Abstract
Nitrile hydratase (NHase) from Rhodococcus rhodochrous J1 is widely used for industrial production of acrylamide and nicotinamide. However, the two types of NHases (L-NHase and H-NHase) from R. rhodochrous J1 were only slightly expressed in E. coli by routine methods, which limits the comprehensive and systematic characterization of the enzyme properties. We successfully expressed the two types of recombinant NHases in E. coli by codon-optimization, engineering of Ribosome Binding Site (RBS) and spacer sequences. The specific activity of the purified L-NHase and H-NHase were 400 U/mg and 234 U/mg, respectively. The molecular mass of L-NHase and H-NHase was identified to be 94 kDa and 504 kDa, respectively, indicating that the quaternary structure of the two types of NHases was the same as those in R. rhodochrous J1. H-NHase exhibited higher substrate and product tolerance than L-NHase. Moreover, higher activity and shorter culture time were achieved in recombinant E. coli, and the whole cell catalyst of recombinant E. coli harboring H-NHase has equivalent efficiency in tolerance to the high-concentration product relative to that in R. rhodochrous J1. These results indicate that biotransformation of nitrile by R. rhodochrous J1 represents a potential alternative to NHase-producing E. coli.
Introduction
Nitrile hydratase (NHase, EC 4.2.1.84) catalyzes the hydration of a nitrile molecule to the corresponding amide [1]. NHase forms a hetero-tetramer that is composed of α- and β-subunits with either a non-heme iron (Fe-NHase) or non-corrin cobalt ion (Co-NHase) in the active center [2]. The α-subunits, which share a characteristic metal-binding motif [CXLC(SO2H)SC(SOH)] containing two modified cysteine residues, cysteine-sulfinic acid (αCys-SO2H) and cysteine-sulfenic acid (αCys-SOH), bind the metal ions in both Co-NHase and Fe-NHase [1, 3–5]. As observed in previous studies on NHase [6] and a related enzyme, the metal free enzyme is likely to be unmodified thiocyanate hydrolase (SCNase) [7].
The transfer of metal ions into NHases is accomplished by the function-associated protein called the “activator proteins” [8]. The activators for Fe-NHases have been shown to act as metallochaperones [9], whereas the “activator proteins” act as “self-subunit swapping chaperones” for cobalt incorporation into most Co-NHases [8, 10, 11]. Self-subunit swapping is one of the post-translational maturation steps of the cobalt-containing NHase (Co-NHase) family of enzymes [10]. The activator protein exists as a complex with the α-subunit of NHase, and cobalt incorporation into NHase is dependent on the α-subunit swapping between the cobalt-free α-subunit of the cobalt-free NHase and the cobalt-containing α-subunit of the complex [8]. Self-subunit swapping is quite different from the currently known general mechanisms of metal center biosynthesis [8, 10, 12]. Self-subunit swapping was first discovered in the low-molecular-weight NHase (L-NHase) of Rhodococcus rhodochrous J1 [8] and, subsequently, in the high-molecular-weight NHase (H-NHase) of the same strain [10]. This swapping is speculated to occur in other Co-NHases and in another NHase family enzyme, thiocyanate hydrolase, which also contains a unique noncorrin cobalt center with two post-translationally modified cysteine ligands [11, 13]. “Self-subunit swapping chaperones” exhibit a surprising protein function in contrast with metallochaperones in metal center biosynthesis and molecular chaperones in protein folding [8, 10].
NHase is widely used for production of acrylamide and nicotinamide in industrial settings. Industrial acrylamide production has been performed with NHase from Rhodococcus sp. N774 as the first-generation catalyst and with NHase from Pseudomonas chlororaphis B23 as the second generation catalyst [14]. Industrial acrylamide is now produced by NHase from R. rhodochrous J1 as the third generation catalyst due to its higher catalytic ability and tolerance to acrylamide [14]. The two NHases (H-NHase and L-NHase) from R. rhodochrous J1 have been expressed in R. rhodochrous ATCC12674 [15] and in R. facians DSM43985 [8], respectively, and the two NHases have been well-characterized. Although H-NHase and L-NHase have been successfully expressed in these two expression systems, both of them have the same defect as that in the R. rhodochrous J1, i.e., long incubation time and unstable expression level. The incubation time for R. rhodochrous J1 and the two expression systems are at least 72 h [16]. Compared to the expression system of Rhodococcus, E. coli has a particular advantage due to its simple cell culture and molecular biological manipulation. E. coli is expected to develop a genetically engineered E. coli for overproduction of NHase from R. rhodochrous J1. However, both H-NHase and L-NHase have not been overexpressed in the E. coli expression system, the H-NHase and L-NHase expressed by the E. coli expression system were only slightly detectable on SDS-PAGE, and the activity was very low [17].
In this study, we successfully expressed the two types of NHases from R. rhodochrous J1 via codon-optimization, substitution of stronger ribosome binding site (RBS), and optimization of spacing sequences between different genes. Especially, the catalytic properties of H-NHase and the applicable procedure for biotransformation using whole-cell catalysts were systematically determined to provide the basis for industrial transformation. The expressions of the two NHases were stable, and culture times were only 20 h, which was considerably faster than the wild-type strain and the expression system of other Rhodococcus (at least 72 h). It is possible that the genetically engineered E. coli could be an alternative to R. rhodochrous J1 for industrial applications.
Materials and methods
Strains, media, and culture conditions
E.coli JM109 was used for recombinant plasmid construction. E.coli BL21 (DE3) was used as the host for recombinant expression in this study. E. coli BL21 (DE3) transformants harboring recombinant plasmids were grown in 2×YT medium (16 g/L tryptone, 10 g/L yeast extract, 5 g/L NaCl) containing CoCl2·6H2O (1 g/L) and kanamycin (50 mg/L) at 37°C, and isopropyl β-D-thiogalactopyranoside (IPTG) was subsequently added to a final concentration of 0.6 mM. The cells were incubated at 200 rpm in the shaker at 24°C for 18 h.
R. rhodochrous J1 was from the stock in our laboratory, which has been used previously [8] and was used as a control in the present study. Pre-culture was performed at 30°C for 48 h in a 500 mL flask containing 50 mL of seed medium containing 0.3 g glucose, 0.3 g yeast extracts, 0.04 g urea, 0.016 g KH2PO4, 0.016 g K2HPO4, and 0.016 g MgSO4. Ten milliliters of the pre-culture was then inoculated into 100 mL fermentation medium of a 500 mL shaking flask containing 2.5 g glucose, 6 g yeast extract, 0.7 g urea, 0.06 g KH2PO4, 0.06 g K2HPO4, 0.06 g MgSO4, 0.1 g sodium glutamate, and 0.027 g CoCl2·6H2O. Incubation was performed at 28°C for 120 h [18].
Construction of plasmids
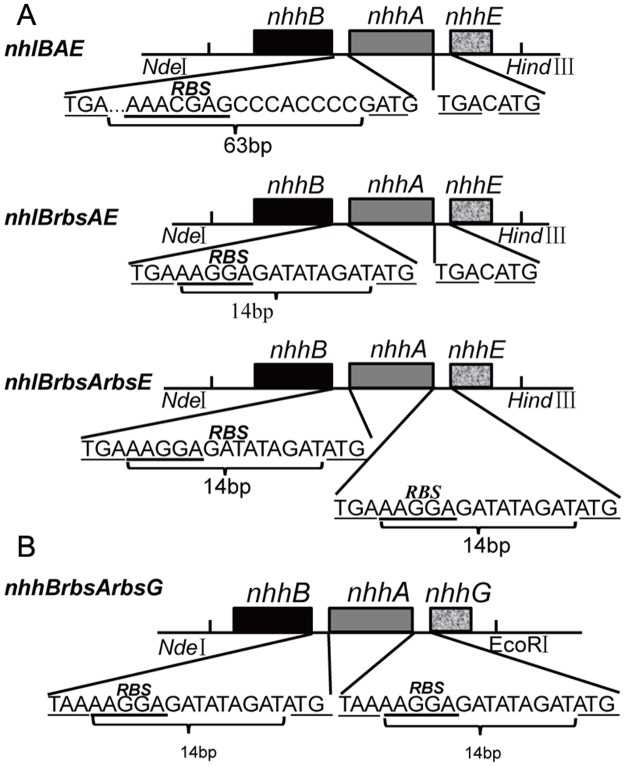
The primers used in this study were listed in Table 1. The plasmid pREIT19-nhlBAE from R. fascians DSM43985 [8] was isolated and used as a template to clone the nhlBAE gene (GenBank: DJ492951.1) with the primer pairs B-up and E-down (Table 1). The PCR product and the vector pET24a(+) were simultaneously digested with NdeI-HindIII, and the two fragments were jointed together, generating pET24a-nhlBAE (Fig 1A). Plasmids pET24a-nhlBrbsAE and pET24a-nhlBrbsArbsE, which contained a strong RBS sequence in front of nhlA and nhlE, respectively, were constructed using pET24a-nhlBAE as a template. Reverse PCRs were performed with primer pairs 1RBS-up and 1RBS-down, 2RBS-up and 2RBS-down, respectively (Fig 1A). To facilitate the purification and separation, Strep-tag was fused into the plasmid pET24a-nhlBrbsArbsE in front of nhlB by reverse PCR with primer Strep-B-up and Strep-B-down, generating plasmid pET24a-(B-Strep)rbsArbsE.
Table 1. Primers used for NHase expression.
| Name | Sequence(5’-3’) | Length (bp) |
|---|---|---|
| B-up | AAGGAGATATACATATGGATGGAATCCAC | 29 |
| E-down | CCCAAGCTTCTACCCGTCGGAGTCAGTGGTGC | 32 |
| 1RBS-up | CGAACCGTATCTGCTACCGGCCTGAAAGGAGATATAGATATGACCGCCCACAATCCCG | 58 |
| 1RBS-down | CGGGATTGTGGGCGGTCATATCTATATCTCCTTTCAGGCCGGTAGCAGATACGGTTCG | 58 |
| 2RBS-up | CACCACACCCAGCAAGGCCTGAAAGGAGATATAGATATGCCCCGACTCAACGAACAA | 57 |
| 2RBS-down | TTGTTCGTTGAGTCGGGGCATATCTATATCTCCTTTCAGGCCTTGCTGGGTGTGGTG | 57 |
| Strep -B-up | GTTTAACTTTAAGAAGGAGATATAGATATGGCAAGCTGGAGCCACCCGCAGTTCGAAAAGGATGGAATCCACGACCTCGGTGGCC | 85 |
| Strep -B-down | GGCCACCGAGGTCGTGGATTCCATCCTTTTCGAACTGCGGGTGGCTCCAGCTTGCCATATGTATATCTCCTTCTTAAAGTTAAAC | 85 |
The underlined sequences are the restriction sites. The bold and underlined sequences are the ribosome binding sites and spacer sequences.
Fig 1. Gene organization for the construction of a set of plasmids.
(A) Organization of the expression cassette for L-NHase. nhlBAE, wild-type L-NHase gene; nhlBrbsAE, the intervening sequences shortened gene with one enhanced RBS ahead of nhlA; nhlBrbsArbsE, the intervening sequences shortened gene with two enhanced RBSes ahead of nhlA and nhlE. (B) Gene organization of the constructed genes used for H-NHase expression.
The H-NHase genes were chemically synthesized with optimized codons based on the original H-NHase genes (GenBank: X86737.1) from R. rhodochrous J1, in which the strong RBS was inserted in front of nhhA and nhhG, resulting in the construction of nhhBrbsArbsG. The genes were inserted into pET24a to construct pET24a-nhhBrbsArbsG. The plasmid pET24a-nhhBAG harboring the native gene of H-NHase (nhhBAG) saved in our lab was used as a control for H-NHase expression in E. coli.
Purification and identification of the recombinant proteins
All purification steps were performed at 4°C. All procedures were carried out by an AKTA purifier (GE Healthcare UK Ltd.). E. coli transformant containing the Strep-tagged L-NHase was collected after centrifugation. Cells were lysed by ultra-sonication after being washed three times with Strep Binding Buffer (SBB) composed of 20 mM Na2HPO4, 280 mM NaCl, 6 mM KCl, and 0.5 mM dithiothreitol (DTT), pH 7.4. Next, the cells were re-suspended in the same volume of SBB prior to ultra-sonication. The supernatant and precipitation were separated by centrifugation at 4°C for 10 min at 18,000×g. The supernatant was loaded onto an affinity column (Strep-tag/Strep-Tactin affinity chromatography, GE Healthcare UK Ltd, USA) equilibrated with SBB and eluted with Strep Elution Buffer (2.5 mM d-Desthiobiotin, SBB, 0.5 mM DTT, pH 7.4) [19].
The purification step of H-NHase was as described in a previously reported method with certain modifications [15]. Potassium Phosphate Buffer (KPB) (10 mM, pH 7.4) containing 0.5 mM DTT was used over the purification process according to the previous report[5]. Briefly, after ultra-sonication, H-NHase was partly purified by ammonium sulfate fractionation (40–65%). Afterward, the dialyzed solution was loaded onto the Hitrap Q HP (GE Healthcare) column for separation and purification by a linear gradient elution with the concentration of KCl from 0.2 to 0.6 M in KPB. The solutions were fractionated automatically. The purity of L-NHase and H-Hase were subsequently determined by SDS-PAGE analysis.
Molecular mass determination of the purified proteins
Gel-filtration chromatograph analysis was performed for molecular mass determination, and MW-MARKER PROTEINS (ORIENTAL YEAST CO.,LTD) were used as a standard, which was in accordance with a previous report [8] with minor modifications. In this study, a Superdex 200 10/300 GL pg column (GE Healthcare UK Ltd.) equilibrated with 10 mM KPB (pH 7.4) was used for estimation of the molecular masses of the enzymes. This step was conducted with an AKTA purifier (GE Healthcare UK Ltd.) at 4°C, with a flow rate of 0.5 mL/min.
Determination of protein concentration and metal content
The purified protein was analyzed with the absorbance value at 280 nm with B-500 ultramicro ultraviolet spectrophotometer (Yuan Analysis Instrument Co., Ltd. Shanghai, China). The metal content of the purified enzymes at a concentration of 0.5 mg/ml were analyzed with a Shimadzu AA-7000 atomic absorption spectrophotometer under the following conditions: wavelength, 240.73 nm; lamp current, 12 mA; and slit width, 0.2 nm.
Enzymatic catalysis assay
The NHase activity was assayed as the procedures used previously [5]. The reaction mixture of a total volume of 0.5 mL was comprised of 10 mM KPB (pH 7.4), 400 mM 3-cyanopyridine as substrate, and 0.1 mg/ml purified enzyme. The reaction was performed at 25°C for 10 min and stopped by the addition of 0.5 mL acetonitrile. The amount of nicotinamide produced in the reaction mixture was determined as previously described [11]. One unit of NHase activity was defined as the amount of enzyme that catalyzed the production of 1 μmol of nicotinamide per min at 25°C. All of the experiments were performed independently in triplicate. The values were presented as mean±sd.
Substrate and product tolerance determination
To determine the substrate tolerance of the enzymes, the reaction was carried out in a mixture of a total volume of 0.5 mL containing 0.1 mg/ml purified enzyme and 0.4, 0.8, 1.0 M of 3-cyanopyridine in 10 mM KPB (pH 7.4), respectively. The activities under different substrate concentration were compared, and the activity using 0.4 M of 3-cyanopyridine was defined as 100%. For the product tolerance determination, 0.5 mL of mixture containing 0.1 mg/ml purified enzyme and 0.5, 1.0, 1.5 M of nicotinamide in 10 mM KPB (pH 7.4) were treated for half an hour at 25°C. Next, 0.1 M of 3-cyanopyridine was added into the mixture for reaction. The decrement of 3-cyanopyridine was measured by HPLC in the same method as nicotinamide and used for assay, and the activity without nicotinamide treatment was defined as 100%. To assay the nicotinamide-tolerance of NHases in recombinant E. coli and wild-type R. rhodochrous, we performed the measurement according to the method in a previous report with minor modification [20]. We prepared cells in 0.5 M nicotinamide for 20 min at 25°C. Next, 3-cyanopyridine was added prior to the enzymatic assay. The activity without the addition of a high concentration amide was defined as 100%. The relative activity was the ratio of which the activity of treated NHases accounted for the untreated NHases.
Results and discussion
Overexpression of L-NHase in E. coli
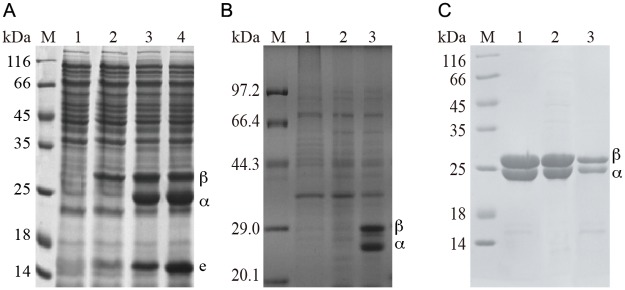
To heterologously express L-NHase in E. coli, the gene cluster of nhlBAE (including β and α-subunit-encoding genes and the self-subunit swapping chaperone-related gene nhlE) were inserted into pET24a resulting in the recombinant plasmid pET24a-nhlBAE. E. coli transformant harboring pET24a-nhlBAE was used for expression of L-NHase. As shown in Fig 2A, lane 2, the band of the β-subunit of L-NHase was clearly seen on SDS-PAGE, while the bands of the α-subunit and NhlE were scarcely observed on the gel. This finding indicates that the routine method for over-expression of nhlBAE in E. coli is essentially adaptively engineered to augment the quantity of the recombinant protein. It has been shown that heterologous expression of proteins in E. coli can be enhanced by employing a synthetically strong ribosome binding site (RBS) [21] and by reducing the distance between the promoter region and target gene [22]. Therefore, strategies involving substitution of strong RBS and space deletion were executed. The most preferred sequence of strong RBS in E. coli is believed to be AAGGA [23]. To enhance the expression of the α-subunit and NhlE, the putative RBS (AAACGAG) between the nhlB and nhlA genes (Fig 1A) was replaced with an enhanced RBS (AAGGA), and the spacing sequences between the two genes were shortened from 63 bp to 14 bp, constructing plasmid pET24a-nhlBrbsAE. However, NhlE is essential for the activity of L-NHase. To enhance the expression of NhlE, the strong RBS (AAGGA) was also inserted between nhlA and nhlE, yielding the expression plasmid pET24a-nhlBrbsArbsE (Fig 1A). E. coli transformants harboring pET24a-nhlBrbsAE and pET24a-nhlBrbsArbsE were used for L-NHase expression. As shown in Fig 2A (lane 3 and lane 4), the bands of the α- and β-subunits and NhlE of L-NHase were clearly detected on SDS-PAGE, and the activity in cell-free extracts of the transformants harboring pET24a-nhlBrbsAE and pET24a-nhlBrbsArbsE were 120 U/mg and 132 U/mg, respectively.
Fig 2. SDS-PAGE analysis of the expression of L-NHases and H-NHase.
(A) The expression of L-NHases. M: Marker; 1: control, BL21(DE3)/pET24a; 2: BL21(DE3)/pET24a-nhlBAE; 3: BL21(DE3)/pET24a-nhlBrbsAE; 4: BL21(DE3)/pET24a-nhlBrbsArbsE. (B) The expression of H-NHase. M: Marker; 1: control, BL21(DE3)/pET24a; 2: BL21(DE3)/pET24a-nhhBAG; 3: BL21(DE3)/pET24a-nhhBrbsArbsG. (C) The purified L-NHases and H-NHase. M: Marker; 1, L-NHase encoded by nhlBrbsAE; 2, L-NHase encoded by nhlBrbsArbsE; 3, H-NHase encoded by nhhBrbsArbsG.
Almost all of the natural mRNAs have RBS upstream of the start codon, which functions at the step of translation initiation. The efficiency for binding of ribosome to the RBS is determined by the complementarity between RBS and the ribosome [24]. The putative RBS sequences of the native α-subunit and the NhlE are AAACGA, which is a weak RBS for recombinant expression in E. coli. The strategy involving substitution of an enhanced RBS (AAGGA) may help the heterologous expression of L-NHase in E. coli. In addition, the spacer between the start codon AUG and RBS, which plays an important role in regulation of translation initiation efficiency, affects the incorporation of fMet-tRNA into the ribosome [25]. In the present study, we reduced the spacer between the start codon AUG and RBS of mRNA of the α-subunit from 63-bp to a 9-bp, which may be another factor enhancing L-NHase expression. These engineering strategies resulted in the successful expression of L-NHase in E. coli.
Overexpression of the type of high-molecular-mass NHase (H-NHase) in E. coli
In addition to the strategies involving substitution of strong RBS and reducing the distance between the promoter and target gene, heterologous expression of proteins in E. coli can also be enhanced by gene codon optimization [26]. With the aforementioned success in overproducing L-NHase, as well as gene codon optimization, we attempted to express H-NHase in E. coli. The gene nhhBAG with the optimized codon (including the β and α-subunit and the self-subunit swapping chaperone nhhG genes) were designed and chemically synthesized, in which the putative RBS upstream of the nhhA gene was replaced by an enhanced RBS (AAGGA). In addition, NhhG is essential for H-NHase activity. To augment the expression of NhhG, the enhanced RBS was also inserted between nhhA and nhhG. The synthesized genes were digested and ligated into pET24a, constructing plasmid pET24a-nhhBrbsArbsG (Fig 1B). E. coli transformant harboring pET24a-nhhBrbsArbsG was used for H-NHase expression. As a control, E. coli transformant harboring pET24a-nhhBAG (the native H-NHase genes) was also used for H-NHase expression. As a result, the bonds corresponding to the α- and β-subunits of H-NHase were obvious on SDS-PAGE of strain harboring pET24a-nhhBrbsArbsG (Fig 2B), and the activity in the cell-free extracts was 85 U/mg, while bonds corresponding to the α- and β-subunits of H-NHase was undetectable on SDS-PAGE of strain harboring the native H-NHase genes.
Characterizations of the recombinant L-NHase and H-NHase
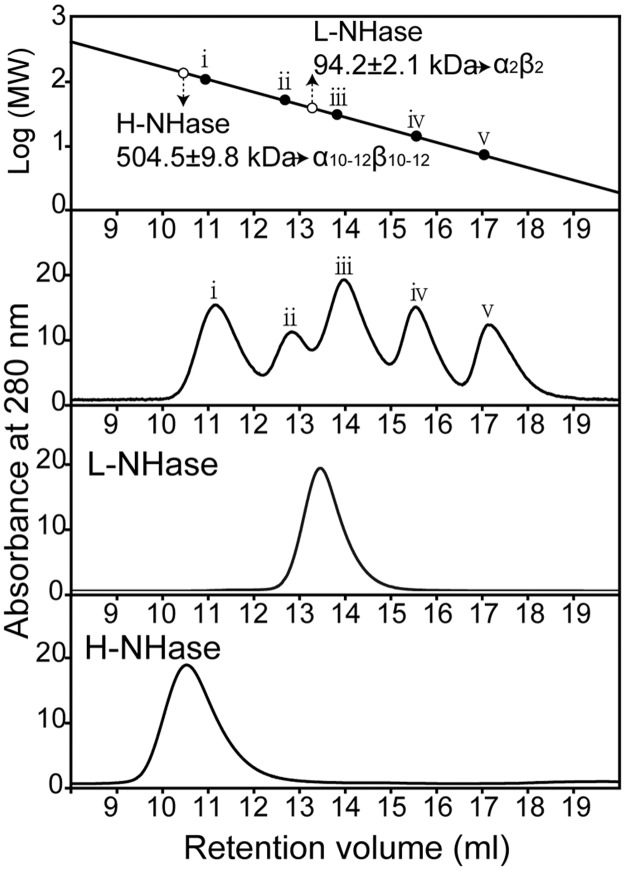
The L-NHases expressed from the transformants harboring pET24a-nhlBrbsAE and pET24a-nhlBrbsArbsE, as well as H-NHase expressed from the transformant harboring pET24a-nhhBrbsArbsG, were purified (Fig 2C) and characterized. The activities are shown in Table 2, and the specific activities of the purified L-NHases were 390 U/mg and 430 U/mg, respectively, which were similar to that from the wild type and expressed in R. facians DSM43985 [8]. The specific activity of the purified H-NHase was 235 U/mg, which was the same level as that from the wild type and that expressed in R. rhodochrous ATCC12674 [15]. The cobalt contents of these NHases were 0.8, 0.9, and 0.8 mol/mol of αβ, indicating that these enzymes were cobalt-containing NHases and that the cobalt incorporation successfully occurred in the E. coli recombinant system. The molecular masses of the recombinant L-NHase and H-NHase were analyzed by gel filtration. As shown in Fig 3, the molecular mass of the L-NHase was determined to be 94 kDa. As the calculated molecular masses of α and β-subunits of L-NHase are 22.8 kDa and 25.2 kDa, respectively, the recombinant L-NHase comprised of α2β2 should be 96.0 kDa, which was the same as the wild-type L-NHase. The molecular mass of the H-NHase was determined to be 504 kDa. Because the calculated molecular masses of the α- and β-subunits of H-NHase are 22.8 kDa and 26.3 kDa, respectively, the recombinant H-NHase comprised of α10β10 should be 490 kDa, which corresponds to that of the wild-type H-NHase (α10–12β10–12) [18]. These findings suggest that both L-NHase and H-NHase from R. rhodochrous J1 are actively expressed in E. coli, and the recombinant enzymes retain the native forms and properties.
Table 2. Characterization of purified NHases.
| Protein | cobalt content (mol of irons/mol of protein) | specific activity (units/mg) |
|---|---|---|
| L-NHase (nhlBrbsAE) | 0.8±0.1(per αβ) | 390±9.5 |
| L-NHase (nhlBrbsAE) | 0.9±0.1(per αβ) | 430±12.1 |
| H-NHase | 0.8±0.1(per αg) | 235±7.6 |
The values represent the means±sd for quadruplicate independent experiments. The corresponding proteins were the same concentration: 0.5 mg/mL for determination of cobalt ion incorporation, 0.1 mg/mL for specific assay.
Fig 3. Determination of the molecular masses of purified proteins.
Determination of the molecular masses and structures of NhlBA and NhhBA by Superdex 200 10/300 GL pg column. The marker proteins were used for gel filtration: (i) glutamate dehydrogenase (yeast) (290 kDa), (ii) lactate dehydrogenase (pig heart) (142 kDa), (iii) enolase (yeast) (67 kDa), (iv) myokinase (yeast) (32 kDa), and (v) cytochrome c (horse heart) (12.4 kDa).
The activity of L-NHase encoded by the expression cassette nhlBrbsArbsE was slightly higher than that by nhlBrbsAE, which might be due to the higher expression level of NhlE in the cassette organization of nhlBrbsArbsE than that of nhlBrbsAE. NhlE acts as a self-subunit swapping chaperone for cobalt incorporation into L-NHase. NhlE functions in a dimer with the α-subunit of L-NHase, forming αe2. The incorporation of cobalt to L-NHase depends on the exchange of the α-subunit between the cobalt-free α-subunit of the L-NHase and the cobalt-containing αe2 [8]. The higher expression level of NhlE increases the formation of αe2, resulting in a more complete α-subunit exchange between the cobalt-free L-NHase and cobalt-containing αe2. This result could be supported by the cobalt content in L-NHases encoded by nhlBrbsArbsE and nhlBrbsAE (0.9, 0.8 mol/mol of αβ, respectively).
Comparison of thermos-stability, substrate and product tolerance of L-NHase and H-NHase
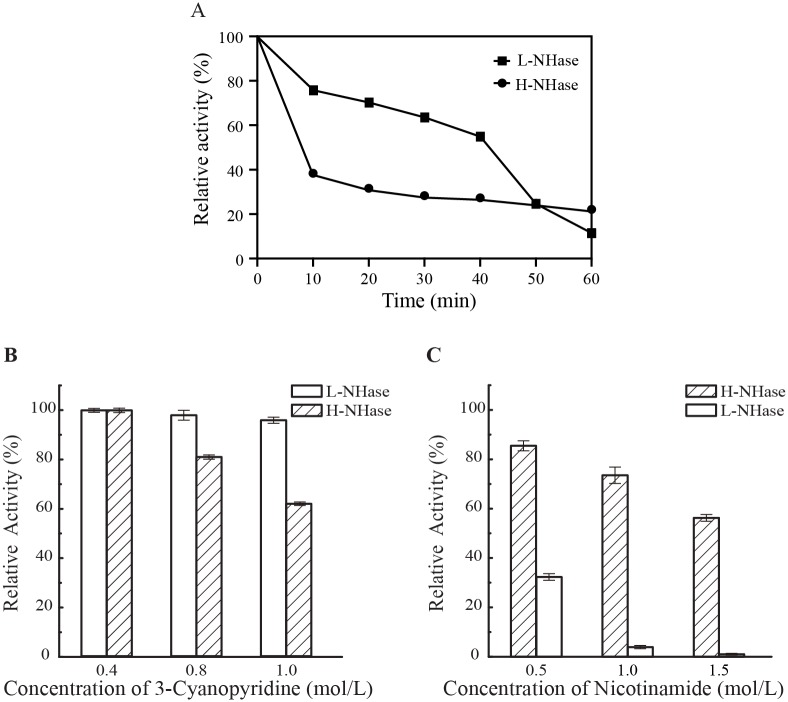
Thermos-stability, substrate and product tolerance of an enzyme are the important factors for the enzyme application. To determine the thermos-stability, we incubated the purified enzymes at 50°C for 60 minutes and measured their activity every ten minutes. As shown in Fig 4A, L-NHase exhibited higher thermos-stability than H-NHase. However, substrate and product tolerance of H-NHase was significantly better than those of L-NHase (Fig 4B and 4C). The activity of L-NHase decreased 40% under the condition of 1 M substrate (3-cyanopyridine), while almost no inhibition could be observed for the H-NHase in the same condition (Fig 4B). The activity of L-NHase was reduced sharply after treatment by 0.5 M product (nicotinamide), in contrast, H-NHase still has 60% of activity after treatment by 1.5 M nicotinamide. H-NHase may be more suitable for industrial application because of its higher product tolerance, which would lead to a high product concentration.
Fig 4. Thermo-stability, substrate and product tolerance of L-NHase and H-NHase.
(A) Thermo-stability of NHases; (B) Substrate tolerance of enzymes; (C) Product tolerance of enzymes.
Production of NHase by recombinant E. coli is superior to that in R. rhodochrous J1
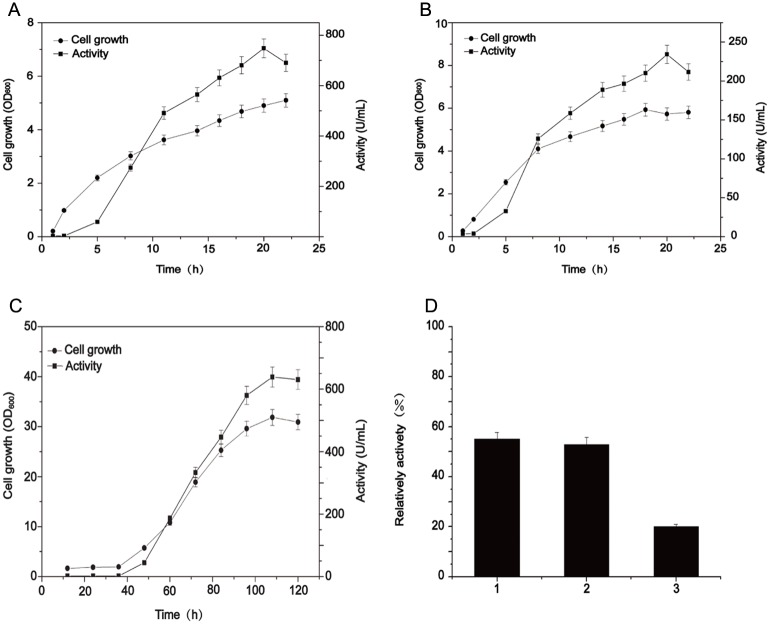
The culture condition and catalytic properties for overproduction of NHase were compared between recombinant E. coli and native R. rhodochrous J1 to highlight which host is superior to produce NHase. As shown in Fig 5, the cell growth of the transformants harboring pET24a-nhlBrbsArbsE (for L-NHase) and pET24a-nhhBrbsArbsG (for H-NHase) were faster than R. rhodochrous J1, and the activities of L-NHase and H-NHase peaked at 20-h cultivation, which was 750 U/ml and 240 U/ml, respectively (Fig 5A and 5B). Regarding the R. rhodochrous J1, the highest total NHase activity was 650 U/ml, which was achieved after 110-h cultivation (excluding the seed culture time for R. rhodochrous J1) (Fig 5C). In parallel, the values of OD600 of E. coli transformants harboring L-NHase and H-NHase genes when reaching maximum enzyme activities were 5.0 and 5.4, respectively, while the OD600 of R. rhodochrous J1 was 30 (Fig 5C). The activity unit per cell of the two recombinant E. colis overproducing L-NHase and H-NHase were 150, 44.4 U/OD600, respectively, which were much higher than that of R. rhodochrous J1(21.7 U/OD600). To compare the product tolerance of NHases in recombinant E. coli and in wild-type R. rhodochrous, we tested the tolerance to nicotinamide in these strains. The reactions were carried out in mixtures containing 400 mM 3-cyanopyridine (substrate) with and without 0.5 M nicotinamide (product) for 20 min. The decrease of 3-cyanopyridine in each reaction was measured, and the reduction ratio (the proportion of the reduced 3-cyanopyridine amount in the reaction with and without 0.5 M nicotinamide) was calculated (Fig 5D). The reduction ratios of L-NHase and H-NHase in genetically engineered E. coli were 20% and 53%, respectively. The reduction ratio for the wild-type strain was 55%, indicating that the H-NHase recombinant E. coli exhibit similar product tolerance to that of the wild-type strain.
Fig 5. Production of NHase in recombinant E. coli and in native R. rhodochrous J1.
Cell growth and NHase activity of (A) the genetically engineered E. coli for H-NHase, (B) L-NHase, and (C) R. rhodochrous J1. (D) Comparison of nicotinamide-tolerance of NHases from the cells of R. rhodochrous J1 (column 1) and the H-NHase, and L-NHase from the genetically engineered E. coli (column 2 and 3). The reduction ratio (the proportion of the reduced 3-cyanopyridine amount in the reaction with and without 0.5 M nicotinamide) was calculated.
Regardless of the immobilized cells or free NHase, the enzyme should be exposed to an environment with a high concentration of product at the late phase during biotransformation. Tolerance to a high concentration of product is an essential property that NHase must possess for industrial applications. Although the product-tolerance for L-NHase-harboring recombinant E. coli was slightly lower than that for R. rhodochrous J1, the H-NHase-harboring recombinant E. coli displayed equivalent product-tolerance compared to that of R. rhodochrous J1. The low activity in the culture of recombinant E. coli harboring H-NHase could be eliminated by augmenting the quantity of biomass through high-density fermentation. In addition, the capacity for product-tolerance of NHase could be enhanced by protein engineering [27]. The best known strategy is to fuse self-assembling peptide to the N-terminus of NHase, which can largely enhance the capacity for tolerance to high-concentration product [27]. This strategy may also elevate the tolerance to product in recombinant E. coli harboring L-NHase.
Conclusions
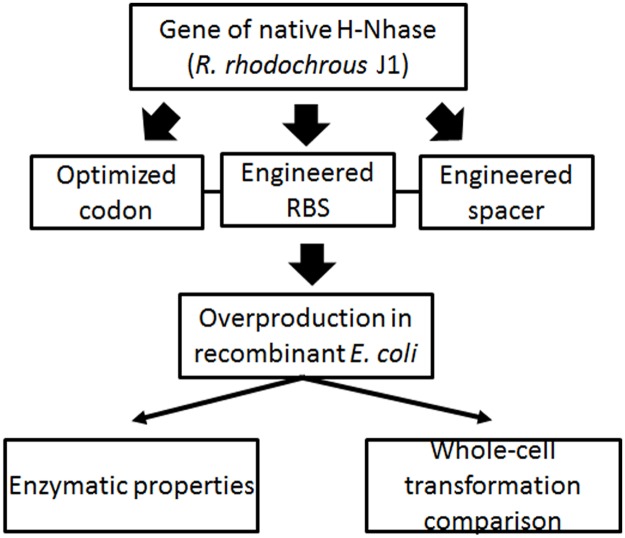
The two NHases from R. rhodochrous J1 were overexpressed in E.coli via codon-optimization, substitution of stronger RBS, and optimization of spacing sequences between different genes. Production of NHase by recombinant E. coli is superior to that in R. rhodochrous J1. It is possible that the genetically engineered E. coli could be used for industrial applications instead of R. rhodochrous J1 (Fig 6).
Fig 6. The flow chart to illustrate the strategy for this study.
The sketch depicted the overall framework for this study. Genetic engineering of native NHase was carried out in two tiers. Firstly, the codons were optimized. Secondly, the genetic elements, RBS and spacer, were modified and optimized. Resorting to these modifications, the optimized NHases were over-produced in recombinant E. coli. Accordingly, the enzymatic properties were characterized and a transformation pipeline by whole-cell catalysis was primarily established.
Data Availability
All relevant data are within the paper.
Funding Statement
National natural science foundation of China, 31400058, www.nsfc.gov.cn/, WC. National natural science foundation of China, 31400078, www.nsfc.gov.cn/, ZL. Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, ZZ. 111 Project, 111-2-06, http://www.dost.moe.edu.cn ZZ. Jiangsu province "Collaborative Innovation Center for Advanced Industrial Fermentation" industry development program ZZ. International S&T Innovation Cooperation Key Project, 2016YFE0127400, www.most.gov.cn/ ZL. Fundamental Research Funds for the Central Universities JUSRP51713B www.moe.gov.cn/ WC. The funder provided support in the form of research materials, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The company Rudong Nantian Nongke Chemical Engineering Co. Ltd and ENNova Health Science Technology Co., Ltd., ENN Group participated in the data analysis and a part of fermentation experiment.
References
- 1.Noguchi T, Nojiri M, Takei K, Odaka M, Kamiya N. Protonation Structures of Cys-Sulfinic and Cys-Sulfenic Acids in the Photosensitive Nitrile Hydratase Revealed by Fourier Transform Infrared Spectroscopy. Biochemistry (Moscow). 2003;42:11642–11650. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi M, Shimizu S. Cobalt proteins. Eur J Biochem. 1999;261:1–9. [DOI] [PubMed] [Google Scholar]
- 3.Nagashima S, Nakasako M, Dohmae N, Tsujimura M, Takio K, Odaka M, et al. Novel non-heme iron center of nitrile hydratase with a claw setting of oxygen atoms. Nat Struct Mol Biol. 1998;5:347–351. [DOI] [PubMed] [Google Scholar]
- 4.Murakami T, Nojiri M, Nakayama H, Dohmae N, Takio K, Odaka M, et al. Post-translational modification is essential for catalytic activity of nitrile hydratase. Protein Sci. 2000;9:1024–1030. doi: 10.1110/ps.9.5.1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y., Cui W., Fang Y., Yu Y., Cui Y., Xia Y., et al. Strategy for successful expression of the Pseudomonas putida nitrile hydratase activator P14K in Escherichia coli. BMC Biotechnol. 2013;13:48 doi: 10.1186/1472-6750-13-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyanaga A, Fushinobu S, Ito K, Shoun H, Wakagi T. Mutational and structural analysis of cobalt-containing nitrile hydratase on substrate and metal binding. Eur J Biochem. 2004;271:429–438. [DOI] [PubMed] [Google Scholar]
- 7.Arakawa T, Kawano Y, Kataoka S, Katayama Y, Kamiya N, Yohda M, et al. Structure of thiocyanate hydrolase: a new nitrile hydratase family protein with a novel five-coordinate cobalt(III) center. J Mol Biol. 2007;366:1497–1509. doi: 10.1016/j.jmb.2006.12.011 [DOI] [PubMed] [Google Scholar]
- 8.Zhou Z, Hashimoto Y, Shiraki K, Kobayashi M. Discovery of posttranslational maturation by self-subunit swapping. Proc Natl Acad Sci U S A. 2008;105:14849–14854. doi: 10.1073/pnas.0803428105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Z, Hashimoto Y, Kobayashi M. Self-subunit swapping chaperone needed for the maturation of multimeric metalloenzyme nitrile hydratase by a subunit exchange mechanism also carries out the oxidation of the metal ligand cysteine residues and insertion of cobalt. J Biol Chem. 2009;284:14930–14938. doi: 10.1074/jbc.M808464200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Z, Hashimoto Y, Cui T, Washizawa Y, Mino H, Kobayashi M. Unique biogenesis of high-molecular mass multimeric metalloenzyme nitrile hydratase: intermediates and a proposed mechanism for self-subunit swapping maturation. Biochemistry. 2010;49:9638–9648. doi: 10.1021/bi100651v [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Cui W, Xia Y, Cui Y, Kobayashi M, Zhou Z. Self-subunit swapping occurs in another gene type of cobalt nitrile hydratase. PLoS One. 2012;7:e50829 doi: 10.1371/journal.pone.0050829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yukl ET, Wilmot CM. Cofactor biosynthesis through protein post-translational modification. Curr Opin Chem Biol. 2012;16:54–59. doi: 10.1016/j.cbpa.2012.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arakawa T, Kawano Y, Katayama Y, Nakayama H, Dohmae N, Yohda M, et al. Structural basis for catalytic activation of thiocyanate hydrolase involving metal-ligated cysteine modification. J Am Chem Soc. 2009;131:14838–14843. doi: 10.1021/ja903979s [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi M, Shimizu S. Metalloenzyme nitrile hydratase: structure, regulation, and application to biotechnology. Nat Biotechnol. 1998;16:733–736. doi: 10.1038/nbt0898-733 [DOI] [PubMed] [Google Scholar]
- 15.Komeda H, Kobayashi M, Shimizu S. Characterization of the gene cluster of high-molecular-mass nitrile hydratase (H-NHase) induced by its reaction product in Rhodococcus rhodochrous J1. Proc Natl Acad Sci U S A. 1996;93:4267–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagasawa T, Takeuchi K, Nardi-Dei V, Mihara Y, Yamada H. Optimum culture conditions for the production of cobalt-containing nitrile hydratase by Rhodococcus rhodococcus J1. Appl Microbiol Biotechnol. 1990;34:783–788. [Google Scholar]
- 17.Kobayashi M, Nishiyama M, Nagasawa T, Horinouchi S, Beppu T, Yamada H. Cloning, nucleotide sequence and expression in Escherichia coli of two cobalt-containing nitrile hydratase genes from Rhodococcus rhodochrous J1. Biochim Biophys Acta. 1991;1129:23–33. [DOI] [PubMed] [Google Scholar]
- 18.Nagasawa T, Takeuchi K, Yamada H. Characterization of a new cobalt-containing nitrile hydratase purified from urea-induced cells of Rhodococcus rhodochrous J1. Eur J Biochem. 1991;196:581–589. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Cui W, Fang Y, Yu Y, Cui Y, Xia Y, et al. Strategy for successful expression of the Pseudomonas putida nitrile hydratase activator P14K in Escherichia coli. BMC Biotechnol. 2013;13:48 doi: 10.1186/1472-6750-13-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui Y, Cui W, Liu Z, Zhou L, Kobayashi M, Zhou Z. Improvement of stability of nitrile hydratase via protein fragment swapping. Biochem Biophys Res Commun. 2014;450:401–408. doi: 10.1016/j.bbrc.2014.05.127 [DOI] [PubMed] [Google Scholar]
- 21.Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol. 2009;27:946–950. doi: 10.1038/nbt.1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Icev A, Ruiz C, Ryder E. Distance-enhanced association rules for gene expression. Gene. 2003;10:34–40. [Google Scholar]
- 23.Kuriki Y. The translation start signal region of TEM beta-lactamase mRNA is responsible for heat shock-induced repression of amp gene expression in Escherichia coli. J Bacteriol. 1989;171:5452–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vellanoweth RL, Rabinowitz JC. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol Microbiol. 1992;6:1105–1114. [DOI] [PubMed] [Google Scholar]
- 25.Na D, Lee D. RBSDesigner: software for designing synthetic ribosome binding sites that yields a desired level of protein expression. Bioinformatics. 2010;26:2633–2634. doi: 10.1093/bioinformatics/btq458 [DOI] [PubMed] [Google Scholar]
- 26.Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–353. doi: 10.1016/j.tibtech.2004.04.006 [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Cui W, Liu Z, Cui Y, Xia Y, Kobayashi M, et al. Enhancement of thermo-stability and product tolerance of Pseudomonas putida nitrile hydratase by fusing with self-assembling peptide. J Biosci Bioeng. 2014;118:249–252. doi: 10.1016/j.jbiosc.2014.02.015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.