Abstract
Skin serves as a protective barrier and also harbors numerous microorganisms collectively comprising the skin microbiome. As a result of recent advances in sequencing (Next-Generation Sequencing, NGS), our understanding of microbial communities on skin has advanced substantially. In particular, the 16S ribosomal RNA (rRNA) gene sequencing technique has played an important role in efforts to identify the global communities of bacteria in healthy individuals and patients with various disorders in multiple topographical regions over the skin surface. Here, we describe basic principles, study design, and a workflow of 16S rRNA gene sequencing methodology, primarily for investigators who are not familiar with this approach. This article will also discuss some applications and challenges of 16S rRNA sequencing as well as directions for future development.
Background
Human skin forms a protective barrier that limits external invasion, but also provides a natural habitat for a myriad of microbes (bacteria, fungi, viruses), called the ‘skin microbiota’ (Fredricks, 2001; Grice and Segre, 2011; Kong, 2011; Leyden et al., 1987). Commensal microbes and humans have a symbiotic relationship, which can be mutually beneficial with the human host supporting the microbes and microbes contributing to proper organ development and immune homeostasis (Belkaid and Segre, 2014). For example, commensal microbes, such as Propionibacterium species and Staphylococcus epidermidis, can drive the expression of cytokines and anti-microbial peptides, potentially shaping immune responses and preventing colonization by pathogenic bacteria (Cogen et al., 2010; Nagy et al., 2005). Thus, the human host and skin microbes represent a network featuring bidirectional communication. Dysbiosis of skin flora has been associated with skin disorders, such as atopic dermatitis and acne (Kong and Segre, 2012).
Traditional methods to identify and characterize microbes require growth of isolates on culture plates. Although these methods have allowed many essential discoveries, there are limitations. In particular, some microbial species do not grow readily in routine laboratory culture conditions or as mono-cultures, leading to underestimations of the diversity of microbial communities. Next-Generation Sequencing (NGS) technologies have allowed direct and comprehensive sequence-based interrogation of microbial communities, complementing culture-based methods. The 16S rRNA gene is conserved among prokaryotes with specific variable regions that can be used for taxonomic classification, making the 16S rRNA gene a molecular signature to identify members of bacterial communities. This article provides an overview of 16S rRNA gene sequencing techniques and their limitations.
Basics of 16S rRNA gene sequencing
In conducting a microbiome study, multiple factors must be considered in study design and sample collection. Ultimately, the scientific questions will guide the selection of the study population, which include host characteristics such as health status, clinical phenotyping, disease severity, and chronological age/sexual maturity. Given the topographical and temporal heterogeneity of microbial communities over the skin surface, the selection of sampling sites and sampling frequency will vary depending on possible sites of skin disease predilection and on importance of longitudinal sampling over a period of time versus a single time point, respectively (Grice et al., 2009). Consistent sampling of the same skin sites across a study population is important in downstream analyses and in potentially drawing clinically relevant conclusions. Additionally, examples of considerations when enrolling study participants may include recent use of systemic and/or topical antimicrobials, which potentially disrupt the skin microbial communities, use of other medications, and history of dermatological or systemic diseases. At the time of sampling, relevant clinical metadata and medical history should be collected for all subjects. Subjects may also be instructed to participate in specific hygiene routines, e.g. showering/bathing at least 12–24 hours prior to sampling; or to avoid use of topical products, e.g. antimicrobial products, for a time period prior to sample collection to minimize possible confounding factors (Grice et al., 2008). Similarly, when using animal models, housing conditions, handling, and other factors that may influence microbial colonization must be considered in the study design. Given the high potential for contamination of the relatively low-biomass skin samples, a consistent protocol is needed along with proper negative controls to confirm the integrity of sample collection and processing.
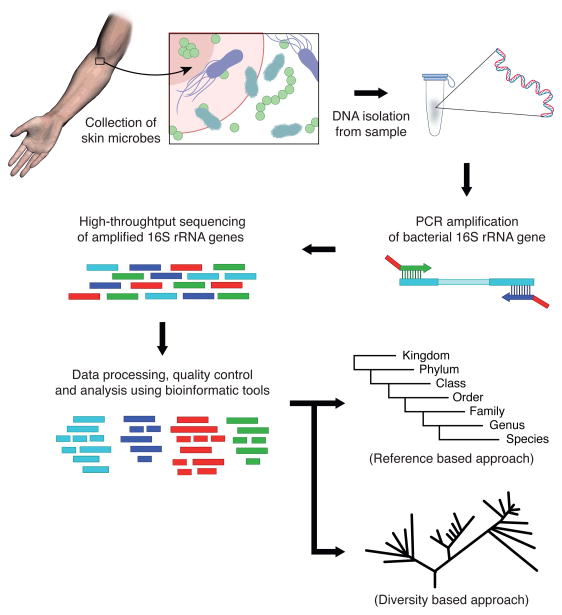
The process of conducting 16S rRNA gene sequencing analyses is depicted in Figure 1. With regards to sample collection, skin microbes can be collected by methods including non-invasive swabs and invasive punch biopsies. Each collection method has a different depth of penetration and results in slightly different microbial profiles (Grice et al., 2008), thus consistency in the sampling method and collection of concurrent negative controls are important. DNA is isolated from the collected skin samples using isolation protocols that minimize the loss of DNA and avoid the introduction of potential contaminants (Salter et al., 2014; Weiss et al., 2014). As part of the DNA isolation process, the cell lysis step should take into account that bacteria differ in their susceptibility to lysing. Generally, Gram positive bacteria are more difficult to lyse than Gram negative bacteria, and incomplete lysis may misrepresent the taxonomic profile of the community (McOrist et al., 2002). Cell lysis is commonly performed with chemical methods such as detergents or enzymes in combination with physical methods such as bead beating.
Figure 1.
Schematic illustration of basic workflow for skin 16S rRNA gene-based sequencing. Adapted and modified from (Kong, 2011).
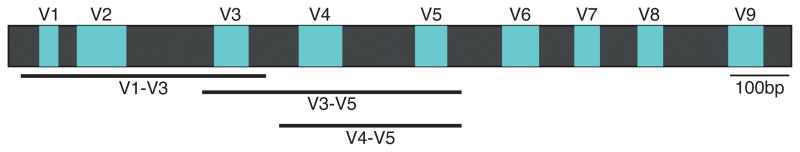
After DNA isolation, the DNA is selectively PCR-amplified using primers targeting the 16S rRNA gene and processed based on protocols specific to the sequencing platform of choice. Common NGS platforms cover 100 to 600 base pairs (bps) per single read with varying degrees of accuracy, but the full-length 16S rRNA gene consists of approximately 1,500 bps. Therefore, primers are chosen to cover only a portion of the 16S rRNA gene. The 16S rRNA gene contains conserved regions interspersed by 9 variable regions (V), which can be used for taxonomic assignments (Fig. 2). In principle, primers should correspond to conserved regions to amplify all 16S rRNA genes, and the amplicons should encompass variable regions for bacterial taxonomic classification. Frequently used primer sets are F27-R534, F9-R534 (encompass V1-V3), F357-R926 (V3-V5), and F515-R926 (V4-V5). Since the V1-V3 region has been identified as the most useful for distinguishing among species of the ubiquitous and clinically important skin bacterial genus Staphylococcus, this region is generally used for skin microbiome studies (Conlan et al., 2012).
Figure 2.
16S rRNA gene structure illustrating variable (blue, V1 to V9) and conserved (dark gray) regions. Common regions (amplicons) for next generation sequencing (NGS)-based microbial community profiling are depicted below.
The next step is sequencing the amplified PCR products. Widely used NGS platforms for 16S rRNA gene analyses (Illumina MiSeq; Roche 454, which is phasing out; and Thermo Fisher Ion Torrent) have distinct features from the traditional Sanger sequencing method, including ‘sequencing during synthesis’ and ‘massive parallel sequencing’. Each sequencing platform utilizes proprietary chemistry/processes to determine the DNA sequence in a high throughput manner. Illumina uses fluorescently labeled nucleotides, which are incorporated during DNA synthesis, detected by a camera, and cleaved to proceed to the next cycle of DNA synthesis and imaging. The 454 technology detects chemi-luminescent signals of pyrophosphate released during nucleotide incorporation (pyrosequencing). Ion Torrent is somewhat similar to pyrosequencing, but detects pH changes when hydrogen ions are released during DNA synthesis. To increase speed and to sequence multiple DNA strands simultaneously in a parallel fashion, 454 and Ion Torrent platforms use micron-sized beads and Illumina platforms use flow cell technology to capture, amplify, and sequence millions of template DNA at the same time. Each platform can generate >400 bps length/read on the order of 1~10 million reads with high performance reagent kits, which is sufficient for high-throughput amplicon-based 16S rRNA gene analysis. However, since each sequencing platform has distinct characteristics, the selection of a specific platform will depend upon the size of the amplicon, sample numbers, desired read numbers, expected sequence accuracy, and budget. For example, if one is interested in sequencing the V1-V3 region (~500bp) to identify skin microbial communities, then selecting a platform that sufficiently covers that length will be important. Since new sequencers continue to be developed, consultation with an experienced sequencing facility or lab is an important consideration of experimental design.
After DNA sequences are obtained, bioinformatic tools such as mothur (Schloss et al., 2009) and QIIME (Caporaso et al., 2010) are commonly used for basic sequence processing, taxonomic assignment, diversity analyses, and community comparisons. A frequently used term in 16S rRNA gene analysis is the Operational Taxonomic Unit (OTU), which is a set of sequences with high similarity/identity (generally >97%) and therefore binned (or categorized) into a single group. Raw NGS data are processed to minimize errors and are subjected to two different, but complementary, analysis strategies. One is a reference-based approach, where sequences, or OTUs, are compared and grouped according to their similarity to existing reference sequences, such as those in the SILVA (Pruesse et al., 2007) or greengenes databases (DeSantis et al., 2006). This provides a taxonomic assignment for each sequence and is useful for understanding the composition of certain taxa in a community. Because reference databases do not include all bacterial taxa, reference-based approaches may not accurately model the structures of bacterial communities. A second approach is a diversity-based approach. Sequences are compared and grouped according to their similarity to each other (de novo OTU clustering), and community structure is analyzed by comparing the sequence similarity of OTUs. Although clustered OTUs can also be used for taxonomic assignments, this approach is exclusively focused on examining the diversity and structure of the community.
Applications of 16S rRNA sequencing in skin research
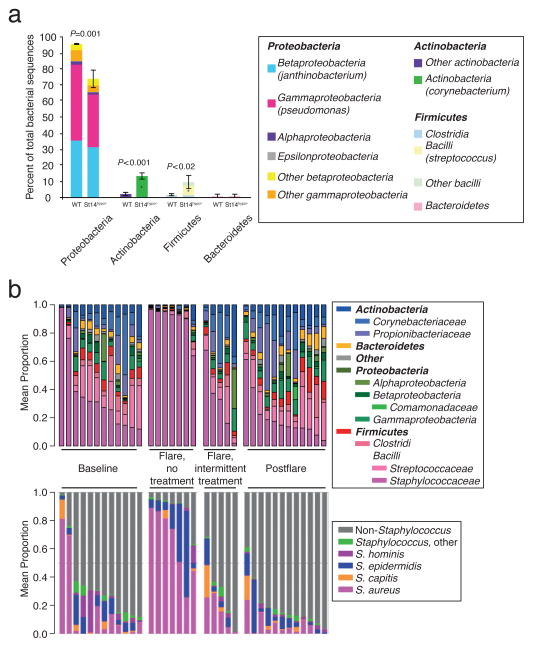
The advantage of 16S rRNA gene sequencing is its direct and culture-independent analysis of the bacterial community at a homeostatic state or in response to various internal or external perturbations. Reports using 16S rRNA gene sequencing have suggested that host factors are closely associated with microbial composition (Oh et al., 2012; Oh et al., 2013). For example, Scharschmidt et al. (2009) have shown decreased Pseudomonas and increased Corynebacterium and Streptococcus on the skin of Matriptase-deficient mice, suggesting that the genetic characteristics of the host influence microbial community structure (Fig. 3a) (Scharschmidt et al., 2009). Researchers have also utilized 16S rRNA gene sequencing to study the linkage between the skin microbiome and clinical human diseases such as atopic dermatitis, psoriasis, and acne. These reports indicate that alterations in skin microbial diversity and community structure are closely correlated with disease states (Fig. 3b) (Kong et al., 2012).
Figure 3.
Skin microbial structure is closely correlated with host status. (a) Relative abundance (phylum and order level) for wild-type (WT) and hypomorphic matriptase (St14hypo/−) mice. Adapted from (Scharschmidt et al., 2009). (b) Relative genus (upper) and Staphylococcus species (lower) abundance in the antecubital and popliteal creases for atopic dermatitis status: baseline, flare (with or w/o intermittent treatment), and postflare. Each bar represents individual patients. Adapted from (Kong et al., 2012).
Limitations and Future directions
Although 16S rRNA gene sequencing is a powerful tool for understanding the linkage between the microbial community and skin homeostasis or disease, there are limitations. Chimera generation and the intrinsic error rate of sequencing are major considerations. A chimera, which is an artifact created during the PCR process, is a single sequence that is comprised of pieces from two or more different origins. Chimeras are believed to arise when prematurely terminated amplicons are used as primers in later PCR cycles. Both chimera and sequencing errors result in nonsense sequences that may be incorrectly classified or incorrectly identified as new species. In an effort to overcome these technical limitations, algorithms that identify and remove chimera sequences (ex. UCHIME) and sequencing errors (ex. denoising) have been developed.
16S rRNA gene sequencing results are relative rather than absolute, such that the actual quantity of a particular bacteria is uncertain. While more direct and comprehensive than culture-based methods, 16S rRNA gene sequencing also has biases: each 16S rRNA gene may not amplify with equal efficiency during PCR reactions due to differential primer affinity and GC content. In addition, taxonomy assignment is reliant on the completeness of reference databases. The quality and quantity of references determine the accuracy and resolution of the taxonomic classification, and results may differ depending upon the choice of reference database. Yet another significant challenge is discriminating cause-and-effect relationships. 16S rRNA gene sequencing captures microbial profiles at a particular moment in time. Thus, it is difficult to conclude if the altered microbial community caused, or resulted from, the disease. Therefore, additional hypothesis-testing mechanistic investigations are required.
16S rRNA gene sequencing methodology is likely to improve rapidly in terms of accuracy and reliability in accordance with advances in sequencing technology. As longer and more accurate sequence reads are made, more sophisticated classification and clustering will be possible. 16S rRNA gene sequencing is a highly versatile assay, providing higher dimensional information when combined with other high-throughput analyses, such as proteomics, lipidomics, transcriptomics, and metagenomics. Robust bioinformatic pipelines are under active development to analyze multi-dimensional high-throughput data.
Our knowledge of the skin microbiome continues to increase, but many questions remain, such as how are microbial communities maintained or altered, and how do microbiota communicate with the host? 16S rRNA gene sequencing can expand our insights into host-microbe and microbe-microbe interactions, which may have substantial implications for many issues in dermatology, including skin inflammation, skin cancer, skin aging, and antibiotic development.
Summary points.
Advantages of 16S rRNA gene sequencing
16S rRNA gene sequencing provides extensive and in-depth information about microbial communities on skin.
Allows interrogation of information about microbial communities without culturing.
Limitations
Results are relatively rather than absolutely quantitative.
16S rRNA sequencing can be biased, due to varying PCR amplification frequencies and incomplete reference databases used for sequence analysis.
Does not determine cause-and-effect relationships.
Acknowledgments
We thank Mark C. Udey, Julia A. Segre, and Carmen Contreras-Sesvold for their helpful discussions. This work was supported by NIH NCI Intramural Research Program and a grant of the Korean Health Technology R&D Project, Ministry of Health and Welfare, Republic of Korea (HI15C1095; J-HJ).
Footnotes
Conflict of interest statement
The authors state no conflict of interest.
References
- Belkaid Y, Segre JA. Dialogue between skin microbiota and immunity. Science. 2014;346:954–9. doi: 10.1126/science.1260144. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogen AL, Yamasaki K, Sanchez KM, et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. The Journal of investigative dermatology. 2010;130:192–200. doi: 10.1038/jid.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan S, Kong HH, Segre JA. Species-level analysis of DNA sequence data from the NIH Human Microbiome Project. PloS one. 2012;7:e47075. doi: 10.1371/journal.pone.0047075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and environmental microbiology. 2006;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredricks DN. Microbial ecology of human skin in health and disease. The journal of investigative dermatology Symposium proceedings / the Society for Investigative Dermatology, Inc [and] European Society for Dermatological Research. 2001;6:167–9. doi: 10.1046/j.0022-202x.2001.00039.x. [DOI] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Renaud G, et al. A diversity profile of the human skin microbiota. Genome research. 2008;18:1043–50. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–2. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Segre JA. The skin microbiome. Nature reviews Microbiology. 2011;9:244–53. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong HH. Skin microbiome: genomics-based insights into the diversity and role of skin microbes. Trends in molecular medicine. 2011;17:320–8. doi: 10.1016/j.molmed.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong HH, Oh J, Deming C, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome research. 2012;22:850–9. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong HH, Segre JA. Skin microbiome: looking back to move forward. The Journal of investigative dermatology. 2012;132:933–9. doi: 10.1038/jid.2011.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyden JJ, McGinley KJ, Nordstrom KM, et al. Skin microflora. The Journal of investigative dermatology. 1987;88:65s–72s. doi: 10.1111/1523-1747.ep12468965. [DOI] [PubMed] [Google Scholar]
- McOrist AL, Jackson M, Bird AR. A comparison of five methods for extraction of bacterial DNA from human faecal samples. Journal of microbiological methods. 2002;50:131–9. doi: 10.1016/s0167-7012(02)00018-0. [DOI] [PubMed] [Google Scholar]
- Nagy I, Pivarcsi A, Koreck A, et al. Distinct strains of Propionibacterium acnes induce selective human beta-defensin-2 and interleukin-8 expression in human keratinocytes through toll-like receptors. The Journal of investigative dermatology. 2005;124:931–8. doi: 10.1111/j.0022-202X.2005.23705.x. [DOI] [PubMed] [Google Scholar]
- Oh J, Conlan S, Polley EC, et al. Shifts in human skin and nares microbiota of healthy children and adults. Genome medicine. 2012;4:77. doi: 10.1186/gm378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Freeman AF, Program NCS, et al. The altered landscape of the human skin microbiome in patients with primary immunodeficiencies. Genome research. 2013;23:2103–14. doi: 10.1101/gr.159467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic acids research. 2007;35:7188–96. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC biology. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharschmidt TC, List K, Grice EA, et al. Matriptase-deficient mice exhibit ichthyotic skin with a selective shift in skin microbiota. The Journal of investigative dermatology. 2009;129:2435–42. doi: 10.1038/jid.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and environmental microbiology. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S, Amir A, Hyde ER, et al. Tracking down the sources of experimental contamination in microbiome studies. Genome biology. 2014;15:564. doi: 10.1186/s13059-014-0564-2. [DOI] [PMC free article] [PubMed] [Google Scholar]