Abstract
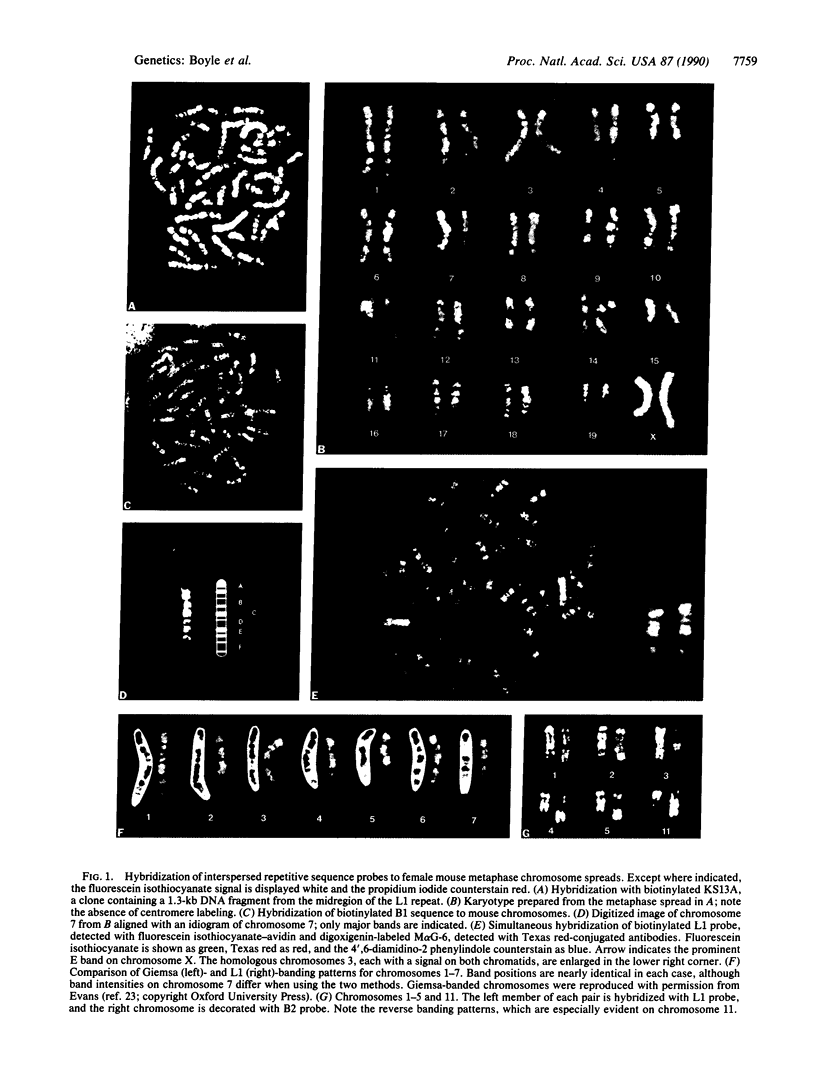
Fluorescence in situ hybridization has been used to demonstrate the differential distribution of interspersed repetitive elements in the genome of Mus musculus domesticus. Hybridization with a mouse long interspersed element sequence results in a sharp, highly reproducible banding pattern on metaphase chromosomes, which is quite similar to Giemsa banding for all chromosomes except 7 and X. The families of short interspersed elements, B1 and B2, preferentially cluster in the R, or reverse, bands. There is no evidence of any interspersed repeat present in the centromeric heterochromatic regions. Both the long interspersed element and B2 probes give banding patterns suitable for karyotype analysis. Simultaneous hybridization of the biotinylated long interspersed element probe and a digoxigenin-labeled cosmid to metaphase spreads allows rapid localization of a probe of interest to a particular cytogenetic band on a chromosome.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertson D. G., Fishpool R., Sherrington P., Nacheva E., Milstein C. Sensitive and high resolution in situ hybridization to human chromosomes using biotin labelled probes: assignment of the human thymocyte CD1 antigen genes to chromosome 1. EMBO J. 1988 Sep;7(9):2801–2805. doi: 10.1002/j.1460-2075.1988.tb03135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett K. L., Hill R. E., Pietras D. F., Woodworth-Gutai M., Kane-Haas C., Houston J. M., Heath J. K., Hastie N. D. Most highly repeated dispersed DNA families in the mouse genome. Mol Cell Biol. 1984 Aug;4(8):1561–1571. doi: 10.1128/mcb.4.8.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt B., Burns J., Flannery D., McGee J. O. Direct visualization of single copy genes on banded metaphase chromosomes by nonisotopic in situ hybridization. Nucleic Acids Res. 1988 May 11;16(9):3951–3961. doi: 10.1093/nar/16.9.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle A. L., Lichter P., Ward D. C. Rapid analysis of mouse-hamster hybrid cell lines by in situ hybridization. Genomics. 1990 May;7(1):127–130. doi: 10.1016/0888-7543(90)90529-4. [DOI] [PubMed] [Google Scholar]
- Brigati D. J., Myerson D., Leary J. J., Spalholz B., Travis S. Z., Fong C. K., Hsiung G. D., Ward D. C. Detection of viral genomes in cultured cells and paraffin-embedded tissue sections using biotin-labeled hybridization probes. Virology. 1983 Apr 15;126(1):32–50. doi: 10.1016/0042-6822(83)90460-9. [DOI] [PubMed] [Google Scholar]
- Chandler M. E., Yunis J. J. A high resolution in situ hybridization technique for the direct visualization of labeled G-banded early metaphase and prophase chromosomes. Cytogenet Cell Genet. 1978;22(1-6):352–356. doi: 10.1159/000130970. [DOI] [PubMed] [Google Scholar]
- Chen T. L., Manuelidis L. SINEs and LINEs cluster in distinct DNA fragments of Giemsa band size. Chromosoma. 1989 Nov;98(5):309–316. doi: 10.1007/BF00292382. [DOI] [PubMed] [Google Scholar]
- Cherif D., Bernard O., Berger R. Detection of single-copy genes by nonisotopic in situ hybridization on human chromosomes. Hum Genet. 1989 Mar;81(4):358–362. doi: 10.1007/BF00283691. [DOI] [PubMed] [Google Scholar]
- Cowell J. K. A photographic representation of the variability in the G-banded structure of the chromosomes in the mouse karyotype. A guide to the identification of the individual chromosomes. Chromosoma. 1984;89(4):294–320. doi: 10.1007/BF00292478. [DOI] [PubMed] [Google Scholar]
- Fanning T. G., Singer M. F. LINE-1: a mammalian transposable element. Biochim Biophys Acta. 1987 Dec 8;910(3):203–212. doi: 10.1016/0167-4781(87)90112-6. [DOI] [PubMed] [Google Scholar]
- Johnson G. D., Davidson R. S., McNamee K. C., Russell G., Goodwin D., Holborow E. J. Fading of immunofluorescence during microscopy: a study of the phenomenon and its remedy. J Immunol Methods. 1982 Dec 17;55(2):231–242. doi: 10.1016/0022-1759(82)90035-7. [DOI] [PubMed] [Google Scholar]
- Kent R. B., Fallows D. A., Geissler E., Glaser T., Emanuel J. R., Lalley P. A., Levenson R., Housman D. E. Genes encoding alpha and beta subunits of Na,K-ATPase are located on three different chromosomes in the mouse. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5369–5373. doi: 10.1073/pnas.84.15.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenberg J. R., Rykowski M. C. Human genome organization: Alu, lines, and the molecular structure of metaphase chromosome bands. Cell. 1988 May 6;53(3):391–400. doi: 10.1016/0092-8674(88)90159-6. [DOI] [PubMed] [Google Scholar]
- Kramerov D. A., Grigoryan A. A., Ryskov A. P., Georgiev G. P. Long double-stranded sequences (dsRNA-B) of nuclear pre-mRNA consist of a few highly abundant classes of sequences: evidence from DNA cloning experiments. Nucleic Acids Res. 1979 Feb;6(2):697–713. doi: 10.1093/nar/6.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krayev A. S., Markusheva T. V., Kramerov D. A., Ryskov A. P., Skryabin K. G., Bayev A. A., Georgiev G. P. Ubiquitous transposon-like repeats B1 and B2 of the mouse genome: B2 sequencing. Nucleic Acids Res. 1982 Dec 11;10(23):7461–7475. doi: 10.1093/nar/10.23.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landegent J. E., Jansen in de Wal N., Dirks R. W., Baao F., van der Ploeg M. Use of whole cosmid cloned genomic sequences for chromosomal localization by non-radioactive in situ hybridization. Hum Genet. 1987 Dec;77(4):366–370. doi: 10.1007/BF00291428. [DOI] [PubMed] [Google Scholar]
- Lawrence J. B., Villnave C. A., Singer R. H. Sensitive, high-resolution chromatin and chromosome mapping in situ: presence and orientation of two closely integrated copies of EBV in a lymphoma line. Cell. 1988 Jan 15;52(1):51–61. doi: 10.1016/0092-8674(88)90530-2. [DOI] [PubMed] [Google Scholar]
- Lichter P., Cremer T., Borden J., Manuelidis L., Ward D. C. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum Genet. 1988 Nov;80(3):224–234. doi: 10.1007/BF01790090. [DOI] [PubMed] [Google Scholar]
- Lichter P., Tang C. J., Call K., Hermanson G., Evans G. A., Housman D., Ward D. C. High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science. 1990 Jan 5;247(4938):64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- Loeb D. D., Padgett R. W., Hardies S. C., Shehee W. R., Comer M. B., Edgell M. H., Hutchison C. A., 3rd The sequence of a large L1Md element reveals a tandemly repeated 5' end and several features found in retrotransposons. Mol Cell Biol. 1986 Jan;6(1):168–182. doi: 10.1128/mcb.6.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L., Ward D. C. Chromosomal and nuclear distribution of the HindIII 1.9-kb human DNA repeat segment. Chromosoma. 1984;91(1):28–38. doi: 10.1007/BF00286482. [DOI] [PubMed] [Google Scholar]
- Miller O. J., Miller D. A., Kouri R. E., Allderdice P. W., Dev V. G., Grewal M. S., Hutton J. J. Identification of the mouse karyotype by quinacrine fluorescence, and tentative assignment of seven linkage groups. Proc Natl Acad Sci U S A. 1971 Jul;68(7):1530–1533. doi: 10.1073/pnas.68.7.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyzis R. K., Torney D. C., Meyne J., Buckingham J. M., Wu J. R., Burks C., Sirotkin K. M., Goad W. B. The distribution of interspersed repetitive DNA sequences in the human genome. Genomics. 1989 Apr;4(3):273–289. doi: 10.1016/0888-7543(89)90331-5. [DOI] [PubMed] [Google Scholar]
- Nesbitt M. N., Francke U. A system of nomenclature for band patterns of mouse chromosomes. Chromosoma. 1973;41(2):145–158. doi: 10.1007/BF00319691. [DOI] [PubMed] [Google Scholar]
- Pardue M. L., Gall J. G. Chromosomal localization of mouse satellite DNA. Science. 1970 Jun 12;168(3937):1356–1358. doi: 10.1126/science.168.3937.1356. [DOI] [PubMed] [Google Scholar]
- Sawin V. L., Skalka A. M., Wray W. Simultaneous observation of quinacrine bands and silver grains on radiolabeled metaphase chromosomes. Histochemistry. 1978 Dec 28;59(1):1–8. doi: 10.1007/BF00506472. [DOI] [PubMed] [Google Scholar]
- Sawyer J. R., Hozier J. C. High resolution of mouse chromosomes: banding conservation between man and mouse. Science. 1986 Jun 27;232(4758):1632–1635. doi: 10.1126/science.3715469. [DOI] [PubMed] [Google Scholar]
- Sawyer J. R., Moore M. M., Hozier J. C. High resolution G-banded chromosomes of the mouse. Chromosoma. 1987;95(5):350–358. doi: 10.1007/BF00293182. [DOI] [PubMed] [Google Scholar]
- Singer M. F. Highly repeated sequences in mammalian genomes. Int Rev Cytol. 1982;76:67–112. doi: 10.1016/s0074-7696(08)61789-1. [DOI] [PubMed] [Google Scholar]
- Somssich I., Hameister H., Winking H. The pattern of early replicating bands in the chromosomes of the mouse. Cytogenet Cell Genet. 1981;30(4):222–231. doi: 10.1159/000131613. [DOI] [PubMed] [Google Scholar]
- Soriano P., Meunier-Rotival M., Bernardi G. The distribution of interspersed repeats is nonuniform and conserved in the mouse and human genomes. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1816–1820. doi: 10.1073/pnas.80.7.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask B., Pinkel D., van den Engh G. The proximity of DNA sequences in interphase cell nuclei is correlated to genomic distance and permits ordering of cosmids spanning 250 kilobase pairs. Genomics. 1989 Nov;5(4):710–717. doi: 10.1016/0888-7543(89)90112-2. [DOI] [PubMed] [Google Scholar]
- Ullu E., Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984 Nov 8;312(5990):171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]