016). Contrary to results reported by Zhang et al., we did not find any significant association between mitochondrial haplogroups and depressive symptoms in our study. Several hypotheses could explain our findings. First, the studies cited before (except the last two) were made among populations affected by mitochondrial diseases and without a control group. Therefore, it is difficult to say whether a particular mitochondrial haplogroup is associated or not with depression. Second, since our knowledge regarding the possible effect of mitochondrial haplotypes on depression derives from the reported similarities with other psychiatric diseases and from some indirect findings (such as people with depression have reduced blood flow and metabolism in the prefrontal cortex compared to healthy controls) (Videbech, 2000), thus it is possible that mitochondrial haplotypes do not play a role in the development of this disease. It is noteworthy, for example, that Stine et al. in postmortem samples found no significant alterations in mtDNA in nine depressed people who completed suicide compared to controls (Stine et al., 1993). However our study suggests further studies are required to disentangle the potential relationship between mitochondria haplogroups and depression. Moreover, Zhang et al. (2016) compared patients with a full diagnosis of MDD with healthy controls, thus including people with more pronounced depressive illness than ours which included people with depressive symptoms. Also, while we focused on a Caucasian population, Zhang et al. (2016) focused on a Chinese sample, which might have different mithocondrial haplotypes’ distribution. Furthermore, we did not account for stress in our analyses; since it has been suggested that stress can modify amount of mtDNA, our findings may have been biased by neglecting stress as confounding factor. Finally, it may be that there were too few participants within each mitochondria haplogroups and thus some of our analyses were underpowered.
A special consideration should also account for the close relation between mithocondrial functioning and ageing, in elderly population. Ageing is per sè associated with a weakened mitochondrial efficiency (Chistiakov et al., 2014), and mitochondrial age-related impairment may also play a role in depression onset (Harper et al., 2016), beyond haplotypes’ distribution.
4.1. Limitations
The findings of our paper should be interpreted within its limitations. First, the diagnosis of depressive symptoms was made only using the CES-D that, however, includes many symptoms defined by the (DSM-V) for a major depressive episode and therefore is somewhat justifiable (Veronese et al., 2016). Second, we did not assess the role of medications used for depression and this could introduce an important bias. Third, the number of people in some haplogroups was limited. Fourth, the medical diseases in the OAI are self-reported, and this could introduce another potential bias. Fifth, data was only available of mitochondrial haplogroups among Caucasians and the results do not extend beyond this ethnicity. Sixth, the OAI did not contain detailed information regarding other psychiatric diseases that seem to be associated with mitochondrial haplogroups and this could affect our results. Finally, the OAI included only people with or at high risk of knee osteoarthritis and so it could be not fully representative of the general population.
4.2. Conclusion
In conclusion, in this large cohort study we did not find a relationship between any mitochondrial haplogroups with depressive symptoms. Further research with different diagnostic criteria for depression and with a longitudinal design are needed to disentangle if any mitochondrial haplotype is associated with depression.
Acknowledgments
Funding sources
The OAI is a public-private partnership comprised of five contracts (N01-AR-2–2258; N01-AR-2–2259; N01-AR-2–2260; N01-AR-2–2261; N01-AR-2–2262) by the National Institutes of Health, a branch of the Department of Health and Human Services, and conducted by the OAI Study Investigators. Private funding partners include Merck Research Laboratories; Novartis Pharmaceuticals Corporation, GlaxoSmithKline; and Pfizer, Inc. Private sector funding for the OAI is managed by the Foundation for the National Institutes of Health. This manuscript was prepared using an OAI public use data set and does not necessarily reflect the opinions or views of the OAI investigators, the NIH, or the private funding partners.
Role of founding source
The funding sources did not have any role in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Footnotes
Authors’ contribution
NV, AK: made the statistical analysis; NV, BS, MS: wrote the paper; AV, JD, TT: analysis of the data; AFC, MZ, SM: critical revision of the paper. All authors have approved the final article.
References
- Bano G, Trevisan C, Carraro S, Solmi M, Luchini C, Stubbs B, Manzato E, Sergi G, Veronese N. Inflammation and sarcopenia: a systematic review and meta-analysis. Maturitas. 2017;96:10–15. doi: 10.1016/j.maturitas.2016.11.006. [DOI] [PubMed] [Google Scholar]
- Cai N, Chang S, Li Y, Li Q, Hu J, Liang J, Song L, Kretzschmar W, Gan X, Nicod J, Rivera M, Deng H, Du B, Li K, Sang W, Gao J, Gao S, Ha B, Ho HY, Hu C, Hu J, Hu Z, Huang G, Jiang G, Jiang T, Jin W, Li G, Li K, Li Y, Li Y, Li Y, Lin YT, Liu L, Liu T, Liu Y, Liu Y, Lu Y, Lv L, Meng H, Qian P, Sang H, Shen J, Shi J, Sun J, Tao M, Wang G, Wang G, Wang J, Wang L, Wang X, Wang X, Yang H, Yang L, Yin Y, Zhang J, Zhang K, Sun N, Zhang W, Zhang X, Zhang Z, Zhong H, Breen G, Wang J, Marchini J, Chen Y, Xu Q, Xu X, Mott R, Huang GJ, Kendler K, Flint J. Molecular signatures of major depression. Curr Biol. 2015;25:1146–1156. doi: 10.1016/j.cub.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistiakov DA, Sobenin IA, Revin VV, Orekhov AN, Bobryshev YV. Mitochondrial aging and age-related dysfunction of mitochondria. Biomed Res Int. 2014;2014:238463. doi: 10.1155/2014/238463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay HB, Sillivan S, Konradi C. Mitochondrial dysfunction and pathology in bipolar disorder and schizophrenia. Int J Dev Neurosci. 2011;29:311–324. doi: 10.1016/j.ijdevneu.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland WC, Longley MJ. Mitochondrial genome maintenance in health and disease. DNA Repair. 2014;19:190–198. doi: 10.1016/j.dnarep.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner A, Johansson A, Wibom R, Nennesmo I, von Döbeln U, Hagenfeldt L, Hällström T. Alterations of mitochondrial function and correlations with personality traits in selected major depressive disorder patients. J Affect Disord. 2003;76:55–68. doi: 10.1016/s0165-0327(02)00067-8. [DOI] [PubMed] [Google Scholar]
- Grigoroiu-Serbanescu M, Martinez M, Nothen MM, Propping P, Milea S, Mihailescu R, Marinescu E. Patterns of parental transmission and familial aggregation models in bipolar affective disorder. Am J Med Genet. 1998;81:397–404. [PubMed] [Google Scholar]
- Harper DG, Joe EB, Jensen JE, Ravichandran C, Forester BP. Brain levels of high-energy phosphate metabolites and executive function in geriatric depression. Int J Geriatr Psychiatry. 2016;31:1241–1249. doi: 10.1002/gps.4439. [DOI] [PubMed] [Google Scholar]
- Horan MP, Cooper DN. The emergence of the mitochondrial genome as a partial regulator of nuclear function is providing new insights into the genetic mechanisms underlying age-related complex disease. Hum Genet. 2014;133:435–458. doi: 10.1007/s00439-013-1402-4. [DOI] [PubMed] [Google Scholar]
- Karabatsiakis A, Bock C, Salinas-Manrique J, Kolassa S, Calzia E, Dietrich DE, Kolassa IT. Mitochondrial respiration in peripheral blood mononuclear cells correlates with depressive subsymptoms and severity of major depression. Transl Psychiatry. 2014;4:e397. doi: 10.1038/tp.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T. The other, forgotten genome: mitochondrial DNA and mental disorders. Mol Psychiatry. 2001;6:625–633. doi: 10.1038/sj.mp.4000926. [DOI] [PubMed] [Google Scholar]
- Katz JN, Chang LC, Sangha O, Fossel AH, Bates DW. Can comorbidity be measured by questionnaire rather than medical record review? Med Care. 1996;34:73–84. doi: 10.1097/00005650-199601000-00006. [DOI] [PubMed] [Google Scholar]
- Kirk R, Furlong RA, Amos W, Cooper G, Rubinsztein JS, Walsh C, Paykel ES, Rubinsztein DC. Mitochondrial genetic analyses suggest selection against maternal lineages in bipolar affective disorder. Am J Hum Genet. 1999;65:508–518. doi: 10.1086/302507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist D, Fernström J, Grudet C, Ljunggren L, Träskman-Bendz L, Ohlsson L, Westrin Å. Increased plasma levels of circulating cell-free mitochondrial DNA in suicide attempters: associations with HPA-axis hyperactivity. Transl Psychiatry. 2016;6:e971. doi: 10.1038/tp.2016.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnane AW, Marzuki S, Ozawa T, Tanaka M. Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet. 1989;1:642–645. doi: 10.1016/s0140-6736(89)92145-4. [DOI] [PubMed] [Google Scholar]
- Luchini C, Veronese N, Yachida S, Cheng L, Nottegar A, Stubbs B, Solmi M, Capelli P, Pea A, Barbareschi M, Fassan M, Wood LD, Scarpa A. Different prognostic roles of tumor suppressor gene BAP1 in cancer: a systematic review with meta-analysis. Genes Chromosomes Cancer. 2016 doi: 10.1002/gcc.22381. [DOI] [PubMed] [Google Scholar]
- Mafficini A, Simbolo M, Parisi A, Rusev B, Luchini C, Cataldo I, Piazzola E, Sperandio N, Turri G, Franchi M, Tortora G, Bovo C, Lawlor RT, Scarpa A. BRCA somatic and germline mutation detection in paraffin embedded ovarian cancers by next-generation sequencing. Oncotarget. 2016;7:1076–1083. doi: 10.18632/oncotarget.6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mink JW, Blumenschine RJ, Adams DB. Ratio of central nervous system to body metabolism in vertebrates: its constancy and functional basis. Am J Physiol. 1981;241:R203–R212. doi: 10.1152/ajpregu.1981.241.3.R203. [DOI] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, Sukernik RI, Olckers A, Wallace DC. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA. 2003;100:171–176. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicod J, Wagner S, Vonberg F, Bhomra A, Schlicht KF, Tadic A, Mott R, Lieb K, Flint J. The amount of mitochondrial DNA in blood reflects the course of a depressive episode. Biol Psychiatry. 2016;80:e41–e42. doi: 10.1016/j.biopsych.2015.12.019. [DOI] [PubMed] [Google Scholar]
- Pearce N. Analysis of matched case-control studies. Br Med J. 2016;352:i969. doi: 10.1136/bmj.i969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard M, Wallace DC, Burelle Y. The rise of mitochondria in medicine. Mitochondrion. 2016;30:105–116. doi: 10.1016/j.mito.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff LS. The CES-D scale: a self-report depression scale for research in the general population. Appl Psychol Meas. 1977;1:385–401. [Google Scholar]
- Rajasekaran A, Venkatasubramanian G, Berk M, Debnath M. Mitochondrial dysfunction in schizophrenia: pathways, mechanisms and implications. Neurosci Biobehav Rev. 2015;48:10–21. doi: 10.1016/j.neubiorev.2014.11.005. [DOI] [PubMed] [Google Scholar]
- Rego-Perez I, Fernandez-Moreno M, Fernandez-Lopez C, Arenas J, Blanco FJ. Mitochondrial DNA haplogroups: role in the prevalence and severity of knee osteoarthritis. Arthritis Rheum. 2008;58:2387–2396. doi: 10.1002/art.23659. [DOI] [PubMed] [Google Scholar]
- Rollins B, Martin MV, Sequeira PA, Moon EA, Morgan LZ, Watson SJ, Schatzberg A, Akil H, Myers RM, Jones EG, Wallace DC, Bunney WE, Vawter MP. Mitochondrial variants in schizophrenia, bipolar disorder, and major depressive disorder. PloS One. 2009;4:e4913. doi: 10.1371/journal.pone.0004913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soysal P, Isik AT, Carvalho AF, Fernandes BS, Solmi M, Schofield P, Veronese N, Stubbs B. Oxidative Stress And Frailty: A Systematic Review And Best Evidence Synthesis. Maturitas. doi: 10.1016/j.maturitas.2017.01.006. [DOI] [PubMed] [Google Scholar]
- Soysal P, Stubbs B, Lucato P, Luchini C, Solmi M, Peluso R, Sergi G, Isik AT, Manzato E, Maggi S, Maggio M, Prina AM, Cosco TD, Wu YT, Veronese N. Inflammation and frailty in the elderly: a systematic review and meta-analysis. Ageing Res Rev 2016 [Google Scholar]
- Stine OC, Luu SU, Zito M, Casanova M. The possible association between affective disorder and partially deleted mitochondrial DNA. Biol Psychiatry. 1993;33:141–142. doi: 10.1016/0006-3223(93)90317-7. [DOI] [PubMed] [Google Scholar]
- Suomalainen A, Majander A, Haltia M, Somer H, Lonnqvist J, Savontaus ML, Peltonen L. Multiple deletions of mitochondrial DNA in several tissues of a patient with severe retarded depression and familial progressive external ophthalmoplegia. J Clin Investig. 1992;90:61–66. doi: 10.1172/JCI115856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Taniyama M, Muramatsu T, Atsumi Y, Hosokawa K, Asahina T, Shimada A, Murata C, Matsuoka K. Diabetes mellitus associated with 3243 mitochondrial tRNA(Leu(UUR)) mutation: clinical features and coenzyme Q10 treatment. Mol Asp Med. 1997;18(Suppl):S181–S188. doi: 10.1016/s0098-2997(97)00041-1. [DOI] [PubMed] [Google Scholar]
- Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R, Obinu D, Savontaus ML, Wallace DC. Classification of European mtDNAs from an analysis of three European populations. Genetics. 1996;144:1835–1850. doi: 10.1093/genetics/144.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese N, Stubbs B, Solmi M, Smith TO, Noale M, Cooper C, Maggi S. Association between lower limb osteoarthritis and incidence of depressive symptoms: data from the osteoarthritis initiative. Age Ageing. 2016 doi: 10.1093/ageing/afw216. [DOI] [PubMed] [Google Scholar]
- Videbech P. PET measurements of brain glucose metabolism and blood flow in major depressive disorder: a critical review. Acta Psychiatr Scand. 2000;101:11–20. doi: 10.1034/j.1600-0447.2000.101001011.x. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial DNA variation in human radiation and disease. Cell. 2015;163:33–38. doi: 10.1016/j.cell.2015.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb Perspect Biol. 2013;5:a021220. doi: 10.1101/cshperspect.a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Brinton RD. Triad of risk for late onset Alzheimer’s: mitochondrial haplotype, APOE genotype and chromosomal sex. Front Aging Neurosci. 2016;8:232. doi: 10.3389/fnagi.2016.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn RA, McAuley E, Katula J, Mihalko SL, Boileau RA. The physical activity scale forthe elderly (PASE): evidence for validity. J Clin Epidemiol. 1999;52:643–651. doi: 10.1016/s0895-4356(99)00049-9. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Ni J, Zhang J, Tang W, Li X, Wu Z, Zhang C. A haplotype in the 5′-upstream region of the NDUFV2 gene is associated with major depressive disorder in Han Chinese. J Affect Disord. 2016;190:329–332. doi: 10.1016/j.jad.2015.10.034. [DOI] [PubMed] [Google Scholar]


