Abstract
Photocrosslinked, biodegradable hydrogels have been extensively investigated for biomedical applications, including drug delivery and tissue engineering. Here, dextran (DEX) was chemically modified with mono(2-acryloyloxyethyl) succinate (MAES) via an esterification reaction, resulting in macromers that could be photocrosslinked to form hydrolytically degradable hydrogels. Hydrogel swelling ratio and degradation rate were controlled by varying the degree of MAES modification. Thiolated cell adhesion peptides (GRGDSPC) were conjugated to acrylated dextran via thiol-acrylate reaction to regulate the interactions of human mesenchymal stem cells (hMSCs) with the photocrosslinkable hydrogels. The hydrogels permitted sustained release of short interfering RNA (siRNA) over 7 weeks and were cytocompatible with hMSCs. Sustained presentation of siRNA from these photocrosslinked DEX hydrogels enhanced the osteogenic differentiation of encapsulated hMSCs. These DEX hydrogels with tunable siRNA delivery and cell adhesive properties may provide an excellent platform for bioactive molecule delivery and tissue regeneration applications.
Introduction
RNA interference (RNAi) is a biological process in which small interfering RNA (siRNA) and microRNA (miRNA) silence cell gene expression post-transcriptionally.1–5 These small RNA molecules function in the cytoplasm after entering cells, where they are incorporated into an RNA-induced silencing complex.3, 5 The siRNA complex degrades complementary messenger RNA (mRNA), whereas the miRNA complex can inhibit the translation of many different mRNA sequences.5 Delivery of these small RNAs to regulate cell production of specific proteins has therapeutic potential to, for example, treat diseases such as cancer6, 7 and direct tissue formation in regeneration strategies.8, 9 Owing to their hydrophilic, biomacromolecular and ionic character, it is challenging for siRNA and miRNA to interact with and cross the negatively charged cell membrane. To facilitate their cellular uptake, they have been chemically modified with chemicals, such as 2′-OMethyl10, 11, 2′-Fluoro12 (substitution for 2′-OH), phosphorothiolate13 or their combination13, 14, ionically bound to cationic polymers into nanocomplexes15, 16 or encapsulated in liposomes.17–19 It is, however, challenging to target these modified RNAs and nanoparticles to specific cell populations upon administration3, 20 either systemically or locally. siRNA has also been incorporated into microparticles to prolong its release locally to target cells, but microparticles can also disperse rapidly from the site of application in vivo.3, 21 Alternatively, macroscopic hydrogels, highly hydrated three-dimensional polymeric networks, have been widely investigated for localized delivery of bioactive factors.21–23 Localized delivery of siRNA from macroscopic hydrogels allows for release at the target site of interest and can reduce necessary dosage while maintaining efficacy and potentially avoiding side effects from treatment of off-target cells. We previously reported on photocrosslinked alginate and DEX hydrogels for localized and/or controlled release of chemically modified siRNA21, 24 and other hydrogels and scaffolds have been developed to deliver RNA/cationic polymer nanoparticles.8, 25–32
Photocrosslinked macroscopic hydrogels have attracted increasing interest for therapeutics delivery and tissue engineering applications because the aqueous macromer solutions used to form them can be injected at a desired site in a minimally invasive manner and then photopolymerized in situ to form hydrogels at physiological conditions.22, 33, 34 Photocrosslinkable polymer solutions containing photoinitiator can be crosslinked upon short exposure to visible or ultraviolet (UV) light. During exposure to these light sources, the photoinitiator is decomposed, producing free radicals that can induce the crosslinking of acrylate and/or methacrylate terminated polymeric macromers.33 The photocrosslinking process has minimal adverse effects on cells and/or bioactive factors when an appropriate photoinitiator is used in conjunction with low-intensity light at specific wavelength(s).35 The UV intensity, photoinitiator concentration, crosslinking time, the number of available crosslinkable moieties, and macromer concentration and molecular weight22, 36–38 can be adjusted to control hydrogel mechanical properties, swelling kinetics and degradation rate.
Dextran (DEX) is a bacterial polysaccharide composed of α(1→6) linked D-glucopyranosyl residues connecting with α(1→3) linked side chains.39–42 Since it is biodegradable and biocompatible, DEX has been widely used for many biomedical purposes, such as in drug delivery24, 41, 43, 44 and tissue engineering45–47 applications. Each glucose unit of DEX contains three hydroxyl groups that permit chemical modification with various functional groups for covalent crosslinking.42 To prepare photocrosslinkable macromers, DEX has been modified with glycidyl methacrylate (GMA)48, 49 or methacrylic anhydride (MA)40 to covalently conjugate methacrylate groups to the hydroxyl groups of the glucose residues. However, these methacrylated DEX macromers result in stable hydrogels in aqueous solutions at physiological conditions.43, 50, 51 In addition, DEX has also been modified with acrylate groups by conjugating vinyl acylate to the hydroxyl groups of DEX52, and this resulting DEX-acrylate macromer also resulted in non-biodegradable hydrogels.53 To prepare hydrolytically degradable hydrogels, imidazolyl carbamate-2-hydroxyl ethyl methacrylate (IC-HEMA) has been covalently conjugated to hydroxyl groups of DEX.43, 51 However, this synthesis route requires two lengthy steps. Here, we proposed to prepare one step synthesized, hydrolytically degradable DEX macromer via an esterification reaction of mono(2-acryloyloxyethyl) succinate (MAES) with hydroxyl groups of DEX.
Control over the cell adhesion properties of biomaterials can affect cell behaviors such as viability,54 attachment,55–57 migration,58 proliferation55, 57, 59 and differentiation.57, 59–62 In addition, regulating the density of covalently coupled peptides containing cell adhesion motifs (e.g., arginyl-glycyl-aspartic acid (RGD)) has been shown to influence the degree of gene knockdown in cells cultured in two-dimensions on alginate hydrogels treated exogenously in the media with siRNA.63 However, how the cell adhesive properties of a hydrogel affect efficacy of siRNA delivered from the scaffold itself to encapsulated cells in three-dimensions (3D) is currently unknown. Thus, developing biomaterial hydrogels with tunable cell adhesivity would be valuable for investigating siRNA-mediated gene knockdown in stem cells encapsulated within a 3D polymer network. Here, a new cytocompatible, hydrolytically degradable, photocrosslinkable DEX hydrogel system with controlled cell adhesivity is reported for tunable, prolonged siRNA delivery. Hydrogel degradation profiles, mechanical properties, cell adhesivity and siRNA release profiles were investigated. In addition, it was assessed whether controlled release of siRNA against noggin, a bone morphogenetic protein (BMP) antagonist that decreases osteogenic differentiation in human adipose-derived stem cells (hADSCs)9 and human mesenchymal stem cells (hMSCs)8, to encapsulated hMSCs in vitro could enhance their osteogenic differentiation.
Experimental
Synthesis of DEX macromers
4-(dimethylamino)pyridinium 4-toluenesulfonate (DPTS) catalyst was synthesized as previously described.8 DEX (Sigma, St. Louis, MO) was chemically modified with different theoretical degrees (20, 30 and 40%) of MAES (TIC America, Portland, OR). For example, to synthesize DEX-MAES40%, DEX (10 g) was dissolved in a dry 250 ml round bottom flask containing 180 ml dimethyl sulfoxide (DMSO) (Sigma). After complete dissolution of DEX, dicyclohexylcarbodiimide (DCC) (7.70 g, Sigma), MAES (5.50 g) and DPTS (1.16 g) were added to the DEX solution under stirring. To synthesize DEX-MAES20 and 30%, the molar ratios of DCC, MAES and DPTS were kept constant. The reaction was allowed to occur at room temperature for 1 day. The solution was then passed through filter paper to remove formed urea salts, and the supernatant was precipitated in 1.8 l acetone. The resulting white powder was collected, rehydrated in ultrapure deionized H2O (diH2O) and dialyzed using 3,500 Da cutoff membrane (Fisher Scientific, Pittsburgh, PA) for 3 days at 4 °C. The DEX solution was frozen and lyophilized until dry. The DEX-MAES product was characterized with proton NMR in D2O. DEX-hydroxyl ethyl methacrylate with 20% theoretical (actual 16%) HEMA modification (DEX-HEMA16) was synthesized as previously described.24
Peptide conjugation to DEX-MAES
Thiolated RGD (GRGDSPC, Genscript, Piscataway, NJ) at various concentrations (5, 10, 20 mg peptide/g DEX-MAES) was mixed with the 40% theoretical MAES (16% actual) -modified DEX macromer solution containing 0.05% Igracure D-2959 for 0.25 – 3h at room temperature to examine conjugation efficiency and kinetics. The peptide conjugation efficiency was determined using Ellman’s assay (Thermo Fisher Scientific, Rockford, IL) per the manufacturer’s instruction.64 In addition, conjugation efficiency of the peptide to acrylated DEX (DEX-MAES16) and methacrylated DEX (DEX-HEMA16) with similar degrees of acrylate and methacrylate modification, respectively, were compared.
Hydrogel preparation
DEX-MAES macromer (12 %w/w) was dissolved in phosphate buffered saline (PBS) containing 0.05% w/v photoinitiator (Irgacure-D2959, Sigma), and then 100 μl of the macromer solution was pipetted onto the inner surface of a petri dish lid. The hydrogels were formed upon exposure of DEX-MAES solutions to 320–500 nm UV light at 2.5 mW/cm2 for 2 min using an Omnicure S1000 UV Spot Cure System (Lumen Dynamics Group, Mississauga, Ontario, Canada).
Swelling and degradation
To determine swelling kinetics of the photocrosslinked hydrogels, their initial dry weight (Wdi) and their swollen weights (Wst) over time were measured. After weighing each lyophilized gel at time t = 0 (Wdi), they were placed into 15 ml conical tubes with 10 ml PBS or Low Glucose Dulbecco’s Modified Eagle’s Medium (DMEM-LG, Sigma) pH 7.4 and incubated at 37°C. The PBS was replaced every 3 days and the swollen gels were collected and weighed at each predetermined time point. The swelling kinetics was determined by Q = Wst / Wdi. N = 3 for each condition at each time point.
To measure degradation profiles of the hydrogels, the lyophilized gels were similarly put into a 15 ml conical tube with 10 ml PBS, incubated at 37°C and, at predetermined time points, the swollen gels were removed, rinsed with diH2O overnight at 4°C and lyophilized until dry to obtain their dry weight (Wdt). The mass loss of the hydrogels was calculated by (Wdt − Wdi) / Wdi * 100. N = 3 for each condition at each time point. For swelling, degradation and rheology experiments with incorporated RGD peptide and siRNA complexes, 10 mg peptide/1g DEX-MAES and 4 μg siRNA/100 μl hydrogel were used.
Rheology
Rheological measurement of the photocroslinkable hydrogels was performed on a Haake Mars III Rotational Rheometer (Thermo Fisher Scientific Inc., Waltham, MA). PBS solutions of DEX-MAES containing 0.05% w/v Igracure D-2959 photoinitiator were placed between a glass plate and a quartz plate separated by two 0.75 mm spacers followed by photocrosslinking via the application of UV light (2.5 mW/cm2) for 2 min. Hydrogel discs were then punched out with a 0.8 cm biopsy punch. To measure their rhelogical properties, each gel disc was loaded between two stainless steel parallel plates (0.8 cm in diameter). A dynamic frequency sweep test with a constant maximum shear strain amplitude (0.1%) over a frequency range of 0.1 – 10.47 rad/sec was used to measure the storage (G′) modulus of each hydrogel at room tempererature (N = 3).
siRNA release
siRNA targeting green fluorescent protein (siGFP, Thermo Scientific Dharmacon, Lafayette, CO) was used to assess siRNA release kinetics from the photocrosslinked DEX hydrogels. The siRNA sequence is shown in Table 1. siRNA was complexed with branched polyethylenemine (PEI, 25 kDa, Sigma) at an N/P ratio of 10, and the resulting complexes were then mixed with DEX-MAES precursor PBS solutions containing 0.05% photoinitiator (4 μg siRNA/100 μl gels). Hydrogels containing siRNA/PEI complexes formed upon exposure of the siRNA/DEX solutions to UV light as described above. Each formed hydrogel (100 μl) was placed into a microcentrifuge tube containing 1 ml of nuclease free PBS (Life Technologies, Grand Island, NY), and at predetermined time points, 1 ml of release sample was removed and replaced with 1 ml of fresh PBS. siRNA/PEI release samples were dissociated by incubation with heparin solutions in water (10 mg/ml, Sigma) for 20 min at room temperature (10 μl release samples with 5 μl heparin solution) and quantified using a Ribogreen assay (Life Technologies) with the standard prepared using fresh siRNA/PEI complexes. siRNA fluorescence value was measured using a plate reader (fmax, Molecular Devices, Sunnyvale, CA) set at excitation/emission of 485 nm/538 nm (N = 3).
Table 1.
Sequences of RNA interfering molecules.
| RNA name | Sense sequence |
|---|---|
| siGFP | 5′-GCA AGC UGA CCC UGA AGU UC-3′ |
| siNoggin | 5′-AAC ACU UAC ACU CGG AAA UGA UGG G-3′ |
| Negative control | 5′-UUC UCC GAA CGU GUC ACG UTT-3′ |
hMSCs culture on the RGD-DEX hydrogels
hMSCs were obtained from human bone marrow harvested under a protocol approved from the University of Cleveland Institutional Review board as previously described.65, 66 Briefly, marrow was aspirated from the posterior iliac crest of healthy donors and washed with growth medium, which was comprised of DMEM-LG containing 10% prescreened Fetal Bovine Serum (FBS, Sigma). Mononuclear cells were isolated via centrifugation using a Percol (Sigma) density gradient. Cells were then plated at 1.8 x 105 cells/cm2 in growth medium, which was replaced every 3 days. After 14 days of culture, the cells were subcultured and passaged at a density of 5 x 103 cells/cm2. Cells at passage 3 were used for all experiments. DEX-MAES was conjugated to the peptide (10 mg/g DEX-MAES) for 1h as mentioned above, and the polymer solution was placed between a glass plate and a quartz plate separated by two 0.75 mm spacers and photopolymerized by exposure to UV light. Photocrosslinked hydrogel disks were cut using a 6-mm diameter biopsy punch and placed in wells of 24 well plates. hMSCs in 0.5 ml of growth media consisting of DMEM-LG with 10% FBS and 1% penicillin/streptomycin (Mediatech Inc., Herndon, VA) were seeded onto the hydrogels at a density of 10,000 cells/well. The cells were allowed to adhere to the gels for 4 h in a humidified incubator at 37°C with 5% CO2. The gels with cells were then transferred into a new 24 well plate and cultured in growth media. After 2 days of culture, hMSCs were stained with a live/dead assay containing fluorescein diacetate (FDA, Sigma) and ethidium bromide (EB, Fisher Scientific). 20 μl of live/dead staining solution consisting of 1 ml of FDA (1.5 mg/ml in DMSO) and 0.5 ml of EB solution (1 mg/ml in PBS) with 0.3 ml of PBS (pH 8.0) was added to each well and incubated for 2–5 min. A fluorescence microscope (ECLIPSE TE 300; Nikon, Tokyo, Japan) equipped with a digital camera (Retiga-SRV; Qimaging, Burnaby, BC, Canada) was used to image the hMSCs on the hydrogels.
Viability and osteogenic differentiation of encapsulated hMSCs in hydrogels
Hydrogels (50 μl) were prepared by applying UV light to RGD-modified DEX-MEAS solutions (5 mg RGD/1g DEX-MAES) containing no siRNA, negative control siRNA (siNegative control) or siRNA targeting noggin (siNoggin; Insight Genomics, Falls Church, VA) (40 μg/ml gel) complexed with PEI at N/P ratio of 10, and hMSCs at a concentration of 5 x 106 cells/ml. siNegative control was used as non-targeting control and siNoggin was used to induce hMSC osteogenesis. The siRNA sequences are listed in Table 1. The hydrogels were cultured in 24 well plates with 0.5 ml of ostegenic media (10 mM β-glycerophosphate (CalBiochem, Billerica, MA), 50 μM ascorbic acid (Wako USA, Richmond, VA), 100 nM dexamethasone (MP Biomedicals, Solon, OH) and 100 ng/ml BMP-2 (Department of Developmental Biology, University of Würzburg, Germany). Media was replaced every 2 days, and at specific points the hydrogels were placed in 1 ml of alkaline phosphatase (ALP) lysis buffer [1mM MgCl2 (Sigma), 20 μM ZnCl2 (Sigma), 0.1% octyl-beta-glucopyranoside (Sigma)] followed by homogenization at 35,000 rpm for 60 s using a TH homogenizer (Omni International, Marietta, GA). Supernatants of the homogenized solutions were collected post-centrifugation at 500 g with a Sorval Legent RT plus centrifuge (Thermo Fisher Scientific Inc., Waltham, MA) for ALP, calcium and DNA quantification (N=3). p-nitrophenylphosphate (pNPP, 100 μl, Sigma) substrate was added to the supernatant (100 μl), and then the mixture was quenched with 0.1 N NaOH (50 μl) to measure ALP activity. The absorbance was measured at 405 nm with a plate reader (VersaMax, Molecular Devices, Sunnyvale, CA). The supernatant was also used to measure calcium content using a calcium assay kit (Pointe Scientific, Canton, MI). 4 μl of the supernatant was added to the color and buffer reagent (250 μl) and the absorbance was measured at 570 nm with a plate reader (VersaMax, Molecular Devices). The Picogreen assay kit (Life Technologies) was used to quantify DNA from the supernatant on a plate reader (fmax, Molecular Devices) at excitation/emission of 485 nm/538 nm to normalize ALP and calcium measurements (N = 3). To further examine mineralization of the hydrogel constructs, the constructs were fixed in neutral buffered formalin solution, embedded in paraffin, sectioned at a 10 μm thickness, and stained with Alizarin red. The stained samples were imaged using an Olympus BX61VS microscope (Olympus, Center Valley, PA) with a Pike F-505 camera (Allied Vision Technologies, Stadtroda, Germany).
A live/dead assay was used to determine the viability of hMSCs encapsulated within the hydrogels using FDA and EB. 20 μl of live/dead staining solution was added to each well containing the hMSC-hydrogel constructs. After 5 min incubation at room temperature, cell microphotographs were imaged using the ECLIPSE TE 300 (Nikon, Tokyo, Japan) fluorescence microscope equipped with a Retiga-SRV digital camera (Qimaging, Burnary, BC, Canada).
Statistical analysis
The data is presented as mean ± standard deviation. Statistical comparisons were performed with Tukey-Kramer Multiple Comparisons Test with one-way analysis of variance (ANOVA) using InStat software (GraphPad Software, La Jolla, CA). p < 0.05 was considered statistically significant.
Results
Macromer characterization
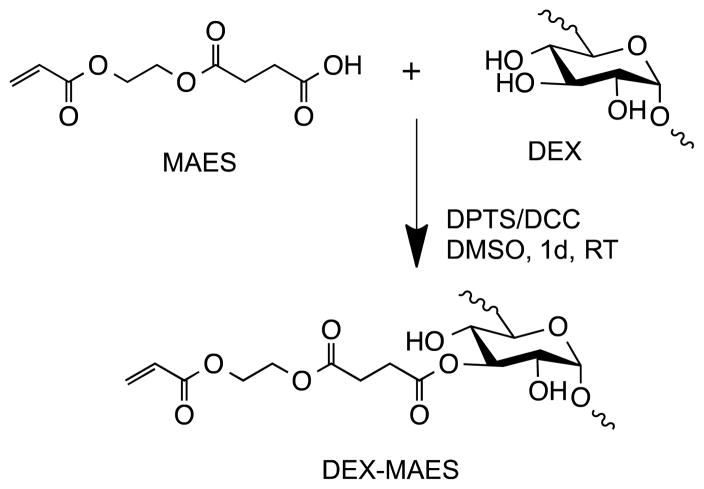
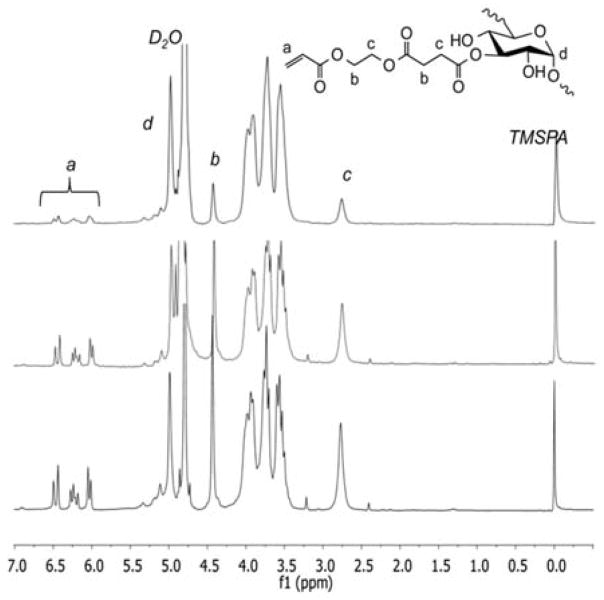
DEX-MAES with various degrees of MAES modification was synthesized via an esterification of carboxylic acid groups of MAES and hydroxyl groups of DEX, as illustrated in Figure 1. The resulting DEX-MAES was characterized with 1H NMR and its proton NMR spectra are shown in Figure 2. DEX-MAES with 20, 30 and 40% theoretical MAES modification resulted in 6, 11 and 16% actual acrylation (DEX-MAES6, DEX-MAES11 and DEX MAES16), respectively, calculated using the proton NMR spectra as previously described.48 The acrylate peaks (a, b, c) increased when the degree of modification increased from 6 to 16%.
Figure 1.
Schematic illustration of DEX-MAES synthesis.
Figure 2.
Proton NMR spectra of DEX-MAES with various degrees of MAES modification.
Peptide conjugation kinetics and efficiency
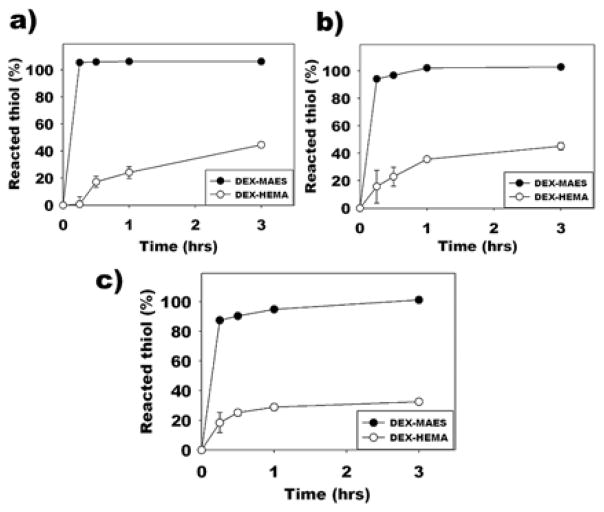
To determine the conjugation kinetics and efficiency of thiolated RGD (GRGDSPC) peptide to DEX-MAES16, various GRGDSPC concentrations (i.e., 5, 10 and 20 mg/1 g DEX-MAES16) were conjugated to the acrylated DEX macromer over time (0.25, 0.5, 1 and 3h) in PBS at pH 7.8 and the free thiol groups of unreacted peptides were quantified using Ellman’s assay. In addition, the reaction kinetics of the thiol-peptide to acrylated (DEX-MAES16) and methacrylated (DEX-HEMA16) macromers were compared. As early as 15 min conjugation, with 5, 10 and 20 mg of GRGDSPC peptide/1 g modified DEX, the peptide conjugation efficiencies with DEX-MAES were 105.40, 94.10 and 87.45%, respectively, while for the reaction with the DEX-HEMA they were 0.73, 15.78 and 18.42%, respectively (Figure 3). After 1h, the GRGDSPC conjugation with DEX-MAES was completed with the peptide concentration of 10 mg, but only 35.66% of the thiol groups of the peptide reacted with DEX-HEMA (Figure 3b). The reaction kinetics were also monitored at 3 h of conjugation, and all of the 20 mg GRGDSPC peptide reacted with acrylated DEX compared to only 32.53% for the methacrylated DEX at this time point (Figure 3c).
Figure 3.
Quantification of reacted thiol after reacting (a) 5 mg, (b) 10 mg and (c) 20 mg GRGDSPC to 1g DEX-MAES and DEX-HEMA macromers.
Characterization of hydrogel physical properties
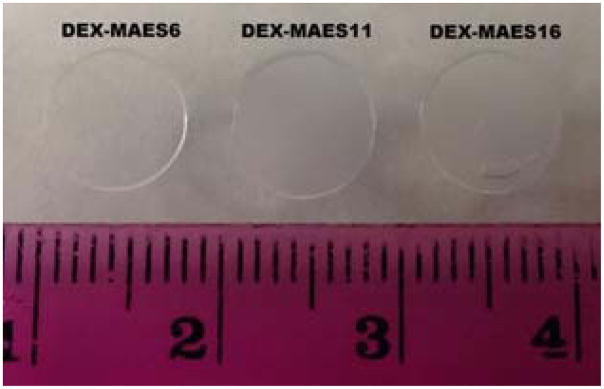
DEX-MAES hydrogels were formed when macromer solutions containing photoinitiator were exposed to low-intensity UV light (Figure 4). While the DEX-MAES6% hydrogels were transparent, the DEX-MAES11 and 16 hydrogels were more cloudy, likely due to the increased density of hydrophobic MAES groups.
Figure 4.
Morphology of DEX-MAES hydrogels following photocrosslinking in PBS.
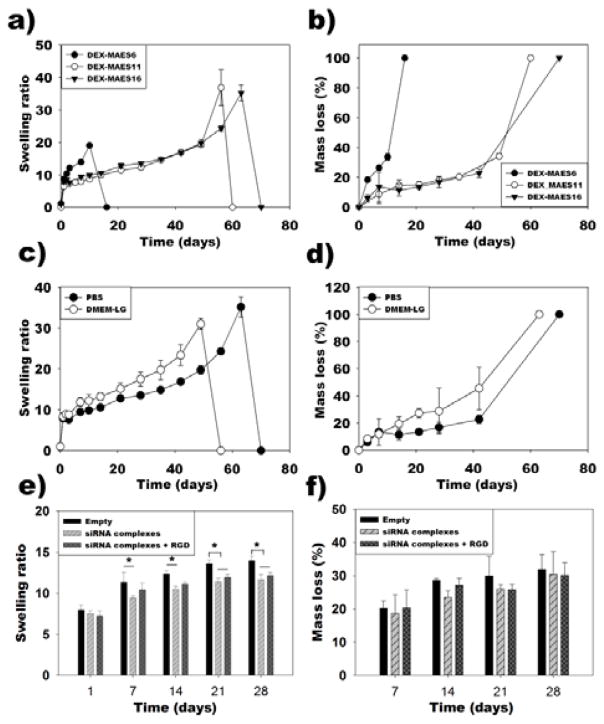
Swelling ratio in PBS over time reflects the changes in physical and chemical structure of the hydrogels. All the gels swelled rapidly and attained equilibrium swelling after one day incubation in PBS at 37°C (Figure 5a). The DEX-MAES6 gels swelled more rapidly than DEX-MAES11 and 16 and degraded completely by day 17. In contrast, the swelling ratio of DEX-MAES11 and 16 gels increased more slowly with maximum values at days 56 and 63, respectively. Hydrogel degradation profiles were determined by measuring mass loss of the hydrogels in PBS at 37°C over time. The DEX-MAES6 hydrogels degraded completely by day 17 as a result of their lower crosslinking density (Figure 5b). The DEX-MAES11 and 16 degradation rates were similar until they fully degraded by days 60 and 70, respectively.
Figure 5.
Swelling (a) and degradation (b) profiles of photocrosslinked DEX-MAES hydrogels in PBS. Swelling (c) and degradation (d) profiles of photocrosslinked DEX-MAES16 in PBS and DMEM-LG. Swelling (e) and degradation (f) profiles of the DEX-MAES16 hydrogels with and without incorporated siRNA complexes and/or conjugated RGD peptide in DMEM-LG. “Empty” condition does not contain siRNA complexes or RGD peptide. * p < 0.05.
In addition, the swelling ratio and degradation of DEX-MAES16% was examined in both PBS and DMEM-LG to determine the influence of varying media on hydrogel structure. The hydrogels reached equilibrium swelling by day 1 of incubation in both solutions (Figure 5c). The hydrogels swelled at a similar rate in the first week and then swelled more rapidly prior to reaching the swelling peak at day 49 in DMEM-LG compared to day 63 in PBS. These hydrogels degraded at the same rate during the first week in both solutions, and then their degradation increased more rapidly in DMEM-LG, with complete degradation occurring by day 64 in DMEM-LG compared to 70 days in PBS (Figure 5d).
The influence of encapsulated siRNA complexes and conjugated RGD peptide on swelling and degradation of the hydrogels in DMEM-LG was also tested. DEX-MAES16 hydrogels without siRNA complexes or conjugated RGD peptide (“Empty”) swelled slightly more than the hydrogels with incorporated siRNA complexes only (“siRNA complexes”) at days 7, 14, 21 and 28, and than the hydrogels with both incorporated siRNA complexes and conjugated RGD peptide (“siRNA complexes + RGD”) at days 21 and 28 (Figure 5e). However, hydrogel degradation rate was not significant between any of these groups over the course of 4 weeks (Figure 5f).
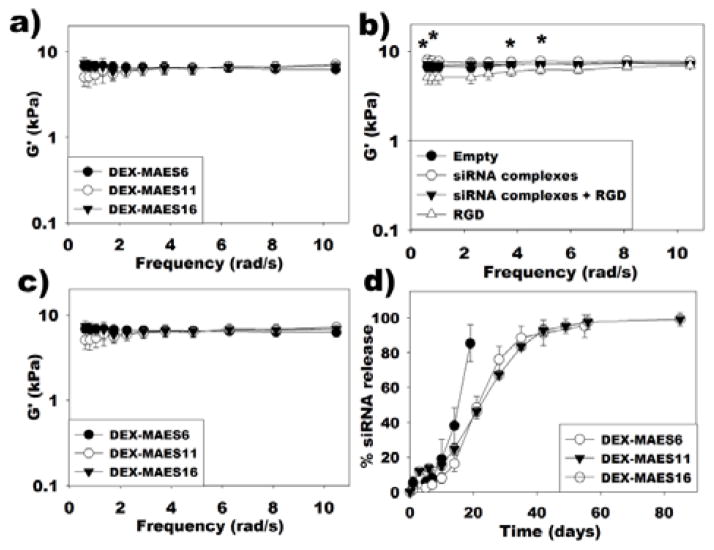
To determine the effect of degree of MAES modification on the hydrogel rheological properties, the storage (G′) modulus of the DEX-MAES6, 11 and 16 hydrogels was assessed under oscillatory strain over a range of frequencies using a rheometer (Figure 6a). Increasing the degree of acrylate modification from 6% to 16% did not alter the rheological properties of the hydrogels in the frequency range from 1 to 10 rad/s. The influence of incorporated siRNA complexes and adhesion ligand conjugation on G′ was also examined. Moduli of “Empty” hydrogels were not significantly different with those of “siRNA complexes”, “RGD”, and “siRNA complexes + RGD” hydrogels at the tested frequencies. However, G′ was significantly greater in “siRNA complexes” group compared to the “RGD” condition at frequencies of 0.63, 0.81, 3.77 and 4.87 rad/s (Figure 6b). In addition, moduli of DEX-MAES6, 11 and 16 hydrogels containing siRNA complexes and conjugated peptide were not significantly different from each other (Figure 6c).
Figure 6.
Storage moduli (G′) of (a) photocrosslinked DEX-MAES hydrogels without incorporated siRNA complexes or conjugated RGD peptide, (b) DEX-MAES16 hydrogels with conjugated RGD peptide and/or incorporated siRNA complexes (* p < 0.05, “siRNA complexes” group is significantly different from “RGD” group at corresponding frequencies), and (c) DEX-MAES hydrogels with conjugated RGD peptide and incorporated siRNA complexes. d) siRNA release profiles from DEX-MAES hydrogels without conjugated RGD peptide.
siRNA release profiles
To test the capacity of the photocrosslinked DEX hydrogels to sustain and tailor the release of siRNA, PEI was complexed with siRNA followed by the incorporation of the resulting complexes into the hydrogels. The release kinetics over time in PBS pH 7.4 at 37°C was regulated by varying the degree of MAES modification (Figure 6d). While most of the siRNA/PEI complexes were released from the DEX-MAES6 hydrogels after 21 days, the DEX-MAES11 and 16 hydrogels exhibited similar profiles until they were completely degraded with more prolonged release for up to 55 days.
hMSC behavior on the surface of the hydrogels
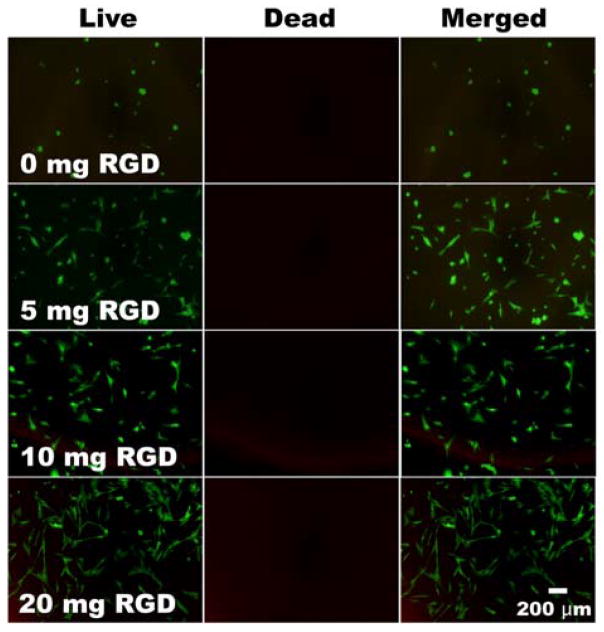
To examine cell adhesivity, viability and morphology, hMSCs were seeded on the surface of hydrogels containing various GRGDSPC amounts (0, 5, 10 and 20 mg peptide/1 g DEX-MAES16), and after two days of culture, cells were visualized with fluorescence staining of the live/dead assay. While fewer hMSCs attached to the hydrogel surface without peptide, an increased number of hMSCs were observed attached to hydrogels with increasing density of covalently conjugated peptide (Figure 7). The fluorescence photomicrographs revealed that adherent cell viability remained high in all groups. The hMSCs on the hydrogels without peptide had a rounded morphology, while those on the hydrogels containing covalently coupled adhesion ligands displayed a spread morphology. However, there were more hMSCs with a rounded morphology on the hydrogels formed with 5 and 10 mg RGD than on hydrogels prepared with 20 mg RGD.
Figure 7.
Cell attachment and morphology of hMSCs on the surface of photocrosslinked DEX-MAES16 hydrogels containing different concentrations of covalently coupled GRGDSPC peptide (0, 5, 10, 20 mg peptide/g DEX-MAES). The cells were stained with a live/dead assay containing FDA and EB at day 2. Green and red depict live and dead cells, respectively.
Viability and osteogenic differentiation of encapsulated hMSCs
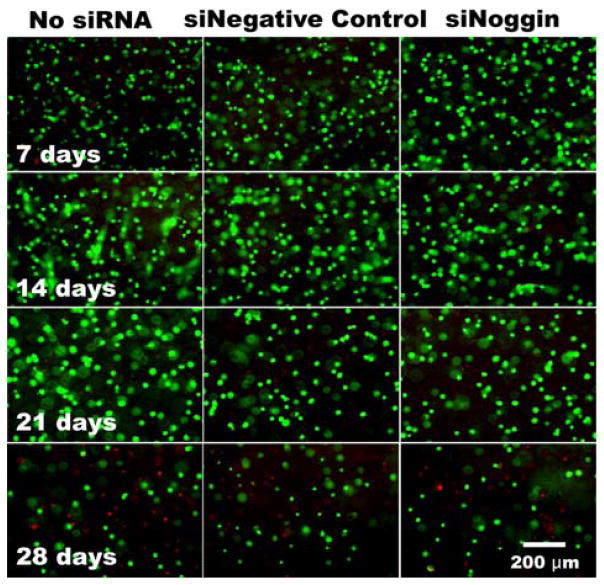
After demonstrating the capacity of the hydrogel system to release siRNA over a prolonged period of time, the capacity of the biomaterial with siRNA to regulate gene expression of encapsulated hMSCs was investigated, which would be useful for tissue regeneration including bone tissue engineering. Therefore, in this study, the sustained, localized delivery of siNoggin to hMSCs encapsulated within the photocrosslinkable DEX-MAES16 hydrogels was examined to determine its ability to enhance osteogenic differentiation. The hydrogel constructs containing hMSCs and siRNA were cultured in osteogenic media supplemented with BMP-2. The osteogenic differentiation of encapsulated hMSCs presented with siNoggin was compared to that of hMSCs in hydrogels without siRNA and to those with negative control siRNA. Cytocompatibility of biomaterials is crucial for tissue engineering application; hence, after co-photoencapsulation of siRNA and hMSCs within the hydrogels, cell viability was measured by fluorescence staining with a live/dead assay to determine whether the siRNA, hydrogels and/or photopolymerization process have a deleterious effect on the hMSCs. Most of the encapsulated hMSCs were highly viable during the first 3 weeks of culture, but the number of cells decreased by day 28 with an increase in dead cells (Figure 8).
Figure 8.
Viability of hMSCs photo-encapsulated within DEX-MAES16 hydrogels with and without siRNA over time. The cells were stained with a live/dead assay containing FDA and EB. Green and red depicts live and dead cells, respectively.
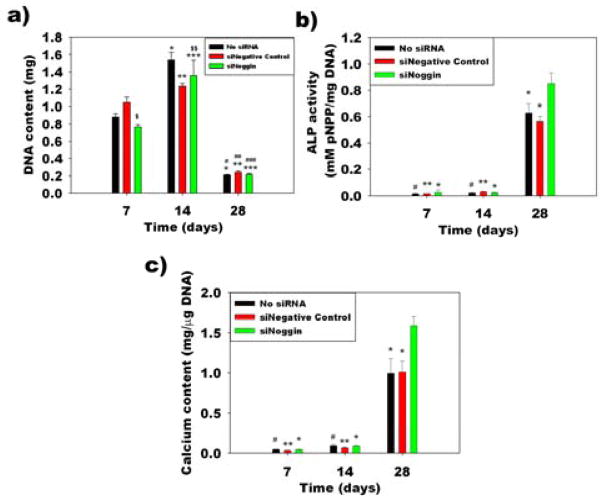
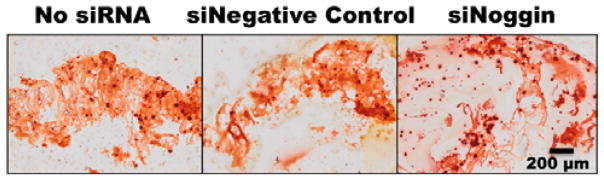
Cell number within the hydrogels was indirectly measured over time by quantifying the amount of DNA in the constructs. DNA content within the hydrogels increased from day 7 to day 14 and decreased at day 28 (Figure 9a). To investigate whether the delivery of siNoggin to the encapsulated hMSCs within the photocrosslinked hydrogels could enhance their osteogenic differentiation compared to the control groups, alkaline phosphatase (ALP) activity, an early osteogenic differentiation marker, was measured. ALP activity normalized to DNA content in all groups increased over time and reached maximum activity at day 28 (Figure 9b). ALP activity in the siNoggin group was significantly higher than that in the no siRNA and siNegative control groups at day 28. When hMSCs undergo osteogenic differentiation, the cells produce mineralized bone tissue. The mineral is in the form of hydroxyapatite, of which calcium is a critical component. Calcium deposition was evaluated by calcium content quantification within the constructs over 4 weeks. Calcium content normalized to DNA increased over time in all groups and was significantly higher in the siNoggin group compared to the control groups at day 28 (Figure 9c). Alizarin red staining for calcium at the 3 weeks revealed slightly darker staining in the siNoggin group compared to the No siRNA and siNegative control groups, supporting that the delivery of siNoggin enhanced calcium deposition (Figure 10).
Figure 9.
(a) Quantification of DNA content in hMSC-hydrogel constructs. *p < 0.05 compared to the no siRNA group at day 7, **p < 0.05 compared to the siNegative control group at day 7, ***p < 0.05 compared to the siNoggin group at day 7, #p < 0.05 compared to the no siRNA group at day 14, ##p < 0.05 compared to the siNegative control group at day 14, ###p < 0.05 compared to the siNoggin day 14, $p < 0.05 compared to the negative control group at day 7 and $$p < 0.05 compared to the no siRNA group at day 14. (b) ALP activity and (c) calcium content in the hMSC-hydrogel constructs. *p < 0.05 compared to the siNoggin group at day 28, **p < 0.05 compared to the siNegative control group at day 28 and #p < 0.05 compared to the no siRNA group at day 28.
Figure 10.
Photomicrographs of Alizarin red stained histologic sections of hydrogels cultured in osteogenic media for 3 weeks with encapsulated hMSCs with No siRNA, siNegative control or siNoggin.
Discussion
The main objective of this work was to develop novel hydrolytically degradable, photopolymerizable acrylated-DEX hydrogels with independently tunable cell adhesivity for prolonged siRNA delivery to encapsulated hMSCs for their enhanced osteogenic differentiation. While the previously reported modification of DEX with vinyl acrylate in the literature occurred at 50°C for 3 days,52, 53 DEX-MAES macromers were synthesized via an esterification reaction of MAES with hydroxyl groups of DEX, which occurred at room temperature for one day. The degree of MAES modification was easily regulated by changing the amounts of MAES used in the reaction, and the hydrogels could be formed by the application of a low-intensity UV light in the presence of a low photoinitiator concentration.
DEX modified with methacrylate groups, such as DEX-HEMA, also formed hydrolytically degradable hydrogels. However, the synthesis process utilized requires two steps with a long overall reaction time (i.e., 5 days in total).24 The synthesis of DEX-MAES is a one-step process with shorter reaction time of only 1 day. DEX modification with vinyl acrylate in previous reports resulted in DEX-acrylate hydrogels that may be challenging to use in tissue engineering due to their slow degradation rate in aqueous solution,53 likely due to the presence of only one ester linkage between each acrylate and DEX molecule. MAES conjugation to DEX creates three ester groups between each acrylate and DEX molecule, which accelerates the hydrolysis of the resulting photocrosslinked hydrogels. Regulation of the MAES modification permits control over the swelling ratio and degradation profiles of hydrogels. The DEX-MAES6 hydrogels swelled and degraded faster than the DEX-MAES11 and 16 hydrogels, due to their lower degree of crosslinking. The DEX-MAES16 hydrogels swelled and degraded more rapidly in DMEM media than PBS at the same incubation conditions. These differences in swelling and degradation properties are likely due to the presence of other components in DMEM, such as vitamins, amino acids and glucose, and/or different salt contents compared to PBS. Hydrogels without incorporated siRNA complexes and/or conjugated RGD peptide (“Empty”) swelled slightly more than the “siRNA complexes” and “siRNA complexes + RGD” hydrogels, but the addition of siRNA complexes and RGD did not affect hydrogel degradation rate, likely due to the small amounts of siRNA and RGD used. The more rapid and tunable degradation of the DEX-MAES hydrogels is valuable for (1) tailoring their mass loss rate to match that of encapsulated cell proliferation and the deposition of new extracellular matrix and (2) creating new pore space to enhance diffusion of oxygen and nutrients into and clearance of metabolic waste out of the constructs.
Due to its hydrophilic, macromolecular and anionic characteristics, siRNA cannot bind to and cross the negatively charged cell membrane.67 In this study, a cationic polymer, PEI, was used to condense siRNA into cationic nanoparticles via electrostatic interactions, enabling cellular uptake,8, 68, 69 and their release kinetics from the photocrosslinked hydrogels were examined. siRNA release was tailored via increasing the level of MAES modification from 6 to 16%. Release from the DEX-MAES6 hydrogels was most rapid. Interestingly, the release profiles of siRNA complexes from DEX-MAES11 and 16 hydrogels were similar, which was probably due to the similar degradation rate of these hydrogels. The release mechanism of the siRNA nanoparticles from the hydrogels is likely a combination of diffusion and hydrogel degradation. Previously, cationic DEX microgels and nanogels were prepared for the intracellular delivery of siRNA43, 70 and larger mm-scale DEX hydrogels were used for the delivery of chemically-modified siRNA for up to 17 days.24 This is the first report of more prolonged release of siRNA/cationic polymer complexes from the photocrosslinked DEX hydrogels for up to 55 days.
The ability of cells to adhere to and interact with biomaterials enables and/or can enhance behaviors such as migration,58 proliferation55 and differentiation;60–62 hence, it can be valuable to endow biomaterials with cell adhesive moieties via modification with specific ligands identified from natural extracellular matrix molecules or through library screens to control cell function for tissue engineering applications.71, 72 DEX is not cell adhesive in its native form,73 but laminin-derived peptides (i.e., CGDPGYIGSR and CQAASIKVAV) have been incorporated into hydrogels formed with maleimide-modified DEX macromer to regulate cell adhesion.74 Similarly, peptides containing the RGD amino acid sequence, a cell binding domain found in fibronectin and type I collagen,75 have previously been conjugated to DEX hydrogel networks. Since DEX contains hydroxyl groups on its glucose residues that cannot be directly modified with amine groups of RGD sequences, DEX hydrogels were photofunctionalized with acrylate-PEG-RGD.76 However, this approach requires the use of multifunctional PEG that is complicated to synthesize, and the synthesis and purification of acrylate-PEG-RGD conjugates may alter the bioactivity of the peptide. In this study, GRGDSPC peptides which contain thiol groups were used to simply conjugate them directly to DEX-MAES macromers via thiol-acrylate Michael reaction. The measurement of free thiol groups of the peptides using Ellman’s assay revealed that the conjugation efficiency increased with time and reduced RGD concentration. In addition, the conjugation of the thiol-containing peptide to DEX acrylate occurred with a higher reaction rate than that to DEX methacrylate. This simple chemistry has been also used to conjugate a thiol-modified RGD peptide to PEG-acrylate hydrogels in the presence of 0.3M triethanolamine.77 Unlike other approaches that require an overnight reaction,74 in this work, the thiol peptide was rapidly conjugated to DEX macromers in PBS pH 7.8 and without the use of any additional chemicals. When cells were seeded on the surface of these hydrogels, fewer hMSCs were able to adhere to hydrogels without conjugated RGD compared to those with the peptide, and increasing the concentration of thiolated RGD peptide in the photocrosslinked DEX hydrogels enhanced hMSC attachment and spreading. These findings indicate that varying the cell adhesion peptide concentration in the hydrogels permits control over these hMSC behaviors. This strategy for cell adhesive peptide conjugation provides an approach for regulating cell interactions with DEX hydrogels and cell function on or in the biomaterials.
Hydrogels used for tissue engineering applications must be cytocompatible. hMSCs retained high cell viability when cultured on the surface of peptide-modified DEX-MAES16 hydrogels after 2 days of culture. In addition, encapsulated hMSC viability remained high at days 7, 14 and 21, revealing that the photocrosslinking process, encapsulation of siRNA/PEI complexes, and DEX-MAES hydrogels themselves did not have a deleterious effect on the hMSCs. However, a visual decrease in encapsulated cell number was observed at day 28, which correlated with the decreased DNA content at the same time point. The hMSC/hydrogel constructs also became very weak and hard to manipulate by day 28. The weakening of hydrogels was likely a result of their near complete degradation and high degree of swelling by this time point. Cell loss may have been due to bulk hydrogel degradation, degradation by-products and/or potential cell movement out of the hydrogels to the bottom of the tissue culture plate wells. It was also qualitatively observed that the hydrogels containing hMSCs in the osteogenesis study were much weaker after one month of culture in osteogenic media compared to the hydrogels without encapsulated hMSCs cultured in DMEM-LG only in the degradation study. It is possible that the hMSCs may decrease the degree of hydrogel crosslinking during photopolymerization, leading to more rapid hydrolytic degradation.
After demonstrating that the photocrosslinked DEX hydrogels were hydrolytically degradable and could sustain the release of siRNA over a prolonged period of time, and that peptide modification of the hydrogels enhanced cell adhesion to the hydrogels, it was then shown that the siRNA-containing, adhesion ligand-modified hydrogels could also enhance the osteogenic differentiation of encapsulated hMSCs. Noggin has been reported to decrease BMP signaling in rat calvarial osteoblasts78 and also to reduce bone formation in a transgenic mouse model over-expressing noggin.79 Studies have also demonstrated that inhibiting noggin expression enhances osteogenic differentiation in MC3T3 preosteoblasts, primary mouse calvarial osteoblasts,80 hADSCs9, 81 and hMSCs.8 Therefore, the effect of siNoggin delivery on osteogenic differentiation of hMSCs was then examined in this study. The sustained, localized delivery of siNoggin to hMSCs encapsulated within the DEX hydrogels cultured in osteogenic media increased ALP activity and calcium content within the constructs compared to the control hydrogels without siRNA and with negative control siRNA, indicating their enhanced osteogenic differentiation. Slightly darker Alizarin red staining also suggests enhanced osteogenesis when incorporated siNoggin was delivered to encapsulated hMSCs. Constructs cultured for 3 weeks were used for the staining as they were too soft for paraffin embedding at 4 weeks.
Previously, bone formation has been promoted by using a thermoresponsive hydrogel to deliver naked siNoggin followed by electroporation to rat muscle,82 which failed to examine the effect of sustained siRNA delivery on bone regeneration. Recently, sustained siNoggin presentation from in situ forming PEG hydrogels was shown to enhance the osteogenesis of encapsulated hMSCs in vitro.8 While PEG hydrogels have been widely reported as a promising biomaterial for bone tissue engineering,56, 83 PEG contains fewer functional hydroxyl groups for further chemical modifications with functional moieties such as cell adhesion ligands and crosslinkable groups compared to DEX at the same molar concentration. A gelatin, hyaluronic acid, PEG and heparin composite hydrogel delivery system was also investigated to deliver miRNA-26a and hMSCs for increased bone regeneration in a mouse critical-sized calvarial bone defect model, but the biomaterial provided little control over delivery of the miRNA.27 In addition, cell-free, nanofibrous polylactic acid scaffolds containing miRNA-26a loaded polylactic-co-glycolic acid (PLGA) microspheres could also regenerate bone in a similar mouse calvarial defect model,29 but the effects of the system on seeded cells was not examined. In this study, a photocrosslinked hydrogel system permitting tunable and prolonged siRNA release profiles via regulating the hydrolytically degradable ester group density within the biomaterial network was examined for osteogenic differentiation of encapsulated hMSCs.
Recently, a report has shown that the effect of siRNA presentation on the gene expression of cells cultured on the 2D surface of alginate hydrogels was regulated via varying the density of cell adhesion ligands containing RGD.63 Therefore, it would be informative to investigate the role of adhesion ligand type and concentration in conjunction with siRNA delivery from within the DEX hydrogels reported here on siRNA transfection efficacy and osteogenic differentiation of hMSCs encapsulated in this 3D system. Such studies would enhance our understanding of the role of cell-biomaterial interactions on RNA delivery to cells in 3D matrices for tissue engineering therapeutics.
It has been demonstrated that controlling degradation rate of hydrogels could regulate the rate of tissue formation by encapsulated cells.57 The degradation rate of these DEX-MAES photocrosslinked hydrogels is tunable by changing the degree of DEX acrylation, enabling the effect of degradation rate of these materials and thus different siRNA release profiles on the osteogenic differentiation of encapsulated cells to be examined. The influence of varying DEX molecular weight and hydrogel concentration on hydrogel degradation rate and siRNA release profiles may also be valuable to investigate in the future. In addition, it is known that different siRNA concentrations can suppress gene expression at different levels.21 While in this study, only a single siRNA concentration was used for all the experiments, future studies may examine the effect of varying the siRNA concentration within the hydrogels on encapsulated cell gene expression and subsequent osteogenic differentiation. Finally, alternatively to siRNA delivery, this DEX biomaterial system is versatile and may also be used for the delivery of other genetic molecules, such as microRNA, antisense oligonucleotides and DNA plasmids.
Conclusions
In this paper, a novel photocrosslinkable degradable hydrogel system with controllable swelling ratio and hydrolytic degradation properties was engineered by synthesizing acrylated DEX macromers. The hydrogels permitted controllable cell adhesivity, and tailorable, prolonged release of siRNA. hMSCs encapsulated within the peptide-modified hydrogels exhibited high viability for at least 3 weeks. In addition, the sustained delivery of siNoggin from the hydrogels enhanced the osteogenic differentiation of encapsulated hMSCs. These photocrosslinked DEX-MAES hydrogels with controlled cell adhesion properties may provide a valuable platform for the prolonged delivery of genetic material for a wide range of biomedical applications such as tissue engineering and disease therapeutics.
Acknowledgments
The authors gratefully acknowledge funding from the National Institute of Dental & Craniofacial Research (R56DE022376) and the National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01AR069564) of the National Institutes of Health and the Department of Defense Congressionally Directed Medical Research Programs (OR110196).
References
- 1.Gary DJ, Puri N, Won YY. Journal of Controlled Release. 2007;121:64–73. doi: 10.1016/j.jconrel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 2.Monaghan M, Pandit A. Advanced Drug Delivery Reviews. 2011;63:197–208. doi: 10.1016/j.addr.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Krebs MD, Alsberg E. Chemistry – A European Journal. 2011;17:3054–3062. doi: 10.1002/chem.201003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwon YJ. Accounts of Chemical Research. 2011;45:1077–1088. doi: 10.1021/ar200241v. [DOI] [PubMed] [Google Scholar]
- 5.Lam JKW, Chow MYT, Zhang Y, Leung SWS. Mol Ther Nucleic Acids. 2015;4:e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Resnier P, Montier T, Mathieu V, Benoit JP, Passirani C. Biomaterials. 2013;34:6429–6443. doi: 10.1016/j.biomaterials.2013.04.060. [DOI] [PubMed] [Google Scholar]
- 7.Teo J, McCarroll JA, Boyer C, Youkhana J, Sagnella SM, Duong HTT, Liu J, Sharbeen G, Goldstein D, Davis TP, Kavallaris M, Phillips PA. Biomacromolecules. 2016 doi: 10.1021/acs.biomac.6b00185. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen MK, Jeon O, Krebs MD, Schapira D, Alsberg E. Biomaterials. 2014;35:6278–6286. doi: 10.1016/j.biomaterials.2014.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramasubramanian A, Shiigi S, Lee G, Yang F. Pharm Res. 2011;28:1328–1337. doi: 10.1007/s11095-011-0406-9. [DOI] [PubMed] [Google Scholar]
- 10.Czauderna F, Fechtner M, Dames S, Aygün H, Klippel A, Pronk GJ, Giese K, Kaufmann J. Nucleic Acids Research. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. The American Journal of Pathology. 170:1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prakash TP, Allerson CR, Dande P, Vickers TA, Sioufi N, Jarres R, Baker BF, Swayze EE, Griffey RH, Bhat B. Journal of Medicinal Chemistry. 2005;48:4247–4253. doi: 10.1021/jm050044o. [DOI] [PubMed] [Google Scholar]
- 13.Choung S, Kim YJ, Kim S, Park HO, Choi YC. Biochemical and Biophysical Research Communications. 2006;342:919–927. doi: 10.1016/j.bbrc.2006.02.049. [DOI] [PubMed] [Google Scholar]
- 14.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 15.Tzeng SY, Hung BP, Grayson WL, Green JJ. Biomaterials. 2012;33:8142–8151. doi: 10.1016/j.biomaterials.2012.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim SH, Jeong JH, Kim T-i, Kim SW, Bull DA. Molecular Pharmaceutics. 2008;6:718–726. doi: 10.1021/mp800161e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang T, Bantegui T, Pike K, Bloom R, Phipps R, Bai S. Journal of Liposome Research. 2014:1–10. doi: 10.3109/08982104.2014.907306. [DOI] [PubMed] [Google Scholar]
- 18.Wan C, Allen TM, Cullis PR. Drug Deliv and Transl Res. 2014;4:74–83. doi: 10.1007/s13346-013-0161-z. [DOI] [PubMed] [Google Scholar]
- 19.Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DW, Stebbing D, Crosley EJ, Yaworski E, Hafez IM, Dorkin JR, Qin J, Lam K, Rajeev KG, Wong KF, Jeffs LB, Nechev L, Eisenhardt ML, Jayaraman M, Kazem M, Maier MA, Srinivasulu M, Weinstein MJ, Chen Q, Alvarez R, Barros SA, De S, Klimuk SK, Borland T, Kosovrasti V, Cantley WL, Tam YK, Manoharan M, Ciufolini MA, Tracy MA, de Fougerolles A, MacLachlan I, Cullis PR, Madden TD, Hope MJ. Nature biotechnology. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 20.Xu J, Ganesh S, Amiji M. International Journal of Pharmaceutics. 2012;427:21–34. doi: 10.1016/j.ijpharm.2011.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krebs MD, Jeon O, Alsberg E. Journal of the American Chemical Society. 2009;131:9204–9206. doi: 10.1021/ja9037615. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen MK, Alsberg E. Progress in Polymer Science. 2014;39:1235–1265. doi: 10.1016/j.progpolymsci.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen MK, Lee DS. Macromolecular Bioscience. 2010;10:563–579. doi: 10.1002/mabi.200900402. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen K, Dang PN, Alsberg E. Acta Biomaterialia. 2013;9:4487–4495. doi: 10.1016/j.actbio.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim YM, Park MR, Song SC. ACS Nano. 2012;6:5757–5766. doi: 10.1021/nn300842a. [DOI] [PubMed] [Google Scholar]
- 26.Takahashi H, Wang Y, Grainger DW. Journal of Controlled Release. 2010;147:400–407. doi: 10.1016/j.jconrel.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Fan L, Liu S, Liu W, Zhang H, Zhou T, Wu D, Yang P, Shen L, Chen J, Jin Y. Biomaterials. 2013;34:5048–5058. doi: 10.1016/j.biomaterials.2013.03.052. [DOI] [PubMed] [Google Scholar]
- 28.Nelson CE, Kim AJ, Adolph EJ, Gupta MK, Yu F, Hocking KM, Davidson JM, Guelcher SA, Duvall CL. Advanced materials. 2014;26:607–614. 506. doi: 10.1002/adma.201303520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Li Y, Chen YE, Chen J, Ma PX. Nat Commun. 2016:7. doi: 10.1038/ncomms10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hill MC, Nguyen MK, Jeon O, Alsberg E. Advanced Healthcare Materials. 2015;4:714–722. doi: 10.1002/adhm.201400458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huynh CT, Nguyen MK, Tonga GY, Longé L, Rotello VM, Alsberg E. Advanced Healthcare Materials. 2016;5:305–310. doi: 10.1002/adhm.201500778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huynh CT, Nguyen MK, Naris M, Tonga GY, Rotello VM, Alsberg E. Nanomedicine. 2016;11:1535–1550. doi: 10.2217/nnm-2016-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen KT, West JL. Biomaterials. 2002;23:4307–4314. doi: 10.1016/s0142-9612(02)00175-8. [DOI] [PubMed] [Google Scholar]
- 34.Jeon O, Wolfson DW, Alsberg E. Advanced materials. 2015;27:2216–2223. doi: 10.1002/adma.201405337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fisher JP, Dean D, Engel PS, Mikos AG. Annual Review of Materials Research. 2001;31:171–181. [Google Scholar]
- 36.Nettles DL, Vail TP, Morgan MT, Grinstaff MW, Setton LA. Ann Biomed Eng. 2004;32:391–397. doi: 10.1023/b:abme.0000017552.65260.94. [DOI] [PubMed] [Google Scholar]
- 37.Smeds KA, Pfister-Serres A, Miki D, Dastgheib K, Inoue M, Hatchell DL, Grinstaff MW. Journal of Biomedical Materials Research. 2001;55:254–255. doi: 10.1002/1097-4636(200101)54:1<115::aid-jbm14>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 38.Jeon O, Bouhadir KH, Mansour JM, Alsberg E. Biomaterials. 2009;30:2724–2734. doi: 10.1016/j.biomaterials.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 39.Ifkovits JL, Burdick JA. Tissue Engineering. 2007;13:2369–2385. doi: 10.1089/ten.2007.0093. [DOI] [PubMed] [Google Scholar]
- 40.Kim SH, Chu CC. Journal of Biomedical Materials Research. 2000;49:517–527. doi: 10.1002/(sici)1097-4636(20000315)49:4<517::aid-jbm10>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 41.Van Tomme SR, Hennink WE. Expert Review of Medical Devices. 2007;4:147–164. doi: 10.1586/17434440.4.2.147. [DOI] [PubMed] [Google Scholar]
- 42.Sun G, Mao JJ. Nanomedicine. 2012;7:1771–1784. doi: 10.2217/nnm.12.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raemdonck K, Van Thienen TG, Vandenbroucke RE, Sanders NN, Demeester J, De Smedt SC. Advanced Functional Materials. 2008;18:993–1001. [Google Scholar]
- 44.Kim SH, Chu CC. Journal of Biomaterials Applications. 2000;15:23–46. doi: 10.1106/ML30-FMLG-1HRU-7C4B. [DOI] [PubMed] [Google Scholar]
- 45.Jukes JM, van der Aa LJ, Hiemstra C, van Veen T, Dijkstra PJ, Zhong Z, Feijen J, van Blitterswijk CA, de Boer J. Tissue Engineering Part A. 2009;16:565–573. doi: 10.1089/ten.TEA.2009.0173. [DOI] [PubMed] [Google Scholar]
- 46.Maire M, Chaubet F, Mary P, Blanchat C, Meunier A, Logeart-Avramoglou D. Biomaterials. 2005;26:5085–5092. doi: 10.1016/j.biomaterials.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 47.Jin R, Moreira Teixeira LS, Dijkstra PJ, van Blitterswijk CA, Karperien M, Feijen J. Journal of Controlled Release. 2011;152:186–195. doi: 10.1016/j.jconrel.2011.01.031. [DOI] [PubMed] [Google Scholar]
- 48.van Dijk-Wolthuis WNE, Franssen O, Talsma H, van Steenbergen MJ, Kettenes-van den Bosch JJ, Hennink WE. Macromolecules. 1995;28:6317–6322. [Google Scholar]
- 49.De Smedt SC, Lauwers A, Demeester J, Van Steenbergen MJ, Hennink WE, Roefs SPFM. Macromolecules. 1995;28:5082–5088. [Google Scholar]
- 50.van Dijk-Wolthuis WNE, van Steenbergen MJ, Underberg WJM, Hennink WE. Journal of Pharmaceutical Sciences. 1997;86:413–417. doi: 10.1021/js9604220. [DOI] [PubMed] [Google Scholar]
- 51.van Dijk-Wolthuls WNE, Tsang SKY, Kettenes-van den Bosch JJ, Hennink WE. Polymer. 1997;38:6235–6242. [Google Scholar]
- 52.Ferreira L, Gil MH, Dordick JS. Biomaterials. 2002;23:3957–3967. doi: 10.1016/s0142-9612(02)00132-1. [DOI] [PubMed] [Google Scholar]
- 53.Ferreira L, Rafael A, Lamghari M, Barbosa MA, Gil MH, Cabrita AMS, Dordick JS. Journal of Biomedical Materials Research Part A. 2004;68A:584–596. doi: 10.1002/jbm.a.20102. [DOI] [PubMed] [Google Scholar]
- 54.Nuttelman CR, Tripodi MC, Anseth KS. Matrix Biology. 2005;24:208–218. doi: 10.1016/j.matbio.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 55.Jeon O, Alsberg E. Tissue Engineering Part A. 2013;19:1424–1432. doi: 10.1089/ten.tea.2012.0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burdick JA, Anseth KS. Biomaterials. 2002;23:4315–4323. doi: 10.1016/s0142-9612(02)00176-x. [DOI] [PubMed] [Google Scholar]
- 57.Alsberg E, Kong HJ, Hirano Y, Smith MK, Albeiruti A, Mooney DJ. Journal of Dental Research. 2003;82:903–908. doi: 10.1177/154405910308201111. [DOI] [PubMed] [Google Scholar]
- 58.Maheshwari G, Brown G, Lauffenburger DA, Wells A, Griffith LG. Journal of Cell Science. 2000;113:1677–1686. doi: 10.1242/jcs.113.10.1677. [DOI] [PubMed] [Google Scholar]
- 59.Alsberg E, Anderson KW, Albeiruti A, Rowley JA, Mooney DJ. Proceedings of the National Academy of Sciences. 2002;99:12025–12030. doi: 10.1073/pnas.192291499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kang SW, Cha BH, Park H, Park KS, Lee KY, Lee SH. Macromolecular Bioscience. 2011;11:673–679. doi: 10.1002/mabi.201000479. [DOI] [PubMed] [Google Scholar]
- 61.Shin H, Temenoff JS, Bowden GC, Zygourakis K, Farach-Carson MC, Yaszemski MJ, Mikos AG. Biomaterials. 2005;26:3645–3654. doi: 10.1016/j.biomaterials.2004.09.050. [DOI] [PubMed] [Google Scholar]
- 62.Connelly JT, García AJ, Levenston ME. Biomaterials. 2007;28:1071–1083. doi: 10.1016/j.biomaterials.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 63.Khormaee S, Ali OA, Chodosh J, Mooney DJ. Journal of Biomedical Materials Research Part A. 2012;100A:2637–2643. doi: 10.1002/jbm.a.34202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elbert DL, Hubbell JA. Biomacromolecules. 2001;2:430–441. doi: 10.1021/bm0056299. [DOI] [PubMed] [Google Scholar]
- 65.Haynesworth SE, Goshima J, Goldberg VM, Caplan AI. Bone. 1992;13:81–88. doi: 10.1016/8756-3282(92)90364-3. [DOI] [PubMed] [Google Scholar]
- 66.Lennon D, Haynesworth S, Bruder S, Jaiswal N, Caplan A. In Vitro Cell Dev Biol-Animal. 1996;32:602–611. [Google Scholar]
- 67.Wang J, Lu Z, Wientjes MG, Au JL. The AAPS journal. 2010;12:492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Günther M, Lipka J, Malek A, Gutsch D, Kreyling W, Aigner A. European Journal of Pharmaceutics and Biopharmaceutics. 2011;77:438–449. doi: 10.1016/j.ejpb.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 69.Werth S, Urban-Klein B, Dai L, Höbel S, Grzelinski M, Bakowsky U, Czubayko F, Aigner A. Journal of Controlled Release. 2006;112:257–270. doi: 10.1016/j.jconrel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 70.Naeye B, Raemdonck K, Remaut K, Sproat B, Demeester J, De Smedt SC. European Journal of Pharmaceutical Sciences. 2010;40:342–351. doi: 10.1016/j.ejps.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 71.Flaim CJ, Chien S, Bhatia SN. Nat Meth. 2005;2:119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 72.Zhang D, Kilian KA. Journal of Materials Chemistry B. 2014;2:4280–4288. doi: 10.1039/c4tb00375f. [DOI] [PubMed] [Google Scholar]
- 73.Massia SP, Stark J, Letbetter DS. Biomaterials. 2000;21:2253–2261. doi: 10.1016/s0142-9612(00)00151-4. [DOI] [PubMed] [Google Scholar]
- 74.Lévesque SG, Shoichet MS. Bioconjugate Chemistry. 2007;18:874–885. doi: 10.1021/bc0602127. [DOI] [PubMed] [Google Scholar]
- 75.Gehlsen KR, Argraves WS, Pierschbacher MD, Ruoslahti E. The Journal of cell biology. 1988;106:925–930. doi: 10.1083/jcb.106.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ferreira LS, Gerecht S, Fuller J, Shieh HF, Vunjak-Novakovic G, Langer R. Biomaterials. 2007;28:2706–2717. doi: 10.1016/j.biomaterials.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zustiak SP, Durbal R, Leach JB. Acta Biomaterialia. 2010;6:3404–3414. doi: 10.1016/j.actbio.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gazzerro E, Gangji V, Canalis E. J Clin Invest. 1998;102:2106–2114. doi: 10.1172/JCI3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu X-B, Li Y, Schneider A, Yu W, Rajendren G, Iqbal J, Yamamoto M, Alam M, Brunet LJ, Blair HC, Zaidi M, Abe E. The Journal of Clinical Investigation. 112:924–934. doi: 10.1172/JCI15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wan DC, Pomerantz JH, Brunet LJ, Kim JB, Chou YF, Wu BM, Harland R, Blau HM, Longaker MT. Journal of Biological Chemistry. 2007;282:26450–26459. doi: 10.1074/jbc.M703282200. [DOI] [PubMed] [Google Scholar]
- 81.Cui ZK, Fan J, Kim S, Bezouglaia O, Fartash A, Wu BM, Aghaloo T, Lee M. Journal of Controlled Release. 2015;217:42–52. doi: 10.1016/j.jconrel.2015.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Manaka T, Suzuki A, Takayama K, Imai Y, Nakamura H, Takaoka K. Biomaterials. 2011;32:9642–9648. doi: 10.1016/j.biomaterials.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 83.Nuttelman CR, Tripodi MC, Anseth KS. Journal of Biomedical Materials Research Part A. 2004;68A:773–782. doi: 10.1002/jbm.a.20112. [DOI] [PubMed] [Google Scholar]