Abstract
A wealth of valuable data is locked within the millions of research articles published each year. Reading and extracting pertinent information from those articles has become an unmanageable task for scientists. This problem hinders scientific progress by making it hard to build on results buried in literature. Moreover, these data are loosely structured, encoded in manuscripts of various formats, embedded in different content types, and are, in general, not machine accessible. We present a hybrid human-computer solution for semi-automatically extracting scientific facts from literature. This solution combines an automated discovery, download, and extraction phase with a semi-expert crowd assembled from students to extract specific scientific facts. To evaluate our approach we apply it to a challenging molecular engineering scenario, extraction of a polymer property: the Flory-Huggins interaction parameter. We demonstrate useful contributions to a comprehensive database of polymer properties.
Keywords: Crowdsourcing, Information Extraction, Classification, Flory-Huggins, Materials Science
1 Introduction
The amount of scientific literature published every year is growing at a prolific rate. Some studies count more than 28,000 scientific journals and 1.8 million articles published annually [19]. As a result, the amount of information (e.g., experimental results) embedded within the literature is overwhelming. It has become impractical for humans to read and extract pertinent information. This problem hinders the advancement of science, making it hard to build on existing results buried in the literature. It also makes it difficult to translate results into applications and thus to produce valuable products. In materials science and chemistry, for example, difficulties discovering published materials properties directly affect the design of new materials [6]. Indeed, despite the many publications in this domain, the process of designing new materials is still one of trial and error. Access to a structured, queryable database of materials properties would facilitate the design and model validation of new substances, improving efficiency by enabling scientists and engineers to more quickly discover, query, and compare properties of existing compounds. At the very least, it would transform an avalanche of publications into a machine-accessible and human-consumable source of knowledge.
Historically, materials properties have been collected in human-curated review articles and handbooks (e.g., the Physical Properties of Polymers Handbook [7], the Polymer Handbook [18]). However, this approach is laborious and expensive, and thus such collections are published infrequently. We contend that a better approach is to leverage information extraction techniques to process thousands of papers and output structured content for human consumption. To this end, we have developed a semi-automated system, χDB, which, with moderate input from humans, can extract materials properties for the scientific community.
We initially target extraction of a fundamental thermodynamic property called the Flory-Huggins interaction (or χ) parameter, which characterizes the miscibility of polymer blends. We chose to work with this property as a test case as it is particularly challenging to extract, due to the fact that it is published in heterogeneous data formats (e.g., text, figures, tables) and is represented in several different temperature-dependent expressions. To address these challenges, we developed a workflow consisting of an automated Web information extraction phase followed by a crowdsourced curation phase. The output of this workflow is a high quality human- and machine-accessible digital handbook of polymer properties. We show that we are able, using only a small group of students, to create a high quality database of properties with more χ values than in other notable handbooks. We expect that our approach is likely also to work well for other materials properties and in other scientific domains.
The rest of this paper is organized as follows. Section 2 presents background information related to Flory-Huggins theory and polymer science. Section 3 discusses related approaches that support automated extraction. Section 4 describes the χDB architecture. Section 5 presents the data collected via crowdsourcing. Section 6 explores the application of machine learning algorithms to improve the automatic selection of χ-relevant publications. Finally, we conclude and discuss future work in Section 7.
2 Application Background
The initial focus of our work is the extraction of properties of particular polymers blends (e.g, χ parameter and glassification temperature). Although highly curated properties database exist for hard [8] and metallic [17] materials, no equivalent exists for polymers blends. However, there is a clear need for a trusted, up-to-date, and easily accessible databases of properties within the soft matter community.
Polymers are large molecules (macromolecules) composed of many repeating units. Since polymeric materials are both ubiquitous and typically consist of several polymeric components, which are generally incompatible, the χ parameter represents a key property in the design of next-generation materials. A database of χ values would allow researchers to make informed judgments as to which χ values and thermodynamic analysis to use when predicting and understanding the phase behavior of multi-component polymeric materials. However, while there are thousands of published χ parameters, there is little consensus regarding the values. Different measurement methods yield different values, and different groups have at times reported different values for the same polymers. The χ parameter depends on the temperature and the types of polymer(s) or solvent(s) involved. Consequently, many experimental methods have been developed to quantify the temperature dependence of χ, and tabulated values are commonly found in standard textbooks and polymer data handbooks [7, 18]. However, many of these values have not been updated to include recent findings. Moreover, the list of polymer blends found in textbooks is not exhaustive; for example the previously mentioned handbook contains χ values for only 41 polymer-polymer blends. These considerations motivate our goal to collect and store χ values from materials literature into a digital, searchable database. Each record would also include the source and the measurement methodology.
3 Related Work
We review here current practice for building collections of scientific facts, populating scientific databases, information extraction, and crowdsourcing.
Major scientific databases have emerged in various fields where data is growing at exponential rates and the need for data sharing is recognized by the community, notably in biotechnology [2, 9]. In materials science, the Materials Project [8] provides access to large numbers of computed values. For polymers, the expert-curated Physical Properties of Polymers Handbook [7], last published in 2007, is a valuable source of data. However, while a valuable resource, it lacks recent results from the literature and does not contain an exhaustive list of polymers.
Information extraction (IE) from text has been extensively studied [5]. IE aims to extract structured information from unstructured and semi-structured documents. It often focuses primarily on extracting information from written language via natural language processing [4]. Sub-disciplines include Web IE [1] and IE from PDF documents and images. Web IE leverages the inherent structure in HTML rather than grammatical rules to extract semantically meaningful information. Web IE approaches work well when extracting information from many pages with the same structure (e.g., real estate listings); however, they do not work well for heterogeneous web pages or when page structure changes [10]. Extracting information from other data types, such as images and PDFs, is particularly difficult. In the case of images, variations in texture, contrast, font size, style and color, orientation, alignment, etc., all impact the extraction process. Similarly, PDF files, while easy to understand for humans, are not designed for machine accessibility. Thus, it is challenging to extract information from embedded items—such as tables and equations—due to the lack of structure in the document. For example, extraction of tables from PDF documents typically relies on identifying cell borders and attempting to map text locations relative to these borders. As tables differ significantly between documents, a considerable amount of human assistance is needed to achieve good results.
One solution to the challenges associated with PDF files is to use experts to identify and correct errors [15]. Indeed, given inaccuracies in IE methods, many IE systems rely on teams of people to review and curate extracted information [14]. Such crowdsourcing approaches leverage the fact that humans perform certain tasks better than computers, an idea also exploited in systems such as Galaxy Zoo [12], for image labeling in astronomy; the Amazon Mechanical Turk micro-task marketplace [3], and the Wikipedia online encyclopedia.
4 χDB Architecture and Implementation
Mining the literature for a loosely structured property such as the χ parameter requires extracting values from a variety of objects, including text, figures, tables, and equations; processing the many different forms in which the property occurs, e.g., a single number at a given temperature or a linear equation as a function of temperature; and identifying associated information such as the polymers and solvents involved, their molecular masses, the temperature(s) at which experiments were performed, the methods used, and any error estimates. Thus, the techniques used to find, extract and store χ must be flexible.
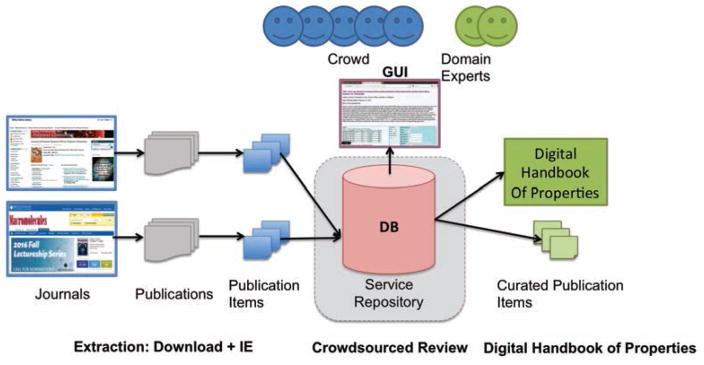
Given these multiple levels of complexity, we have developed χDB—a hybrid machine-human system that leverages both automatic extraction and expert human review via crowdsourcing. The χDB workflow shown in Figure 1 comprises three main phases: automatic download and first-level extraction of publications; crowdsourced extraction and review (the “review process”) of χ values, and finally the exposure of a curated database of χ values (the “Digital Handbook of Properties”). In the rest of this section, we define the χDB data model and then describe the system architecture used to realize each of these workflow phases.
Figure 1.
χDB architecture
4.1 Data Model
The χDB data model is designed to represent (1) the complex extraction and review workflow, (2) the various temperature-dependent formats in which χ occurs, and (3) the complete provenance of each extracted value. To model the different users’ reviews the data model includes a representation of publications before, during, and after reviews, as well as a data model for the multiple representations of χ. The χDB data model includes seven core tables: papers (extracted publications), items (extracted publication items), sources and reviewed sources (reviewed information before and after consensus), chis and reviewed chis (χ values before and after consensus), and reviewed papers (classified papers). One challenge when defining the data model is the need to support different representations in which χ is specified. After reviewing the literature we developed a data model that could include four main representations of χ: 1) a number at a specific temperature; 2) a linear equation in terms of temperature: ; 3) a quadratic equation in terms of temperature: ; 4) a number that combines χ and N, where N is proportional to the degree of polymerization or molecular weight: χN; and a final catch-all class, 5) other representations.
4.2 Extraction
χDB first discovers and downloads relevant publications—in this case publications that contain the keyword Flory-Huggins—from suitable journals. It then uses an HTML tag parser to extract structured publication metadata, including Digital Object Identifier (DOI), title, authors, and date of publication. This information is used to index the publication such that it can be linked to other stored information (e.g., referenced values in other papers). Finally, the publication is parsed into items (e.g., abstract, figures, tables, equations, text) that are separately downloaded and can be reviewed individually. Links between publication items and their originating publication are maintained so that they can be displayed to reviewers in a coherent manner. The full text and the original URL are also stored such that reviewers and users can retrieve the original publication.
We implemented this phase in three components: a Python web crawler (to discover relevant publications), a downloader (to download a copy of the publication), and a WebIE extractor (to extract metadata and items from the publication). We initially focused on Macromolecules, a leading scientific journal on polymers. The crawler is configured to use the Macromolecules search capabilities to prioritize downloads. After discussion with experts, we chose the search term Flory-Huggins and specified a date range from January 2010. The crawler returns a ranked list of publications. The downloader uses these results to download each publication (as an HTML file) using the URL returned by the crawler. The downloader extracts relevant metadata from the structured web page (DOI, title, authors, etc.) Finally, a Python WebIE script parses the HTML to detect and extract items from the publication (e.g., abstract, images, equations, and tables). The abstract and the HTML tables are stored directly in the χDB database. Figures and equations are downloaded and referenced in the database.
4.3 Crowdsourced Review
To assemble a crowd for reviewing extractions we developed a materials science course that combined teaching the fundamentals of polymer chemistry and physics and reviewing the literature containing χ parameters. The reviewing component of the course tasked the students with extracting χ parameters using the χDB system. This involved reviewing the free-text publication, and entering any χ values that they identified.
We implemented this phase as a PHP-based web service and PHP/HTML website. Due to copyright restrictions, the reviewing components of χDB are accessible only within the University of Chicago network. The review interface includes two main pages: a list of all publications with assigned reviewers and a review page for reviewing publications and items. We implemented a consensus-based review process using two reviewers per paper to reduce error. We rely on a second class of reviewers (experts) to resolve conflicting reviews.
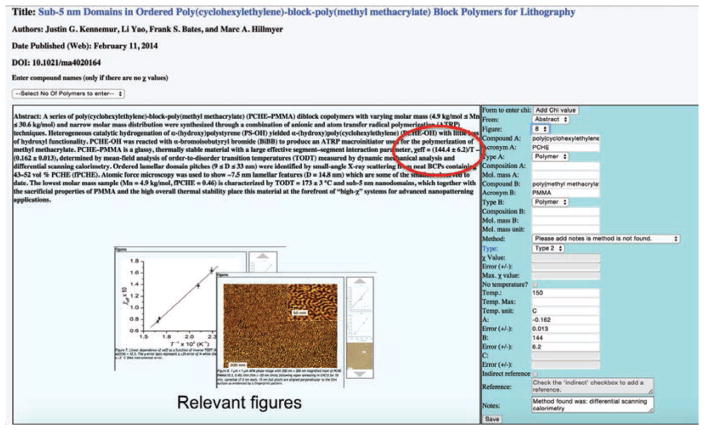
An individual review consists of scanning extracted items for χ values. Once identified, reviewers are asked to extract χ values from all of these items, with the exception of figures as extractions from figures are likely to be inaccurate. The reviewer enters each extracted χ value in an online form. The item from which a value is extracted is marked as relevant. Note: items may be marked as relevant even if they do not contain any χ values. For example, a relevant figure may be a phase diagram or a micrograph of the material; a relevant table may contain supporting information. If a paper contains a single χ value or a single relevant item, it is also marked as relevant. Consequently, a paper that contains neither is classified as irrelevant. Figure 2 shows an example of the review form. To ensure that the resulting database is unambiguous, we define a set of minimum required information for submission of a χ value. Some χ values are embedded directly in the text (rather than in an extracted item); therefore reviewers are able to retrieve the full text article via the link on the review page. If additional χ values are found in the full text, reviewers click the “Add Chi” button next to the abstract with the possibility to indicate in the form that the value was actually extracted from the main text. Second reviews of the same publications consist of a similar process, however second reviewers are able to view the previous reviewers’ input before submitting their own, giving them the opportunity to identify errors or conflicts between reviews. In the case of errors, the interface allows submission of either review; in the case of conflicts it allows the publication to be flagged for expert review.
Figure 2.
Screenshot of the χDB Graphical User Interface with the χ entry form enabled
Students reported an average of 15 minutes to review relevant publications and five minutes to review irrelevant publications. Submissions from second reviewers are automatically stored in our Digital Handbook of χ values.
4.4 Digital Handbook of χ Values
Once a χ value has passed through the review cycle, it is stored in the curated section of the database with associated provenance information that links the value back to the original publication, the item in which it was found, and the reviewers that extracted the value. To facilitate broad access to the database, χDB offers a web service API and HTML website. The website allows users to browse and search the database for specific χ values. The web service API supports ingestion of χ values directly from custom applications, for example to retrieve χ values for a set of specific polymers that may then be used for calculations or visualizations. Both the website and web service are available at http://pppdb.uchicago.edu.
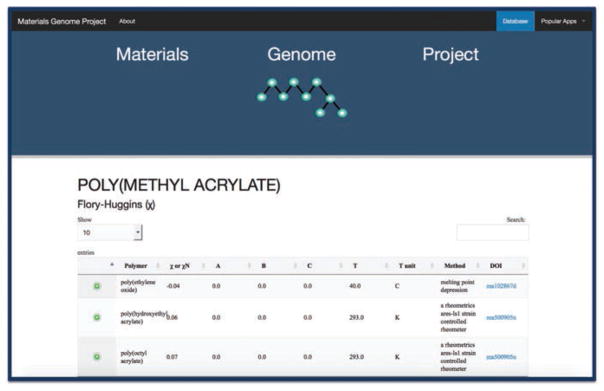
The website allows users to query for information related to a particular polymer. Once the user selects a particular polymer from the search interface, he or she is presented with a table of searchable χ values that relate to that polymer. Each row in the table includes the second compound (polymer or solvent) involved in the interaction, the measurement method used (where available), the temperature at which the parameter was measured (in various forms), and a link to the original publication. Rows can also be expanded to show additional metadata such as molecular masses and concentration. Figure 3 shows an example of χ values for poly(methyl acrylate) in the Digital Handbook.
Figure 3.
Screenshot of the χDB Digital Handbook
The χDB REST API supports querying the Digital Handbook for χ values that relate to a specific polymer-polymer or polymer-solvent pair. The REST API has been used to create a Flory-Huggins phase diagram generator for specific polymer blends. This application determines the liquid-liquid curves for a binary blend of polymers, as well as a polymer solution.
5 Results
During the class and over a two month period immediately thereafter, students reviewed 376 publications from the period 2010–2015 in Macromolecules. We briefly explore here the results of extractions, looking specifically at the characteristics of the χ values, the range of compounds for which χ values were collected, and the methods used to derive χ values.
χ Values
Of the 376 publications reviewed, students deemed 259 (69 %) of the papers relevant, of which 145 (38.5 %) of the papers contained one or more χ values. Our dataset includes 388 χ values, including 237 (61 %) polymer-polymer χ values. Measured χ values account for approximately half (48.5 %) of all χ values extracted, the other half (51.6 %) are cited from other publications. Of these measured values, the dataset includes 84 (21.7 %) measured polymer-polymer χ values. In the most focused case of measured polymer-polymer pairs, we found that 70.9 % of χ values were embedded directly in publication text, and 9.7 % in the abstract. Combined, these values indicate that mining text for χ values would potentially capture about 80 % of χ values. The vast majority (89.0 %) of χ values that we identified were published as type 1 or 2 i.e., a number or a linear function of temperature.
Compounds
Polystyrene (PS) is the most studied polymer by a large margin, with 140 χ values collected. The second and third most frequent, Poly(methyl methacrylate) (PMMA) and Polyisoprene (PI), have 59 and 22 χ values, respectively. The average number of χ values per polymer is 4.74. Not surprisingly, the most frequent polymer pair is PS–PMMA, with 36 χ values.
Methods
One final area of great interest to our experts was evaluating the method used to measure the χ values. Unfortunately the method was not always present (or clear) in publications. Students were unable to identify the method for 62 (16.0 %) of the 388 χ values found and were unsure about 12 others (3.1 %), resulting in a total of 19.1 % χ values with no identified method. Originally, experts provided a list of seven methods that they expected would be commonly used. Analysis of our dataset reveals that, for the target case of measured polymer-polymer values these methods are indeed the most commonly used, with only four of the 84 measured polymer-polymer values not using one of these seven methods.
6 Automated Classification
While our approach has established a rich database of χ values, there is potential for further improvements. For example, only 38.5 % of our selected publications contained χ values; thus, about 62 % of the papers curated by reviewers did not in fact contribute to the digital handbook. As a first step towards improving this ratio we have investigated the application of machine learning techniques to optimize the prioritization and classification of relevant publications.
To undertake this task, we used the Support Vector Classifier (SVC) from Scikit Learn [11], an open source machine learning Python library. SVC is an implementation of Support Vector Machines (SVMs), supervised learning models with associated learning algorithms that analyze data and recognize patterns. The models map data into a feature space to make predictions.
Three performance metrics are commonly used to evaluate the accuracy of classifiers: precision, recall, and F-measure. Precision and recall are expressed in terms of Positive and Negative predictions, i.e., in our case Contains χ and Does not contain χ; True and False predictions correspond to correct and incorrect predictions. Precision measures the percentage of predictions that were correct while recall measures the percentage of items in the test dataset that were correctly predicted. Precision and recall are defined in Equations 1 and 2.
| (1) |
| (2) |
The FX -score is a measure of a test’s accuracy. The traditional F-measure or balanced Fscore (F1 score) is the harmonic mean of precision and recall; it can be interpreted as a weighted average of the precision and recall, with a best value of 1 and worst of 0. The general formula for positive real β is defined in Equation 3.
| (3) |
6.1 Test dataset
Our datasets include two sets of abstracts. The first set is composed of all abstracts of publications reviewed by the students, each of which has been classified by them as either relevant or irrelevant. These 376 publications were selected by the χDB crawler and are therefore biased by the Flory-Huggins keyword search. (However, as previously discussed, only 145 of these publications contained χ values.) To address this bias we downloaded an additional 135 publications from two arbitrarily chosen issues of Macromolecules (January 12, 2010 and January 26, 2010). Table 1 shows the sets of abstracts used in the classification of abstracts; we call the initial and biased set of abstracts “biased abstracts” and the larger set, which contains both the original 376 biased abstracts and the additional 135 unbiased abstracts, “All abstracts.” To classify the additional set of papers we visually inspected the abstracts and full text of each publication and reviewed them for χ values.
Table 1.
Characteristics of the abstracts used as input to the classification process
| Category | Biased abstracts | Unbiased abstracts | All abstracts |
|---|---|---|---|
| Relevant | 145 | 2 | 147 |
| Irrelevant | 231 | 133 | 364 |
| Total | 376 | 135 | 511 |
6.2 Results
We applied Scikit Learn’s Support Vector Classifier to the set of abstracts, varying just the criteria used to identify abstracts as relevant or irrelevant. The features used by the classifier are generated using a word-weighting scheme commonly used in information retrieval [13]. The abstracts are first converted to a matrix of token counts and subsequently transformed into a normalized tf-idf (term frequency-inverse document frequency) representation. The two terms are multiplied in order to reduce the impact of terms that occur frequently in a given corpus and thus are less informative. We used three different definitions of relevancy: includes χ value; includes measured χ value; and includes measured polymer-polymer χ value.
Table 2 shows that the performance of the classifier for both sets of abstracts. Accuracy improves as relevancy becomes more specific. We also see a small (≈3–7 %) improvement in accuracy when using all abstracts. When using all abstracts, the accuracy of classifying measured polymer-polymer relevant papers is 86.9 % precision and 90.9 % recall.
Table 2.
Classification of abstracts in χDB
| Relevancy (contains) | Metric | Biased abstracts | All abstracts |
|---|---|---|---|
|
| |||
| χ Values | Mean F1 score | 0.624 | 0.679 |
| Mean precision score | 60.5 % | 65.1 % | |
| Mean recall score | 64.5 % | 71.2 % | |
|
| |||
| Measured χ values | Mean F1 score | 0.790 | 0.835 |
| Mean precision score | 75.9 % | 80.9 % | |
| Mean recall score | 82.2 % | 86.4 % | |
|
| |||
| Measured polymer-polymer χ values | Mean F1 score | 0.852 | 0.890 |
| Mean precision score | 82.7 % | 86.9 % | |
| Mean recall score | 87.8 % | 90.9 % | |
There is a tradeoff between maximizing the number of relevant publications (and minimizing the number of irrelevant publications) retrieved. Deciding whether these scores are acceptable depends on the cost of errors (false negatives and false positives). Our observed precision score (of 86.9 %) means that 13.1 % irrelevant papers remain; a considerable improvement over the initial 61.5 % of publications that did not contain χ values. The recall score of 90.9 % means that we misclassify ≈9 % of relevant papers. As ideally we would like to capture all such publications, further work should aim at improving this score. Nevertheless, our results demonstrate the potential of capturing a significant portion of targeted publications in the literature.
We observe that the top 25 features (words) used by our classifier in the most focused case of polymer-polymer pairs include a mixture of more or less χ-related terms. For example, terms like “process,” “parameter,” and “form” could refer to various experimental settings. On the other hand, the word “domains” (as in microphase domains) is relevant to measuring χ and is also used for a wide variety of applications in which χ is important. χ is a measure of polymer-polymer “interaction” that is present in the list of features. Microphase “morphologies” are relevant to measuring χ via phase diagrams. This combination represents a challenge in further isolating publications that are specifically related to χ and may require incorporating some domain knowledge into the χDB workflow.
7 Conclusion and Future Work
As part of a long-term project to create a digital handbook of polymer properties, we have developed χDB, a hybrid human computer-system that extracts the Flory-Huggins (or χ) parameter from scientific literature. Our work to date has extracted 388 χ values for 120 polymers and 30 solvents. Our 237 measured χ values for blends of 63 unique polymers exceed the 134 χ values for blends of 41 unique polymers found in the Physical Properties of Polymers Handbook [7]. One reason for our superior performance is that we were able to collect values reported after the 2007 publication of the Handbook (84 of our χ values are from 2010 to 2015); another is that our more exhaustive search leads us to find earlier values not reported in the Handbook. Our results emphasize the potential for using our approach to create and maintain a digital database of χ parameters that is more comprehensive and up to date than any survey publication. The database is currently available at http://pppdb.uchicago.edu.
Using publications marked relevant and machine learning software, we were able to improve the publication selection process considerably, decreasing the number of reviewed publications that do not contribute to the χ database from 61.5 % to 13.1 %. We hope in future work to further improve this classification process by using alternative methods and by integrating polymer science insight gained through exploration of our data collection. For example, we will explore the utility of using more frequently occurring methods as a publication filter prior to running the classifier. We are exploring collaborations with journals in order to gain access to more publications and mine more properties. While this work is focused on χ, the steps required to collect a new property are straightforward; first the crawler must be configured to use a different keyword; the schema for the target property will guide the design of a new input form and the corresponding database table. Future work will also involve addressing crowdsourcing challenges in order to recruit more trained users and experts. Scaling out χDB will also lead us to explore deep learning systems for fact extraction [16].
Acknowledgments
We thank the student contributors to χDB. This work was supported in part by NIST contract 60NANB15D077, the Center for Hierarchical Materials Design, and DOE contract DE-AC02-06CH11357. Certain commercial equipment and/or materials are identified in this report in order to adequately specify the experimental procedure. In no case does such identification imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the equipment and/or materials used are necessarily the best available for the purpose.
References
- 1.Banko M, Cafarella MJ, Soderland S, Broadhead M, Etzioni O. Open information extraction for the web. Proceedings of the 20th International Joint Conference on Artifical Intelligence (IJCAI) 2007:2670–2676. [Google Scholar]
- 2.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL. Genbank. Nucleic Acids Research. 2000;28(1):15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buhrmester M, Kwang T, Gosling SD. Amazon’s Mechanical Turk A new source of inexpensive, yet high-quality, data? Perspectives on Psychological Science. 2011;6(1):3–5. doi: 10.1177/1745691610393980. [DOI] [PubMed] [Google Scholar]
- 4.Cambria Erik, White Bruce. Jumping nlp curves: a review of natural language processing research [review article] Computational Intelligence Magazine, IEEE. 2014;9(2):48–57. [Google Scholar]
- 5.Cowie J, Lehnert W. Information extraction. Communications of the ACM. 1996;39(1):80–91. [Google Scholar]
- 6.de Pablo JJ, Jones B, Kovacs CL, Ozolins V, Ramirez AP. The Materials Genome Initiative, the interplay of experiment, theory and computation. Current Opinion in Solid State and Materials Science. 2014;18(2):99–117. [Google Scholar]
- 7.Eitouni HB, Balsara NP. Physical Properties of Polymers Handbook. Springer; 2007. Thermodynamics of polymer blends; pp. 339–356. [Google Scholar]
- 8.Jain A, Ping Ong S, Hautier G, Chen W, Richards WD, Dacek S, Cholia S, Gunter D, Skinner D, Ceder G. Commentary: The Materials Project: A materials genome approach to accelerating materials innovation. APL Materials. 2013;1(1):011002. [Google Scholar]
- 9.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, et al. The NCBI dbGaP database of genotypes and phenotypes. Nature Genetics. 2007;39(10):1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muslea I. Extraction patterns for information extraction tasks: A survey. The AAAI-99 Workshop on Machine Learning for Information Extraction; 1999. pp. 1–6. [Google Scholar]
- 11.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. Scikit-learn: Machine learning in Python. Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- 12.Raddick MJ, Bracey G, Gay PL, Lintott CJ, Cardamone C, Murray P, Schawinski K, Szalay AS, Vandenberg J. Galaxy Zoo: Motivations of citizen scientists. 2013 arXiv preprint arXiv:1303.6886. [Google Scholar]
- 13.Ramos Juan. Using tf-idf to determine word relevance in document queries. Proceedings of the first instructional conference on machine learning. 2003 [Google Scholar]
- 14.Rzhetsky A, Iossifov I, Koike T, Krauthammer M, Kra P, Morris M, Yu H, Duboué PA, Weng W, Wilbur WJ, Hatzivassiloglou V, Friedman C. GeneWays: A system for extracting, analyzing, visualizing, and integrating molecular pathway data. Journal of Biomedical Informatics. 2004;37(1):43–53. doi: 10.1016/j.jbi.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 15.Seifert C, Granitzer M, Höfler P, Mutlu B, Sabol V, Schlegel K, Bayerl S, Stegmaier F, Zwicklbauer S, Kern R. Human-Computer Interaction and Knowledge Discovery in Complex, Unstructured, Big Data. Springer; 2013. Crowdsourcing fact extraction from scientific literature; pp. 160–172. [Google Scholar]
- 16.Shin J, Wu S, Wang F, De Sa C, Zhang C, Ré C. Incremental knowledge base construction using Deepdive. Proceedings of the VLDB Endowment. 2015;8(11):1310–1321. doi: 10.14778/2809974.2809991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spencer PJ. A brief history of CALPHAD. Calphad. 2008;32(1):1–8. [Google Scholar]
- 18.Van Krevelen Dirk Willem, Nijenhuis Klaas Te. Properties of polymers: their correlation with chemical structure; their numerical estimation and prediction from additive group contributions. Elsevier; 2009. [Google Scholar]
- 19.Ware M, Mabe M. The STM report: An overview of scientific and scholarly journal publishing. Technical report, STM. 2009 [Google Scholar]