Abstract
Histidine phosphorylation is crucial for prokaryotic signal transduction and as an intermediate for several metabolic enzymes, yet its role in mammalian cells remains largely uncharted. This is primarily due to difficulties in studying histidine phosphorylation because of the relative instability of phosphohistidine (pHis) and lack of specific antibodies and methods to preserve and detect it. The recent synthesis of stable pHis analogs has enabled development of pHis-specific antibodies and their use has started to shed light onto this important, yet enigmatic posttranslational modification. We are beginning to understand that pHis has broader roles in protein and cellular function including; cell cycle regulation, phagocytosis, regulation of ion channel activity and metal ion coordination. Two mammalian histidine kinases (NME1 and NME2), two pHis phosphatases (PHPT1 and LHPP), and a handful of substrates were previously identified. These new tools have already led to the discovery of an additional phosphatase (PGAM5) and hundreds of putative substrates. New methodologies are also being developed to probe the pHis phosphoproteome and determine functional consequences, including negative ion mode mass spectroscopy and unnatural amino acid incorporation. These new tools and strategies have the potential overcome the unique challenges that have been holding back our understanding of pHis in cell biology.
Introduction
Histidine phosphorylation was first reported in 1962 when Boyer [1] detected pHis in an enzyme intermediate of oxidative phosphorylation. The enzyme was subsequently revealed to be succinyl-CoA synthetase (SUCLG1), a key TCA cycle enzyme that couples formation of ATP from ADP with conversion of succinyl-CoA to succinate. Phosphorylation of histidine’s imidazole nitrogen atoms (N1 or N3) forms high-energy phosphoramidate (P-N) bonds in contrast to the more stable phosphoester bonds (P-O) formed when serine (Ser), threonine (Thr) and tyrosine (Tyr) are phosphorylated. Thus, a high free energy of hydrolysis [2] makes pHis relatively unstable and the phosphoramidate bond rapidly hydrolyzes at low pH or when exposed to significant heat. A combination of moderate heat and acid (e.g. pH 6 and 60°C for 30 min [3]) or exposure to certain primary amines [2] also efficiently hydrolyzes pHis. Standard biochemical and proteomic procedures for phosphoester amino acids (pSer, pThr and pTyr [4]) fail to preserve and detect pHis. Consequently, the study of histidine phosphorylation over the last half century has lagged behind the study of pSer, pThr and pTyr (Fig. 1A).
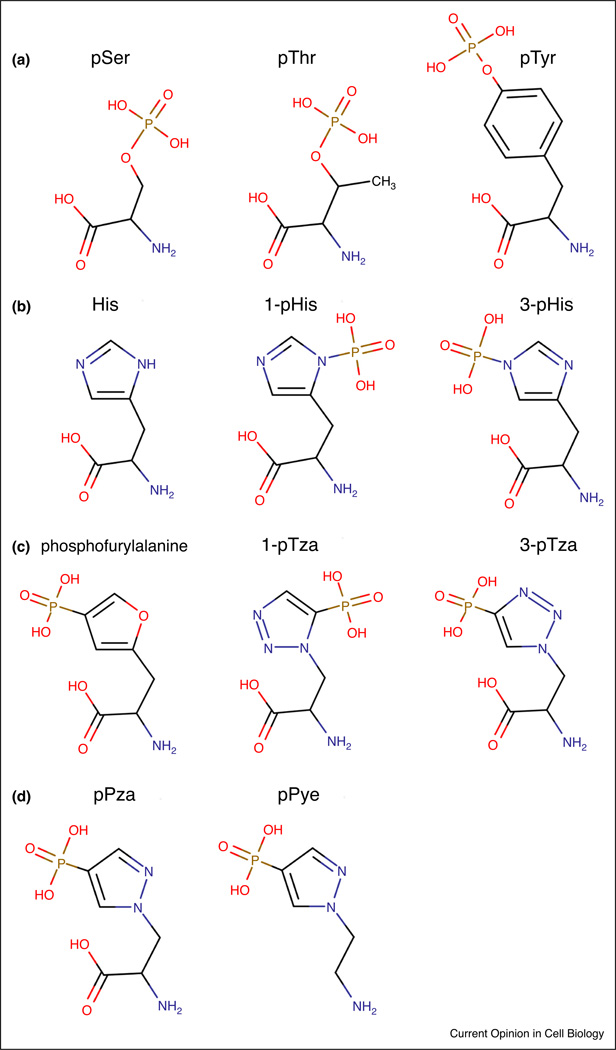
Figure 1. pHis Isomers and Structural Analogs.
Structural drawings of (A) the phosphoester amino acids; pSer, pThr and pTyr are contrasted with (B) histidine, 3-phosphohistidine (3-pHis) and 1-phosphohistidine (1-pHis). Examples of phosphohistidine structural analogs designed for antibody generation include; (C) phosphofurylalanine and the two phosphoryltriazolylalanine analogs (3-pTza) and (1-pTza). (D) Second-generation, pyrazole-based pHis analogs; 4-Phosphopyrazol-2-yl alanine (pPza) and phosphono-pyrazolyl ethylamine (pPye).
The lack of well-established protocols and reagents, including pHis-specific antibodies (Abs) and phosphohistidine phosphatase-specific inhibitors, has also contributed to the sluggish pace of research. Until recently, pHis has been left in the dark; however, the last several years has seen an acceleration in interest and discovery of histidine phosphorylation’s place in cell biology. Light has begun to be shed on its role as a regulator of protein function, in addition to its better known roles as a reactive enzyme intermediate and in prokaryotic signal transduction. This review will focus on recent advances in our understanding of the roles of pHis in mammalian systems that have been stimulated by the availability of new reagents, and the renewed interest and increased appreciation of pHis as an important form of reversible, posttranslational regulation of protein function.
Phosphohistidine Analogs and Antibody Generation
The recent acceleration in pHis research derives from the development of new chemical reagents that facilitated production of pHis-specific Abs. Early attempts at generating pHis Abs were fruitless, since pHis itself is hydrolyzed too rapidly to function as an immunogen. Indeed, our group made an unsuccessful attempt to raise pHis Abs in rabbits by using chemically phosphorylated poly-His [5]. Nevertheless, the fact that an early anti-pTyr Ab (2G8) cross-reacted with pHis in ATP citrate lyase (ACLY [Fig. 2]) gave credence to the idea that generation of pHis Abs should be possible [6]. A strategy of using chemically stable mimetics in place of genuine pHis was therefore needed to circumvent the stability issue. Histidine’s two nitrogens can both be phosphorylated resulting in two distinct isomers; 1-phosphohistidine (1-pHis) and 3-phosphohistidine (3-pHis [Fig. 1B]), which makes pHis unique as a phosphoamino acid. In 1999, Schenkels et al. synthesized a stable phosphofurylalanine analog of 1-pHis (Fig. 1C)[7], but did not try to synthesize peptides or immunize animals with it. In 2010, Muir’s group made a key breakthrough by synthesizing two phosphoryltriazolylalanine analogs (1-pTza and 3-pTza [Fig. 1C]) that mimic 1- and 3-pHis respectively, replacing the unstable phosphoramidate bonds with non-hydrolyzable phosphonate bonds (P-C) and mimicking the electrostatics and geometry of their respective pHis isomer [8,9]. Using a peptide derived from histone H4 (one of the first reported mammalian pHis substrates [10–12]) with His18 replaced by 3-pTza, they immunized rabbits and raised a sequence-specific anti-3-pHis Ab. Shortly thereafter, Webb’s group also reported similar stable analogs compatible with synthetic peptide synthesis [13–15]. Muir subsequently developed “pan-specific” polyclonal 3-pHis Abs, using a 3-pTze analog coupled directly to KLH as antigen, but their usefulness in eukaryotic studies was limited by significant cross-reactivity with pTyr [16], which is structurally most similar to 3-pHis (Fig. 1B).
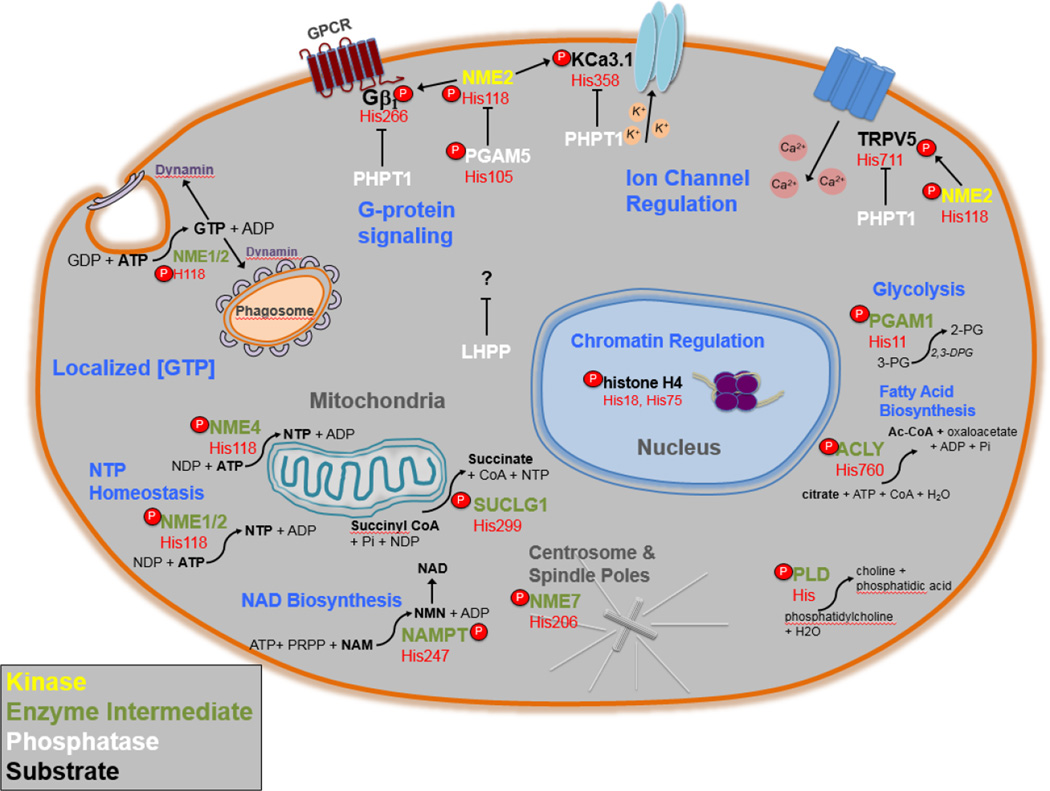
Figure 2. Summary of pHis Cellular Functions.
An illustration of the pHis related proteins discussed in this review and their various functions, enzymatic reactions and subcellular localizations. NME1/2 protein histidine kinase functions are in yellow, pHis enzyme intermediates are in green, phosphohistidine phosphatases are in white and pHis substrates are in bold. Beneath each protein’s gene name is the specific amino acid position number of the pHis residue in red. Cellular functions of specific pHis proteins are in blue. The subcellular localization of pHis related proteins and functions are in grey. Curved arrows represent reactions catalyzed by enzymes that utilize pHis intermediates. For LHPP, phospholysine and 3-phsphohistidine are substrates in vitro, however no known substrates have yet been identified in vivo.
Inspired by this progress, we conceived of a unique approach that used Muir’s pTza analogs embedded in degenerate peptide libraries coupled to KLH as immunogens (Fig. 1C) to promote generation of sequence-independent Abs. Rabbits immunized with either the 1-pTza or 3-pTza library developed high titer polyclonal antibodies; subsequently, we used spleen cells from these rabbits to obtain multiple monoclonal antibody (mAb) clones specific for either 1-pHis or 3-pHis, and validated their usefulness in various immunological assays [3]. These pHis mAbs have been valuable for investigating pHis in mammalian cells [17,18] as well as in bacteria, where they have been used to study auto-phosphorylation of bacterial histidine kinases (unpublished results). Our proteomic and immunofluorescence analyses using these mAbs indicate that pHis has important functions in mammalian biology; 1-pHis appears to play a role in phagocytosis, while 3-pHis signals are elevated during mitosis and detected on specific mitotic structures (i.e. spindle poles, centrosomes and midbodies) [3]. Current efforts are focused on using these mAbs to enrich pHis phosphopeptides from digests of cell lysates followed by mass spectrometry (MS) analysis to define global histidine phosphorylation patterns and specific sites of histidine phosphorylation in proteins. The significant challenges that remain for pHis site detection by MS are discussed below (Phosphohistidine Proteomics).
Attempts to further refine the pHis analog-antibody strategy were recently made by Muimo [19] and Muir [20] who both developed pyrazole-based analogs (pPza and pPye respectively [Fig. 1D]) and used them to raise polyclonal rabbit Abs. It remains to be seen if these “second-generation” analogs will lead to production of pHis Abs with improved attributes and usefulness in detection and enrichment for pHis. We have cloned and expressed recombinant versions of our pHis mAbs and are using them to solve the co-crystal structures of Ab/antigen complexes. The structural and sequence information will inform targeted mutagenesis of key IgG variable domain residues to improve the affinity and sequence-independence attributes of these mAbs. As new pHis substrates and sites are identified and validated, sequence-specific pHis Abs can be raised against peptides containing stable pHis analogs corresponding to these defined sequences, and then used along with genetic analysis (e.g. nonphosphorylatable Asn mutants of pHis sites) to interrogate the functional consequences. Alternatively, non-cleavable 1-pHis or 3-pHis analogs could be incorporated site-specifically into proteins in cells using unnatural amino acid technology [21,22].
Metabolic Enzymes
Nucleoside diphosphate kinases (NDPKs or NMEs) are a highly conserved family of essential enzymes present in bacteria and eukaryotes (NDK is the bacterial homolog) that auto-phosphorylate to form exclusively 1-pHis intermediates that catalyze the transfer of phosphate from nucleoside triphosphates (e.g. ATP) onto nucleoside diphosphates (e.g. GDP)[23] or in some cases other proteins [24–29] (see Mammalian Histidine Kinases). Biossan et al. recently demonstrated that NME1/2 and the mitochondrial specific NME4 provide localized GTP production for dynamin family proteins (e.g. DYN1, DYN2 and OPA1) for membrane remodeling (Fig 2) [23] and this may be true for other GTPases. NME7 localizes to centrosomes (Fig 2) and is part of the gamma-tubulin ring complex (γTuRC), and its kinase activity is required for nucleation of microtubules (Table I) [30]. The observation that NDPK contains a reactive phospho-intermediate was originally made in 1965 [31] and it was subsequently demonstrated to be 1-pHis [32]. Soon after discovery of the 3-pHis isomer in SUCLG1 (Fig. 2), several other 3-pHis intermediates were identified including; phosphoglycerate mutase (PGAM1 [Fig. 2]) in 1970 [33] and ATP citrate lyase (ACLY) in 1971 [34,35]. Subsequently, Dixon’s lab discovered that phospholipase D (PLD [Fig. 2]) and related family members in yeast and E. coli function via a pHis intermediate [36]. Burgos et al. used a crystallographic approach to show that nicotinamide phosphoribosyltransferase (NAMPT), an enzyme that recycles nicotinamide to NAD+, couples ATP hydrolysis with formation of 1-pHis on H247 that dramatically increases catalytic activity [37]. Other enzymes that rely on pHis intermediates may exist that have gone unnoticed due to their instability and difficulty of detection. While these enzymes are selectively phosphorylated at N1 or N3, it is possible that some His residues (not stabilized in a catalytic cleft) can accommodate either isomer. Furthermore, isomerization or phosphotransfer to another acceptor residue, which is known to occur ex vivo/in vitro [38,39], may also occur in vivo after His phosphorylation for certain protein substrates.
Table 1.
Mammalian pHis Related Proteins
| Gene | Uniprot ID | Function | Site | N1 or N3 |
|---|---|---|---|---|
| NME1 | P15531 | NDPK, His kinase | H118 | 1-pHis |
| NME2 | P22392 | NDPK, His kinase | H118 | 1-pHis |
| NME4 | O00746 | NDPK, mitochondrial | H151 | 1-pHis |
| NME7 | Q95YB8 | NDPK, centrosomal | H206 | 1-pHis |
| PGAM1 | P18669 | Glycolysis | H11 | 3-pHis |
| PGAM5 | Q96HS1 | Ser/Thr & His phosphatase | H105 | 3-pHis |
| SUCLG1 | P53597 | Succinyl-CoA ligase | H299 | 3-pHis |
| ACLY | P53396 | ATP-citrate synthase | H760 | 3-pHis |
| PHPT1 | Q9NRX4 | pHis phosphatase | - | - |
| LHPP | Q9H008 | pLys, pHis & pyrophosphate phosphatase | - | - |
| GNB1 | P62873 | GPCR signal transduction | H266 | 3-pHis |
| KCa3.1 | O15554 | Calcium-activated potassium channel | H358 | 3-pHis |
| TRPV5 | Q9NQA5 | Calcium channel, Ca2+ reabsorption | H711 | 3-pHis |
| HIST1H4A | P62805 | Nucleosome, chromatin regulation | H18, | 1/3-pHis |
| PLD | Q13393 | Phospholipid metabolism, signaling | H94 | ? |
| NAMPT | P43490 | NAD+ biosynthesis | H247 | 1-pHis |
For each known pHis related protein discussed in the text of the review; the gene name, Uniprot ID number, a brief functional description, the specific pHis site and the specific pHis isomer (1-pHis [N1] or 3-pHis [N3]) is provided.
What enzymes phosphorylate histidine in proteins?
While there are abundant examples of histidine kinases in prokaryotes, only two, highly related proteins, NME1 and NME2, have been reported to have protein histidine kinase activity in mammalian cells. A histone H4 (Fig. 2) histidine kinase activity was identified in yeast cellular extracts, but the specific protein responsible was never purified [10,11,40]. NME1/2 histidine kinase activity may depend on its usage of the 1-pHis isomer as an intermediate vs. the more thermodynamically stable 3-pHis isomer used by other enzymes. A handful of substrates have been recently identified that are directly and reversibly phosphorylated by NME1/2 and PHPT1 (see Phosphohistidine Phosphphatases) respectively. The best studied of which are the ion channels KCa3.1 [17,28,41,42] and TRPV5 [24]. A G-protein beta subunit, GNB1 [43,44] is also a direct substrate of NME1 kinase activity and PHPT1 phosphatase activity (Fig. 2). Other potential substrates that have not been as well characterized have been reported including; KSR1 [29,45] and annexin-I [46]. We identified over 780 potential pHis substrates using 1- and 3-pHis mAbs to enrich for pHis-containing proteins from HEK293 cell lysates. Although we controlled for non-specific interactions using denaturing lysis conditions and SILAC MS, further validation is required to confirm these substrates and identify specific pHis sites [3]. Given the potentially large size of the pHis proteome, we should be open to the possibility that additional His kinases exist.
What does histidine phosphorylation do?
Phosphorylation of His, other than in enzyme intermediates, could have several functional consequences and some of these are likely to be unique to His, while others are analogous to Ser, Thr and Tyr phosphorylation, and lead to altered protein conformation or the binding of another protein. While no pHis-specific binding domain has yet been identified, such a domain could be used to promote docking of proteins, similar to 14-3-3, WW or SH2 domains for pSer/pThr and pTyr, respectively [47]. Indirect evidence indicates that SH2 from Grb2 can bind pTza [48]; however, there is no evidence that SH2 binds pHis in vivo. Uniquely, His phosphorylation would cause a charge switch from +1 to −1.5 and this large change could in theory be a mechanism to regulate certain electrostatic protein-protein or protein-nucleic acid interactions.
Our SILAC MS data identified 239 out of 786 proteins involved in RNA and DNA/nucleic acid binding/processing [3]. The Skolnik and Hubbard groups [17] recently demonstrated that the ion channel activity of the tetrameric calcium-activated potassium channel KCa3.1 is stimulated by phosphorylation of His358, showing that this prevents chelation of a Cu2+ ion, which stabilizes the KCa3.1 tetramer in the inactive state by coordinating the four His358 residues in the cytoplasmic C-terminal tails, which form a 4-helix bundle. This intimates that cells may use His phosphorylation as a general mechanism to regulate coordination of metal ions by His that are often key to enzyme catalysis by stabilizing the transition state in the active site. Potential proteins of this sort that were enriched by pHis mAbs and are known to use His to coordinate metal ions include; PP1, FHL2, KSR1 [49], DPF2, PKM2, PRUNE and CA2/8 (Table II).
Table II.
Potential Regulation of Metal Ion Coordination by pHis as A General Mechanism
| Gene | Uniprot ID | Function | Site(s) | Ion(s) |
|---|---|---|---|---|
| KCa3.1 | O15554 | Calcium-activated potassium channel | H358 | Cu2+ |
| PPP1CA | P62136 | Ser/Thr Phosphatase, alpha subunit | H66, 125, 173, 248 | Mn2+ |
| PPP1CB | P62140 | Ser/Thr Phosphatase, beta subunit | H65, 124, 172, 247 | Mn2+ |
| FHL2 | Q14192 | Transcriptional regulation | H60, 123, 182, 244 | Zn2+ |
| KSR1 | Q8IVT5 | Ras/Raf/MAPK signaling scaffold | H348, 381 | Zn2+ |
| DPF2 | Q92785 | Transcription factor, hematopoiesis | H227, H232, 303, 353 | Zn2+ |
| PKM/PKM2 | P14618 | Glycolysis, Warburg effect, metabolism | H78 | Cu2+,K+ |
| PRUNE | Q86TP1 | Phosphodiesterase, NME1 inhibitor | H107, 108 | Mn2+,Mg2+ |
| CA2 | P00918 | Carbonic anhydrase | H64, 67, 94, 96, 119 | Zn2+ |
| CA8 | P35219 | Carbonic anhydrase-like | H87, 118, 141 | Zn2+ |
Based on the intriguing finding that phosphorylation of histidine in KCa3.1 regulates its function via coordination of a metal ion (Cu2+)[17], we speculate that other proteins that were enriched by pHis mAbs [3] and have metal ion coordination sites involving histidine(s) may also be functionally regulated by histidine phosphorylation. The gene name, Uniprot ID number, a brief functional description, the potential pHis regulated site(s) and the metal ions coordinated by these sites are provided. The list of similarly regulated proteins could be much longer; however, at this point this list is hypothetical and further, direct evidence is necessary for validation.
Phosphohistidine Phosphatases
Like phosphoester amino acids, pHis is reversible by specific phosphatase enzymes; three such mammalian enzymes are known; PHPT1 [24,25,42,50–53], LHPP [54,55] and the recently identified PGAM5 [18]. Known substrates for PHPT1 include; KCa3.1[42], TRPV5 [24], ACLY [53] and GNB1 (Fig. 2) [25,56]. No specific substrates have yet been identified for LHPP; however, it seems to play an important role in CNS function and disease. A SNP in LHPP has been linked with major depressive disorder (MDD) and alcohol dependence and risky behavior [57–59]. Other PGAM family members could potentially dephosphorylate pHis as well, since they share sequence homology and belong to the “histidine” phosphatase family that function via a pHis intermediate: STS-1 and STS-2 (TULA1/2; UBASH3B/A) [60]. The sole substrate for PGAM5 identified so far is NME2 (also known as NDPK-B), and PGAM5 exhibits selectivity for this NME family member (Fig. 2) [18]. Since we currently lack phosphohistidine phosphatase inhibitors, the best method for preservation of pHis in cell lysates is to use denaturing conditions to prevent all phosphatase activity, including those phosphoester phosphatases with “promiscuous” activity towards phosphoramidate bonds, which include PP1, PP2A and PP2C [61,62]. An open question is whether or not all pHis modifications require a specific phosphatase, or whether pHis can decay spontaneously acting as either an automatic timer or perhaps even a pH sensor? Conversely, some pHis sites may be unusually stable due to local environment.
Phosphohistidine Proteomics
Some time ago it was suggested that pHis represents as much as 6% of phosphorylation sites [63], but in reality we do not have a good estimate of how large the pHis proteome is. Phosphopeptide enrichment and phosphorylation site identification by MS has been essential to the study of pSer, pThr and pTyr. In addition to the common challenges of phosphoproteomics, like low stoichiometry, reduced ionization efficiency and neutral loss of phosphate, identification of pHis phosphopeptides from complex mixtures presents unique challenges. These include; acid-catalyzed hydrolysis under standard liquid chromatography (LC) conditions (pH 3), isomerization of 1-pHis and 3-pHis isomers [39], inter- and intra-peptide phosphotransfer from His to Asp [38] and inability to distinguish between the two pHis isomers. Previous efforts have been made to selectively enrich for pHis phosphopeptides prior to LC-MS using immobilized Cu II ions [64]). An alternative strategy of elevating LC pH to 5 (without enrichment) was employed as a compromise between stabilization of pHis and ionization of peptides. 20 pHis phosphopeptides were identified; however, these have not yet been validated at the protein level [65,66]. Negative electron-transfer dissociation (NETD) developed by the Coon lab, which is run under alkaline conditions, is a promising new technique that has the potential to solve these issues [67–69]. We are currently using pHis mAbs to selectively enrich pHis phosphopeptides from digests of cell lysates and identify phosphorylation of specific His residues to gain a more accurate catalogue of the pHis proteome in mammalian cells.
Conclusions
While pHis currently has well established roles in prokaryotic two-component signal transduction and as a high-energy intermediate in various metabolic pathways, it remains to be seen if pHis plays as significant of a role in regulation of protein function and signal transduction as pSer, pThr and pTyr. There does appear to have been an evolutionary shift in transmembrane signaling and extracellular sensing mechanisms from prokaryotic histidine kinases to eukaryotic receptor tyrosine kinases. Since no pHis-binding domain has been identified, it is unclear if pHis can also serve as a protein-interaction docking site, but we now see that it has roles beyond simply serving as a means for intermolecular phosphate transfer. These roles may be unique to His since it is a common residue in metalloproteins, catalytic triads, and is used as a proton shuttle (e.g. carbonic anhydrase) and pHis could be a common mechanism to regulate these functions by simple interference with protonation of imidazole nitrogens.
Future efforts should focus on; refining tools and methods that have emerged in the last several years including pHis analog-antibody development, developing new reagents including phosphohistidine kinase and phosphatase inhibitors and improving proteomic and genetic methods, including NETD MS and unnatural amino acid technology. Selective pHis phosphopeptide enrichment using pHis-specific mAbs and site identification by MS will be instrumental in providing a more complete catalog of substrates and possibly His kinases and phosphatases. Knowledge of all the players should help give us a better understanding of the game they are playing in the cell.
Highlights.
Histidine phosphorylation (pHis) is reversible by specific kinases and phosphatases
pHis is heat and acid labile, but stable under alkaline conditions
Stable pHis analogs have enabled development of pHis-specific antibodies
pHis antibodies and refined methods have identified new functions and substrates
Enrichment of pHis phosphopeptides for proteomics is the next critical step
Acknowledgments
The work from our laboratory described here was supported by NIH grants CA08100, CA082683, and CA194584. T.H. is an American Cancer Society Professor, and holds the Renato Dulbecco Chair in Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Boyer PD, DeLuca M, Ebner KE, Hultquist DE, Peter JE. Identification of phosphohistidine in digests from a probable intermediate of oxidative phosphorylation. J Biol Chem. 1962;237:3306–3308. [PubMed] [Google Scholar]
- 2.Attwood PV, Piggott MJ, Zu XL, Besant PG. Focus on phosphohistidine. Amino Acids. 2007;32:145–156. doi: 10.1007/s00726-006-0443-6. [DOI] [PubMed] [Google Scholar]
- 3. Fuhs SR, Meisenhelder J, Aslanian A, Ma L, Zagorska A, Stankova M, Binnie A, Al-Obeidi F, Mauger J, Lemke G, et al. Monoclonal 1- and 3-phosphohistidine antibodies: new tools to study histidine phosphorylation. Cell. 2015;162:198–210. doi: 10.1016/j.cell.2015.05.046. This study describes the generation and use of monoclonal antibodies that specifically recongnize either the 1-pHis or 3-pHis isomer. Their characterization and specificity is demonstrated and methods for their use in several immunological assays including immunoblotting, immunofluorescence and immunoaffity purification of pHis proteins and analysis by LC-MS/MS.
- 4.Hunter T, Sefton BM. Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc Natl Acad Sci U S A. 1980;77:1311–1315. doi: 10.1073/pnas.77.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matthews HR, Chan K. Protein histidine kinase. Methods Mol Biol. 2001;124:171–182. doi: 10.1385/1-59259-059-4:171. [DOI] [PubMed] [Google Scholar]
- 6.Frackelton AR, Ross AH, Eisen HN. Characterization and use of monoclonal antibodies for isolation of phosphotyrosyl proteins from retrovirus-transformed cells and growth factor-stimulated cells. Mol Cell Biol. 1983;3:1343–1352. doi: 10.1128/mcb.3.8.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schenkels C, Erni B, Reymond J-L. Phosphofurylalanine, a stable analog of phosphohistidine. Bioorg Med Chem Lett. 1999:1443–1446. doi: 10.1016/s0960-894x(99)00209-7. [DOI] [PubMed] [Google Scholar]
- 8.Kee JM, Villani B, Carpenter LR, Muir TW. Development of stable phosphohistidine analogues. J Am Chem Soc. 2010;132:14327–14329. doi: 10.1021/ja104393t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kee JM, Muir TW. Chasing phosphohistidine, an elusive sibling in the phosphoamino acid family. ACS Chem Biol. 2012;7:44–51. doi: 10.1021/cb200445w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith DL, Bruegger BB, Halpern RM, Smith RA. New histone kinases in nuclei of rat tissues. Nature. 1973;246:103–104. doi: 10.1038/246103a0. [DOI] [PubMed] [Google Scholar]
- 11.Besant PG, Attwood PV. Histone H4 histidine phosphorylation: kinases, phosphatases, liver regeneration and cancer. Biochem Soc Trans. 2012;40:290–293. doi: 10.1042/BST20110605. [DOI] [PubMed] [Google Scholar]
- 12.Huang JM, Wei YF, Kim YH, Osterberg L, Matthews HR. Purification of a protein histidine kinase from the yeast Saccharomyces cerevisiae. The first member of this class of protein kinases. J Biol Chem. 1991;266:9023–9031. [PubMed] [Google Scholar]
- 13.McAllister TE, Nixa MG, Webb ME. Fmoc-chemistry of a stable phosphohistidine analogue. Chem Commun. 2011;47:1297–1299. doi: 10.1039/c0cc04238b. [DOI] [PubMed] [Google Scholar]
- 14.McAllister TE, Hollins JJ, Webb ME. Prospects for stable analogues of phosphohistidine. Biochem Soc Trans. 2013;41:1072–1077. doi: 10.1042/BST20130071. [DOI] [PubMed] [Google Scholar]
- 15.McAllister TE, Webb ME. Triazole phosphohistidine analogues compatible with the Fmoc-strategy. Org Biomol Chem. 2012;10:4043–4049. doi: 10.1039/c2ob25517k. [DOI] [PubMed] [Google Scholar]
- 16.Kee JM, Oslund RC, Perlman DH, Muir TW. A pan-specific antibody for direct detection of protein histidine phosphorylation. Nat Chem Biol. 2013;9:416–421. doi: 10.1038/nchembio.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Srivastava S, Panda S, Li Z, Fuhs SR, Hunter T, Thiele DJ, Hubbard SR, Skolnik EY. Histidine phosphorylation relieves copper inhibition in the mammalian potassium channel KCa3.1. eLife. 2016;5 doi: 10.7554/eLife.16093. These authors previously reported that KCa3.1 is activated by histidine phosphorylation, but the mechanism was unknown. Here they describe that channel activation occurs because phosphorylation prevents copper from coordinating with H358, which locks the channel in the closed state. Preventing metal coordination is a novel mechanism for regulation of protein function by histidine phosphorylation and could be a more general mechanism for regulation of proteins that require metal ion coordination for their function.
- 18. Panda S, Srivastava S, Li Z, Vaeth M, Fuhs SR, Hunter T, Skolnik EY. Identification of PGAM5 as a mammalian protein histidine phosphatase that plays a central role to negatively regulate CD4+ T cells. Mol Cell. 2016;63:457–469. doi: 10.1016/j.molcel.2016.06.021. This paper reveals that PGAM5 has phosphohistidine phosphatase activity that is specific for autophosphorylated NDPK-B/NME2. It demonstrates the utility of recently developed 1-pHis and 3-pHis mAbs for study of the phosphate transfer from NDPK-B H118 (1-pHis) to PGAM-5 H105 (3-pHis) and shows that PGAM5 decreases KCa3.1 activity and cytokine production in CD4+ T cells by dephosphorylating H118 on NDPK-B.
- 19. Lilley M, Mambwe B, Thompson MJ, Jackson RF, Muimo R. 4-Phosphopyrazol-2-yl alanine: a non-hydrolysable analogue of phosphohistidine. Chem Commun. 2015;51:7305–7308. doi: 10.1039/c5cc01811k. This communication describes the synthesis of a "second-generation" stable analog of pHis, 4-phosphopyrazol-2-yl alanine (pPza). Polyclonal antibodies were raised with it and no cross-reactivity with phosphoester amino acids was observed. See also Kee et al., 2015.
- 20. Kee JM, Oslund RC, Couvillon AD, Muir TW. A second-generation phosphohistidine analog for production of phosphohistidine antibodies. Org Lett. 2015;17:187–189. doi: 10.1021/ol503320p. These authors have previously reported their success in developing stable pHis analogs (pTza) and their use as immunogens for raising pHis antibodies. Since their pTza derived antibodies had cross-reactivity with pTyr, they developed a "second-generation", pyrazole-based pHis analog (pPye) to develop polyclonal "pan-pHis" antibodies. See also Muimo et al., 2015.
- 21.Liu WR, Wang YS, Wan W. Synthesis of proteins with defined posttranslational modifications using the genetic noncanonical amino acid incorporation approach. Mol Biosyst. 2011;7:38–47. doi: 10.1039/c0mb00216j. [DOI] [PubMed] [Google Scholar]
- 22.Parrish AR, Wang L. Genetic incorporation of unnatural amino acids into proteins. Comprehensive Natural Products II. 2010:587–617. [Google Scholar]
- 23. Boissan M, Montagnac G, Shen Q, Griparic L, Guitton J, Romao M, Sauvonnet N, Lagache T, Lascu I, Raposo G, et al. Membrane trafficking. Nucleoside diphosphate kinases fuel dynamin superfamily proteins with GTP for membrane remodeling. Science. 2014;344:1510–1515. doi: 10.1126/science.1253768. This paper reports that NDPK/NME proteins are key to the function of dynamin family proteins through their ability to locally replenish GTP from GDP and ATP. They demonstrate that NME1/2 interact with and provide GTP to dynamin, and the mitochondrial NME4 does the same for the dynamin family protein OPA1.
- 24. Cai X, Srivastava S, Surindran S, Li Z, Skolnik EY. Regulation of the epithelial Ca2+ channel TRPV5 by reversible histidine phosphorylation mediated by NDPK-B and PHPT1. Mol Biol Cell. 2014;25:1244–1250. doi: 10.1091/mbc.E13-04-0180. In this article, the authors identify a second ion channel, TRPV5, that is regulated by phosphorylation of a specific histidine residue by NDPK-B/NME2. They also show that the phosphohistidine phosphatase PHPT1 inhbits the channel by dephosphorlating the channel.
- 25.Maurer A, Wieland T, Meissl F, Niroomand F, Mehringer R, Krieglstein J, Klumpp S. The β-subunit of G proteins is a substrate of protein histidine phosphatase. Biochem Biophys Res Commun. 2005;334:1115–1120. doi: 10.1016/j.bbrc.2005.06.200. [DOI] [PubMed] [Google Scholar]
- 26.Lutz S, Hippe HJ, Niroomand F, Wieland T. Nucleoside diphosphate kinase–mediated activation of heterotrimeric G proteins. Meth Enzymol. 2004;390:403–418. doi: 10.1016/S0076-6879(04)90025-0. [DOI] [PubMed] [Google Scholar]
- 27.Wagner PD, Vu ND. Phosphorylation of ATP-citrate lyase by nucleoside diphosphate kinase. J Biol Chem. 1995;270:21758–21764. doi: 10.1074/jbc.270.37.21758. [DOI] [PubMed] [Google Scholar]
- 28.Srivastava S, Li Z, Ko K, Choudhury P, Albaqumi M, Johnson AK, Yan Y, Backer JM, Unutmaz D, Coetzee WA, et al. Histidine phosphorylation of the potassium channel KCa3.1 by nucleoside diphosphate kinase B is required for activation of KCa3.1 and CD4 T cells. Mol Cell. 2006;24:665–675. doi: 10.1016/j.molcel.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 29.Hartsough MT, Morrison DK, Salerno M, Palmieri D, Ouatas T, Mair M, Patrick J, Steeg PS. Nm23-H1 metastasis suppressor phosphorylation of kinase suppressor of Ras via a histidine protein kinase pathway. J Biol Chem. 2002;277:32389–32399. doi: 10.1074/jbc.M203115200. [DOI] [PubMed] [Google Scholar]
- 30.Liu P, Choi YK, Qi RZ. NME7 is a functional component of the γ-tubulin ring complex. Mol Biol Cell. 2014;25:2017–2025. doi: 10.1091/mbc.E13-06-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mourad N, Parks RE., Jr Ndp Kinase: Demonstration of phosphorylated enzyme as the reactive intermediate. Biochem Biophys Res Commun. 1965;19:312–316. doi: 10.1016/0006-291x(65)90460-2. [DOI] [PubMed] [Google Scholar]
- 32.Walinder O. Evidence of the presence of 1-phosphohistidine as the main phosphohistidine as the main phosphorylated component at the active site of bovine liver nucleoside diphosphate kinase. Acta Chem Scand. 1969;23:339–341. doi: 10.3891/acta.chem.scand.23-0339. [DOI] [PubMed] [Google Scholar]
- 33.Rose ZB. Evidence for a phosphohistidine protein intermediate in the phosphoglycerate mutase reaction. Arch Biochem Biophys. 1970;140:508–513. doi: 10.1016/0003-9861(70)90095-0. [DOI] [PubMed] [Google Scholar]
- 34.Fan F, Williams HJ, Boyer JG, Graham TL, Zhao H, Lehr R, Qi H, Schwartz B, Raushel FM, Meek TD. On the catalytic mechanism of human ATP citrate lyase. Biochem. 2012;51:5198–5211. doi: 10.1021/bi300611s. [DOI] [PubMed] [Google Scholar]
- 35.Mardh S, Ljungstrom O, Hogstedt S, Zetterqvist O. Studies on a rat-liver cell-sap protein yielding 3-[32P]-phosphohistidine after incubation with [32P]ATP and alkaline hydrolysis. Identification of the protein as ATP citrate lyase. Biochim Biophys Acta. 1971;251:419–426. doi: 10.1016/0005-2795(71)90131-0. [DOI] [PubMed] [Google Scholar]
- 36.Gottlin EB, Rudolph AE, Zhao Y, Matthews HR, Dixon JE. Catalytic mechanism of the phospholipase D superfamily proceeds via a covalent phosphohistidine intermediate. Proc Natl Acad Sci U S A. 1998;95:9202–9207. doi: 10.1073/pnas.95.16.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burgos ES, Ho MC, Almo SC, Schramm VL. A phosphoenzyme mimic, overlapping catalytic sites and reaction coordinate motion for human NAMPT. Proc Natl Acad Sci U S A. 2009;106:13748–13753. doi: 10.1073/pnas.0903898106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez-Sanchez MB, Lanucara F, Hardman GE, Eyers CE. Gas-phase intermolecular phosphate transfer within a phosphohistidine phosphopeptide dimer. Int J Mass Spectrom. 2014;367:28–34. doi: 10.1016/j.ijms.2014.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalez-Sanchez MB, Lanucara F, Helm M, Eyers CE. Attempting to rewrite history: challenges with the analysis of histidine-phosphorylated peptides. Biochem Soc Trans. 2013;41:1089–1095. doi: 10.1042/BST20130072. [DOI] [PubMed] [Google Scholar]
- 40.Tan E, Besant PG, Zu XL, Turck CW, Bogoyevitch MA, Lim SG, Attwood PV, Yeoh GC. Histone H4 histidine kinase displays the expression pattern of a liver oncodevelopmental marker. Carcinogenesis. 2004;25:2083–2088. doi: 10.1093/carcin/bgh222. [DOI] [PubMed] [Google Scholar]
- 41.Di L, Srivastava S, Zhdanova O, Sun Y, Li Z, Skolnik EY. Nucleoside diphosphate kinase B knock-out mice have impaired activation of the K+ channel KCa3.1, resulting in defective T cell activation. J Biol Chem. 2010;285:38765–38771. doi: 10.1074/jbc.M110.168070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Srivastava S, Zhdanova O, Di L, Li Z, Albaqumi M, Wulff H, Skolnik EY. Protein histidine phosphatase 1 negatively regulates CD4 T cells by inhibiting the K+ channel KCa3.1. Proc Natl Acad Sci U S A. 2008;105:14442–14446. doi: 10.1073/pnas.0803678105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hippe HJ, Lutz S, Cuello F, Knorr K, Vogt A, Jakobs KH, Wieland T, Niroomand F. Activation of heterotrimeric G proteins by a high energy phosphate transfer via nucleoside diphosphate kinase (NDPK) B and Gβ subunits. J Biol Chem. 2003;278:7227–7233. doi: 10.1074/jbc.M210305200. [DOI] [PubMed] [Google Scholar]
- 44.Cuello F, Schulze RA, Heemeyer F, Meyer HE, Lutz S, Jakobs KH, Niroomand F, Wieland T. Activation of heterotrimeric G proteins by a high energy phosphate transfer via nucleoside diphosphate kinase (NDPK) B and Gβ subunits. Complex formation of NDPK B with Gβϒdimers and phosphorylation of His-266 in Gβ. J Biol Chem. 2003;278:7220–7226. doi: 10.1074/jbc.M210304200. [DOI] [PubMed] [Google Scholar]
- 45.Masoudi N, Fancsalszky L, Pourkarimi E, Vellai T, Alexa A, Remenyi A, Gartner A, Mehta A, Takacs-Vellai K. The NM23-H1/H2 homolog NDK-1 is required for full activation of Ras signaling in C. elegans. Development. 2013;140:3486–3495. doi: 10.1242/dev.094011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muimo R, Hornickova Z, Riemen CE, Gerke V, Matthews H, Mehta A. Histidine phosphorylation of annexin I in airway epithelia. J Biol Chem. 2000;275:36632–36636. doi: 10.1074/jbc.M000829200. [DOI] [PubMed] [Google Scholar]
- 47.Deribe YL, Pawson T, Dikic I. Post-translational modifications in signal integration. Nat Struct Mol Biol. 2010;17:666–672. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- 48.McAllister TE, Horner KA, Webb ME. Evaluation of the interaction between phosphohistidine analogues and phosphotyrosine binding domains. Chembiochem. 2014;15:1088–1091. doi: 10.1002/cbic.201402090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoder JH, Chong H, Guan KL, Han M. Modulation of KSR activity in Caenorhabditis elegans by Zn ions, PAR-1 kinase and PP2A phosphatase. EMBO J. 2004;23:111–119. doi: 10.1038/sj.emboj.7600025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu A, Hao J, Zhang Z, Tian T, Jiang S, Hao J, Liu C, Huang L, Xiao X, He D. 14-kDa phosphohistidine phosphatase and its role in human lung cancer cell migration and invasion. Lung Cancer. 2010;67:48–56. doi: 10.1016/j.lungcan.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 51.Busam RD, Thorsell AG, Flores A, Hammarstrom M, Persson C, Hallberg BM. First structure of a eukaryotic phosphohistidine phosphatase. J Biol Chem. 2006;281:33830–33834. doi: 10.1074/jbc.C600231200. [DOI] [PubMed] [Google Scholar]
- 52.Ma R, Kanders E, Sundh UB, Geng M, Ek P, Zetterqvist O, Li JP. Mutational study of human phosphohistidine phosphatase: effect on enzymatic activity. Biochem Biophys Res Commun. 2005;337:887–891. doi: 10.1016/j.bbrc.2005.09.134. [DOI] [PubMed] [Google Scholar]
- 53.Klumpp S, Bechmann G, Mäurer A, Selke D, Krieglstein J. ATP-citrate lyase as a substrate of protein histidine phosphatase in vertebrates. Biochem Biophys Res Commun. 2003;306:110–115. doi: 10.1016/s0006-291x(03)00920-3. [DOI] [PubMed] [Google Scholar]
- 54.Koike E, Toda S, Yokoi F, Izuhara K, Koike N, Itoh K, Miyazaki K, Sugihara H. Expression of new human inorganic pyrophosphatase in thyroid diseases: its intimate association with hyperthyroidism. Biochem Biophys Res Commun. 2006;341:691–696. doi: 10.1016/j.bbrc.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 55.Yokoi F. Molecular cloning of a cDNA for the human phospholysine phosphohistidine inorganic pyrophosphate phosphatase. J Biochem. 2003;133:607–614. doi: 10.1093/jb/mvg078. [DOI] [PubMed] [Google Scholar]
- 56.Klumpp S, Krieglstein J. Reversible phosphorylation of histidine residues in proteins from vertebrates. Sci Signal. 2009;2:pe13. doi: 10.1126/scisignal.261pe13. [DOI] [PubMed] [Google Scholar]
- 57.Neff CD, Abkevich V, Packer JC, Chen Y, Potter J, Riley R, Davenport C, DeGrado Warren J, Jammulapati S, Bhathena A, et al. Evidence for HTR1A and LHPP as interacting genetic risk factors in major depression. Mol Psychiatry. 2009;14:621–630. doi: 10.1038/mp.2008.8. [DOI] [PubMed] [Google Scholar]
- 58. consortium C. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523:588–591. doi: 10.1038/nature14659. This letter descibes the identification of the phosphohistidine phosphatase LHPP and SIRT1 as two loci contributing to risk of major depressive disorder (MDD) using whole-genome sequencing. Previously, no well defined genetic loci have been identified. This could imply that histidine phosphorylation plays a role in normal neurological function and in psychiatric disease.
- 59.Polimanti R, Wang Q, Meda SA, Patel KT, Pearlson GD, Zhao H, Farrer LA, Kranzler HR, Gelernter J. The interplay between risky sexual behaviors and alcohol dependence: genome-wide association and neuroimaging support for LHPP as a risk gene. Neuropsychopharmacology. 2016 doi: 10.1038/npp.2016.153. Epub Aug 17, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Back SH, Adapala NS, Barbe MF, Carpino NC, Tsygankov AY, Sanjay A. TULA-2, a novel histidine phosphatase, regulates bone remodeling by modulating osteoclast function. Cell Mol Life Sci. 2013;70:1269–1284. doi: 10.1007/s00018-012-1203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim Y, Huang J, Cohen P, Matthews HR. Protein phosphatases 1, 2A, and 2C are protein histidine phosphatases. J Biol Chem. 1993;268:18513–18518. [PubMed] [Google Scholar]
- 62.Attwood PV. P-N bond protein phosphatases. Biochim Biophys Acta. 2013;1834:470–478. doi: 10.1016/j.bbapap.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 63.Matthews H. Protein kinases and phosphatases that act on histidine, lysine, or arginine residues in eukaryotic proteins a possible regulator of the MAPK cascade. Pharmac Ther. 1995;67:232–350. doi: 10.1016/0163-7258(95)00020-8. [DOI] [PubMed] [Google Scholar]
- 64.Napper S, Kindrachuk J, Olson DJH, Ambrose SJ, Dereniwsky C, Ross ARS. Selective extraction and characterization of a histidine-phosphorylated peptide using immobilized copper(ii) ion affinity chromatography and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Chem. 2003;5:1741–1747. doi: 10.1021/ac026340f. [DOI] [PubMed] [Google Scholar]
- 65.Lapek JD, Jr, Tombline G, Kellersberger KA, Friedman MR, Friedman AE. Evidence of histidine and aspartic acid phosphorylation in human prostate cancer cells. Naunyn Schmiedebergs Arch Pharmacol. 2015;388:161–173. doi: 10.1007/s00210-014-1063-4. [DOI] [PubMed] [Google Scholar]
- 66.Lapek JD, Tombline G, Friedman AE. Mass Spectrometry detection of Histidine Phosphorylation on NM23-H1. Proteome Res. 2010;10:751–755. doi: 10.1021/pr100905m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Riley NM, Rush MJ, Rose CM, Richards AL, Kwiecien NW, Bailey DJ, Hebert AS, Westphall MS, Coon JJ. The negative mode proteome with activated ion negative electron transfer dissociation (AI-NETD) Mol Cell Proteomics. 2015;14:2644–2660. doi: 10.1074/mcp.M115.049726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rumachik NG, McAlister GC, Russell JD, Bailey DJ, Wenger CD, Coon JJ. Characterizing peptide neutral losses induced by negative electron-transfer dissociation (NETD) J Am Soc Mass Spectrom. 2012;23:718–727. doi: 10.1007/s13361-011-0331-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McAlister GC, Russell JD, Rumachik NG, Hebert AS, Syka JE, Geer LY, Westphall MS, Pagliarini DJ, Coon JJ. Analysis of the acidic proteome with negative electron-transfer dissociation mass spectrometry. Anal Chem. 2012;84:2875–2882. doi: 10.1021/ac203430u. [DOI] [PMC free article] [PubMed] [Google Scholar]