Summary
In this issue, Behkne et al. describe a novel cell-based peptide binding assay and use it to analyze the binding specificities of the endoplasmic reticulum Hsp70 chaperone and its co-chaperones and to probe their different roles in protein quality control.
The Hsp70 molecular chaperone system plays important roles in protein homeostasis in all compartments of the cell. In the endoplasmic reticulum (ER), the site of maturation for the thousands of proteins that traffic through the eukaryotic secretory pathway, the Hsp70 system is central to protein translocation, folding, quality control and degradation (Braakman and Hebert, 2013). In addition to the ER Hsp70 (BiP, immunoglobulin heavy-chain binding protein), there are numerous Hsp70 co-chaperones: eight ER-resident J-domain proteins (ERdj1–8) and two nucleotide exchange factors (Grp170 and Sil1) (Behnke et al., 2015). These co-chaperones are proposed to direct BiP to its diverse array of functions in the ER by controlling its localization and binding selectivity. In an article from Behnke et al. in this issue (Behnke et al., 2016), the substrate-binding specificities for ER Hsp70 system members known to directly bind substrates (BiP, Grp170 and ERdj3–5) were monitored by deploying an in vivo peptide library that effectively used the ER lumen of live cells as a test tube.
The peptide expression library was engineered based on overlapping 25-amino acid long sequences that span two well-known natural clients of BiP; truncated immunoglobulin γ1 heavy chain and NS-1 κ light chain. These 25-mer sequences were placed in an ER-targeting construct that did not significantly interact with any of the tested proteins on its own. COS-1 cells were co-transfected with the peptide library and tagged versions of the various Hsp70 system members to query their binding preferences in their natural environment. Interactions were monitored by co-immunoprecipitation of metabolically labeled proteins with antibodies directed against the peptide construct, or in some cases using immunoprecipitations combined with immunoblots. Peptides that bound multiple factors were subdivided to separate BiP binding from binding to co-chaperones in order to determine their individual preferences. Mutational analysis of interacting peptides was used to identify the key binding determinants by creating or disrupting binding sites, both in the context of the peptide or introduced into full length, well-folded substrates.
Consistent with the low binding specificity of Hsp70s in general and the known allosteric modulation of substrate binding by ATP (Clerico et al., 2015; Mayer et al., 2003), BiP bound to varying extents with 32 of the 33 overlapping peptides tested, and its binding was diminished by the addition of ATP (Behnke et al., 2016). Binding of ERdj3 to the tested substrates required chemical crosslinking, and its binding selectivity was found to largely overlap with that of BiP. The shared promiscuous binding of BiP and ERdj3, along with their association with Sec61 is proposed to reflect their role in Sec61-mediated translocation of nascent chains into the ER lumen (Guo and Snapp, 2013; Hamman et al., 1998).
The most exciting and provocative results from the Beknke et al. study involved the analysis of Grp170, ERdj4 and ERdj5 substrate-binding selectivity (Behnke et al., 2016). Grp170, ERdj4 and ERdj5 bound to a restricted number of overlapping aggregation-prone peptides (6 of the 33 peptides). Peptide binding occurred in the presence of ATP, indicating that the association of these co-chaperones with peptides was not mediated by BiP. Peptides that did not bind these co-chaperones could be modified to support binding by increasing their aggregation propensity as predicted using the TANGO algorithm (Fernandez-Escamilla et al., 2004). Increased co-chaperone binding correlated with a decrease in half-life for some of the peptides, consistent with previously proposed roles of Grp170, ERdj4 and ERdj5 in ER-associated degradation (ERAD) (Guo and Snapp, 2013; Hamman et al., 1998). When the authors’ mutational approach was used to create binding sites in a full-length protein that normally did not bind these co-chaperones, binding indeed occurred. Somewhat puzzlingly, the mutated proteins formed stable aggregates instead of succumbing to degradation. Nonetheless, altogether these results have opened the door to new sequence-encoded specialization of Hsp70 co-chaperones and raise the possibility that their substrate-binding preferences determine their roles in the protein homeostasis choreography of the ER lumen: facilitating translocation, maturation or degradation.
The work by Behnke et al. exploits a clever experimental approach to characterize interactions of substrates with ER chaperone components in their normal physiological context (Figure 1). That said, the strategies used to detect the interactions necessitated some protocol features that may limit the generality of the observations: First, both the interrogated chaperones and the model substrates were transiently overexpressed, leading to higher than physiological concentrations. This favors the formation of bound complexes, displacing other cellular species that are present at lower concentrations and unable to compete for binding. Would these substrates bind the same way if pitted against the array of physiological substrates present at their normal concentrations? Second, BiP was not overexpressed when the interactions of model substrates with the ERdj’s were assayed. While this strategy allows the authors to observe the substrate-binding abilities of the co-chaperones in the ER luminal environment, conclusions can cannot be drawn about how BiP participates in the binding reaction. Would the observed scenarios change if BiP and ERdj’s were both present at physiological concentrations? Third, because substrate binding by ERdj3 was not stable during the detection procedure, its binding was tested in the presence of crosslinkers. Would crosslinking have altered the observations on binding of Grp170, ERdj4 and ERdj5 to the same substrates?
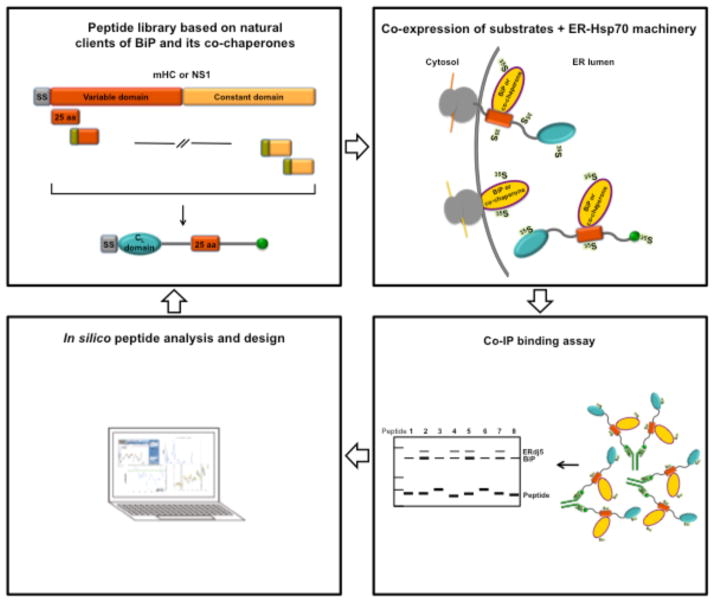
Figure 1. Experimental scheme for the cell-based peptide binding analysis for BiP and its co-chaperones.
Top left panel, ER-targeted constructs were created that contain 25-amino acid long overlapping segments from two bona fide BiP substrates, mHC and NS1. Top right panel, peptide constructs were co-expressed in COS-1 cells with BiP or its co-chaperones and metabolically labelled with [35S]-Met/Cys. Bottom right panel, BiP and co-chaperone binding to the peptides was monitored by co-immunopreciptation and resolved by SDS-PAGE and autoradiography. Bottom left panel, the aggregation propensity of peptides and proteins were analyzed using the TANGO algorithm. Peptides and proteins were mutated to alter aggregation propensity and then re-analyzed for chaperone binding.
A tantalizing outgrowth of this work will be tying the observations on Grp170, ERdj4 and ERdj5 to other processes ongoing in the ER, in particular ERAD. The fact that ERdj4, ERdj5 and Grp170-binding sequences introduced to a domain lacking BiP or co-chaperone binding sequences caused aggregation of the substrates might be because they were jamming the downstream degradation components because of their high concentration. Could the described aggregation-prone sequences cause misfolding or aggregation of the peptides/protein tested and then recruitment of ERdj4, ERdj 5 and Grp170 to rescue misfolding? What is the role of BiP in these processes? Could BiP and Grp170 conspire to act as a disaggregase as has been suggested for their cytoplasmic orthologues Hsp70 and Hsp110 (Gao et al., 2015)?
Moving detailed characterization of chaperone functions and partnerships into their cellular context is a pressing goal, and this work describes an elegant method to test interactions between proteins in the ER. Future studies must build on this work and link substrate-chaperone interactions with secretion, folding, aggregation, and degradation under physiological conditions with the full panoply of associated cellular components.
References
- Behnke J, Feige MJ, Hendershot LM. BiP and its nucleotide exchange factors Grp170 and Sil1: mechanisms of action and biological functions. J Mol Biol. 2015;427:1589–1608. doi: 10.1016/j.jmb.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke J, Mann MJ, Scruggs F-L, Feige MJ, Hendershot LM. Members of the Hsp70 family recognize distinct types of sequences to execute ER quality control. Molecular Cell. 2016 doi: 10.1016/j.molcel.2016.07.012. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I, Hebert DN. Protein folding in the endoplasmic reticulum. Cold Spring Harb Perspect Biol. 2013;5:a013201. doi: 10.1101/cshperspect.a013201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerico EM, Tilitsky JM, Meng W, Gierasch LM. How hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J Mol Biol. 2015;427:1575–1588. doi: 10.1016/j.jmb.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- Gao X, Carroni M, Nussbaum-Krammer C, Mogk A, Nillegoda NB, Szlachcic A, Guilbride DL, Saibil HR, Mayer MP, Bukau B. Human Hsp70 Disaggregase Reverses Parkinson’s-Linked alpha-Synuclein Amyloid Fibrils. Mol Cell. 2015;59:781–793. doi: 10.1016/j.molcel.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Snapp EL. ERdj3 regulates BiP occupancy in living cells. Journal of cell science. 2013;126:1429–1439. doi: 10.1242/jcs.118182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamman BD, Hendershot LM, Johnson AE. BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell. 1998;92:747–758. doi: 10.1016/s0092-8674(00)81403-8. [DOI] [PubMed] [Google Scholar]
- Mayer M, Reinstein J, Buchner J. Modulation of the ATPase cycle of BiP by peptides and proteins. J Mol Biol. 2003;330:137–144. doi: 10.1016/s0022-2836(03)00556-4. [DOI] [PubMed] [Google Scholar]