Abstract
The Golgi is generally recognized for its central role in the secretory pathway to orchestrate protein post-translational modification and trafficking of proteins and lipids to their final destination. Despite the common view of the Golgi as an inert sorting organelle, emerging data demonstrate that important signaling events occur at the Golgi, including those that regulate the trafficking function of the Golgi. The phosphatidylinositol-4-phosphate/GOLPH3/MYO18A/F-actin complex serves as a hub for signals that regulate Golgi trafficking function. Furthermore, the Golgi is increasingly appreciated for its important role in cell growth and in driving oncogenic transformation, as illuminated by the discovery that GOLPH3 and MYO18A are cancer drivers.
Introduction
Recent studies paint a multidimensional picture of the Golgi, revealing dynamic regulatory signaling pathways that act at, on, or emanate from the Golgi. This review highlights a few examples of signaling pathways that function at the Golgi, some of which regulate Golgi trafficking function. We also examine some recent data that implicate the Golgi as a regulator of cell proliferation capable of driving oncogenic transformation.
SREBP, an example of regulation that occurs at the Golgi
Several years ago, Brown and Goldstein elucidated the importance of endoplasmic reticulum (ER)-to-Golgi transport in regulating the activity of sterol regulatory element-binding proteins (SREBPs) [1]. They showed that low cholesterol levels act through a protein called SCAP, resulting in trafficking of SREBP from the ER to the Golgi. Once at the Golgi, two site-specific proteases cleave SREBP, liberating a transcription factor which activates cholesterol biosynthetic genes [2-6]. Thus, the Golgi is critically important for SREBP pathway activation.
Recent studies have identified additional layers of regulation in the SREBP pathway. High glucose levels promote N-glycosylation of SCAP, resulting in trafficking and activation of SREBP, thus linking glucose levels to lipogenesis [7]. In addition, oncogenic PI-3-kinase and K-RAS, through a poorly understood mechanism involving mTORC1, drive SREBP activation to promote lipid synthesis [8].
SREBPs provide an example of proteins whose function depends on regulated trafficking from the ER to the Golgi, but they do not directly affect ER or Golgi function.
Growth factor signaling controls Golgi secretory function
Similar to the SREBPs, SAC1 is a protein whose trafficking from the ER to the Golgi is regulated. However, in contrast to the SREBPs, regulation of SAC1 ER-Golgi localization has important consequences on Golgi trafficking [9]. As discussed further below, phosphatidylinositol-4-phosphate (PtdIns(4)P) plays an important role at the Golgi in enabling Golgi-to-plasma membrane (PM) trafficking. SAC1 dephosphorylates PtdIns(4)P to produce phosphatidylinositol. In growth factor-deprived cells, SAC1 oligomerizes and traffics from the ER to the Golgi, depleting Golgi PtdIns(4)P, thus interfering with secretory trafficking [9]. Since SAC1 is exported from the ER into COPII vesicles at similar levels in starved and non-starved cells, growth factor-dependent ERGolgi trafficking of SAC1 is likely regulated at the level of Golgi retrieval [10]. Stimulation with growth factors such as FGF and PDGF activates the p38/MAPK pathway, triggering dissociation of SAC1 oligomers and COPI-mediated retrograde trafficking of SAC1 to the ER. This leads to a rise in PtdIns(4)P at the Golgi, which drives increased trafficking from the Golgi to the PM.
SAC1 trafficking between the ER and Golgi impacts secretory function by regulating PtdIns(4)P levels at the Golgi. The importance of PtdIns(4)P in Golgi function is mediated by PtdIns(4)P effectors, which bind PtdIns(4)P and serve essential roles in Golgi trafficking.
PtdIns(4)P and its effectors at the Golgi
Known to play a critical role at the Golgi, PtdIns(4)P is highly enriched at the cytosolic face of the trans-Golgi, where it recruits cytosolic proteins that bind to PtdIns(4)P [11,12]. From yeast to humans, PtdIns(4)P is required for Golgi-to-PM trafficking [13-16]. Several proteins have been identified that bind to PtdIns(4)P and function at the Golgi. Here we discuss a few of these proteins, and refer to other recent reviews for a more comprehensive list of PtdIns(4)P effectors [17,18].
Many PtdIns(4)P binding proteins function in non-vesicular lipid transport [19,20]. While some of these bind PtdIns(4)P at the PM (beyond the scope of this review), several bind PtdIns(4)P at the Golgi. Both ceramide transfer protein (CERT) and oxysterol binding protein (OSBP) contain a pleckstrin homology (PH) domain that binds to PtdIns(4)P, contributing to their trans-Golgi localization [20,21]. They also contain an FFAT (Phe-Phe within an Acidic Tract) motif that binds to VAP proteins at the ER [22-24]. CERT transports ceramide from the ER to the trans-Golgi, where ceramide serves as a precursor for sphingomyelin synthesis [20,23]. Similarly, OSBP transports cholesterol from the ER to the trans-Golgi [25,26].
FAPP2 is another non-vesicular lipid transporter. Its N-terminal PH domain binds specifically to both PtdIns(4)P and the Golgi protein ARF, resulting in tight localization to the trans-Golgi [12,21,27]. FAPP2 transports glucosylceramide from the cis-Golgi to the trans-Golgi to promote globotriaosylceramide synthesis [28-30].
It is striking that multiple PtdIns(4)P-binding proteins function as non-vesicular lipid transporters. Given the highly asymmetric distribution of lipids at the cis- versus the trans-Golgi, these PtdIns(4)P-binding lipid transporters appear important for creating the distinct lipid phase that characterizes the trans-Golgi [31,32].
GOLPH3 is a PtdIns(4)P effector that is critical for Golgi-to-PM trafficking
Found by proteomic studies of the Golgi [33,34], GOLPH3 (earlier referred to as GMx33 or GPP34) was identified as a PtdIns(4)P effector through unbiased screening for phosphoinositide-binding proteins [35]. GOLPH3 binds to PtdIns(4)P via its unique GPP34 domain, driving localization to the trans-Golgi that is conserved from yeast (where the ortholog is VPS74) to humans [35,36]. GOLPH3 tightly interacts with an unconventional myosin, MYO18A, linking the Golgi to the F-actin cytoskeleton [35,37,38]. Changes in the morphology of the Golgi upon perturbation of any component of the PtdIns(4)P/GOLPH3/MYO18A/F-actin complex indicate that this linkage serves to apply a stretching force to the Golgi, rendering the trans cisternae flat and stretching the Golgi ribbon around the nucleus, as observed by electron and fluorescence microscopy [35,37]. Measurement of PtdIns(4)P-positive, cargo-positive vesicles exiting the Golgi demonstrates that the GOLPH3 complex is critical for vesicle exit from the Golgi for forward trafficking to the PM [35]. Indeed, measurement of overall or cargo-specific secretory trafficking indicates that the GOLPH3 complex is required for secretion [35,37,39]. The evidence suggests that the tensile force applied to the Golgi membrane by the GOLPH3 complex participates in the process of vesicle budding for forward trafficking [35,37,39,40].
The consequences of reducing Golgi PtdIns(4)P by SAC1, namely, compaction of the Golgi ribbon and inhibition of trafficking, are completely recapitulated by knockdown of GOLPH3 or MYO18A or depolymerization of F-actin [9,35,41]. This implies that the PtdIns(4)P effector, GOLPH3, and its downstream effectors, MYO18A and F-actin, are the major determinants of the Golgi response to PtdIns(4)P.
Other examples of regulation of the Golgi via GOLPH3
Surprisingly little is known about regulation of Golgi trafficking. Growth factor regulation of the Golgi by SAC1 is the first such example. Considering the importance of the GOLPH3 complex in Golgi function, one might predict that the GOLPH3 complex would be a hub for regulation of the Golgi. Interestingly, GOLPH3 is a phosphoprotein with multiple alternative phosphorylation sites [34,42,43]. To begin to identify regulation of the GOLPH3 complex, we mapped phosphorylation sites in GOLPH3 and discovered phosphorylation of Thr 143 [43]. The realization that this site is phosphorylated by the DNA-damage-activated kinase, DNA-PK, led to the discovery of the Golgi response to DNA damage, which results in Golgi fragmentation and impaired trafficking. The Golgi DNA damage response is a consequence of phosphorylation of GOLPH3 on Thr 143 by DNA-PK, resulting in increased interaction of GOLPH3 with MYO18A [43,44].
The existence of GOLPH3L, a paralog of GOLPH3 and an endogenous dominant-negative inhibitor of the GOLPH3 complex, further supports the notion that the GOLPH3 complex represents a central point for regulating Golgi function [37,40]. Only found in vertebrates, GOLPH3L is restricted to highly secretory tissues, such as salivary gland, small intestine, and skin. Like GOLPH3, GOLPH3L binds PtdIns(4)P and localizes to the Golgi. However, GOLPH3L inhibits the GOLPH3 complex because it cannot bind MYO18A [37]. Indeed, overexpression and knockdown of GOLPH3L show opposing effects on Golgi morphology compared to similar perturbation of GOLPH3. The evidence indicates that GOLPH3L acts to suppress excessive secretion in highly secretory cells in order to achieve optimal rates of trafficking.
RAS signaling at the Golgi
RAS signaling is widely recognized for its contribution to cell growth, and as a common target for driver mutations in cancer [45-48]. RAS family small GTPases (N-RAS, H-RAS, K-RAS4A, and K-RAS4B) are activated downstream of receptor tyrosine kinases (RTKs) [49,50]. Activated RAS then signals to activate both the RAF/MEK/ERK and PI-3-kinase/PtdIns(3,4,5)P3/AKT signaling pathways, which serve to drive cell proliferation, motility, and survival [46,51-53]. While it is clear that RAS is predominantly at the PM, a significant proportion of H-RAS and N-RAS reside at the Golgi [54-56].
A number of groups have produced evidence that Golgi-localized Ras might actively signal. For example, RAS activity reporters detect activity at the Golgi [57-60]. RasGRP1, a RAS GTP exchange factor, is recruited to the Golgi upon activation of phospholipase C-γ1 [60-62]. Furthermore, RAS that is tethered to the Golgi is capable of driving signaling and oncogenic transformation [59,63,64]. Recent studies have begun to characterize detailed differences between RAS signaling at the PM versus at the Golgi [59,64].
While it is increasingly apparent that Golgi-localized RAS has unique signaling functions, there is no evidence that RAS plays a role in regulating Golgi function. It appears that RAS, like SREBP, uses the Golgi as a signaling platform but does not significantly alter the trafficking activity of the Golgi.
Trafficking proteins that function as oncogenes: GOLPH3 and MYO18A
Surprisingly, both GOLPH3 and MYO18A have been identified as oncogenic drivers of human cancers. GOLPH3 is the first oncogene discovered to regulate secretory function at the Golgi. Unbiased, genome-wide copy number analysis of human cancer identified frequent amplification of GOLPH3 in several solid tumor types, including lung, ovarian, breast, pancreatic, prostate, melanoma and colon carcinoma [65]. GOLPH3 causes transformation in classical co-transformation cell culture assays and promotes tumor growth in xenograft mouse models, validating GOLPH3 as a true oncogene. More than thirty additional studies confirm GOLPH3's ability to drive transformation, detect overexpression of GOLPH3 in a wide variety of cancers, and correlate high levels of expression with poor patient outcomes (recently reviewed in [44]).
More recently, computational analysis of The Cancer Genome Atlas (TCGA) data identified 17 drivers of breast cancer in humans [66], many of which are expected, such as ERBB2, MYC, and CCND1. Interestingly, MYO18A is one of the genes identified as a driver of breast cancer. TCGA further reports amplification of the MYO18A gene at a high frequency in several cancers, including neuroendocrine prostate, breast, pancreatic, uterine, and bladder cancer [67-73].
The GOLPH3 complex drives cancer through multiple mechanisms
Altogether, the identification of GOLPH3 and MYO18A as drivers of cancer suggests that the GOLPH3 complex that functions in Golgi-to-PM trafficking is capable of driving cancer. A diverse array of cargoes depend on Golgi-to-PM trafficking for their function. Thus, one might predict that perturbations in Golgi function would have pleiotropic effects on cell function. Indeed, a few mechanisms have already been identified that explain how the GOLPH3 complex can drive oncogenic transformation.
One way that GOLPH3 likely drives cancer is through its ability to modulate signaling downstream of mTORC1 and mTORC2. GOLPH3 knockdown results in impaired phosphorylation of p70S6K (at Thr 389) and AKT (at Ser 473) [65]. Likewise, overexpression of GOLPH3 stimulates enhanced phosphorylation of both of these proteins at these sites. Consistent with increased activation of AKT, overexpression of GOLPH3 also leads to enhanced phosphorylation of the AKT substrate FOXO1, thus downregulating its transcriptional activity and promoting cell proliferation in breast cancer [74]. While these consequences of overexpression of GOLPH3 are well documented, the mechanism is in need of further investigation. Nevertheless, it is clear that enhanced signaling through AKT and p70S6K is capable of driving cancer [75,76].
A second way that the GOLPH3 complex drives cancer involves its role in the Golgi DNA damage response. As described previously, the GOLPH3 complex plays an important role in the cellular response to DNA damage [43]. The interaction between GOLPH3 and MYO18A is enhanced in response to DNA damage, a consequence of DNA-PK phosphorylation of GOLPH3 on Thr 143. Furthermore, this pathway is required for normal cellular survival following DNA damage. Interference with the GOLPH3 complex results in increased apoptotic cell death following DNA damage. Likewise, overexpression of GOLPH3 promotes survival following DNA damage, which depends on GOLPH3 localization to the Golgi and its phosphorylation by DNA-PK. Survival in the face of DNA damage is important in cancer progression [77,78]. Additionally, the data demonstrate that enhanced GOLPH3 complex function at the Golgi prevents cancer cell killing by DNA damaging chemotherapeutic agents [43,44].
A third role for the GOLPH3 complex in cancer involves driving cell migration. Several studies have demonstrated that an increase in Golgi PtdIns(4)P levels or overexpression of GOLPH3 each drives increased cell migration, a process which underlies some of the deadliest features of cancer, namely invasion and metastasis [79-81]. Recent work by Xing et al. sheds light on the mechanism, demonstrating that the PtdIns(4)P/GOLPH3/MYO18A/F-actin complex drives cell migration by linking the Golgi to the actin cytoskeleton to promote Golgi reorientation toward the wound edge and Golgi-to-PM trafficking toward the leading edge [81].
Recently, another protein, PITPNC1, was shown to drive oncogenic transformation via GOLPH3 [82]. PITPNC1 drives enhanced secretion of factors that promote tumor angiogenesis, invasion, and metastasis. PITPNC1 regulates GOLPH3 localization to the Golgi and its oncogenic effects are eliminated upon knockdown of GOLPH3 [82].
Altogether, these data indicate that the GOLPH3 complex drives cancer through regulation of Golgi function. Since GOLPH3 represents the first oncogene that acts through Golgi secretory trafficking, its further study is likely to provide insight into novel mechanisms of oncogenesis. Furthermore, genetic experiments in culture and xenograft mouse models suggest that the GOLPH3 complex may be a good target for inhibitors with therapeutic potential against cancer [40,44,65].
Conclusion
The oversimplified picture of the Golgi as a constitutive organelle divorced from the rest of the cell is misguided. Rather, the Golgi hosts incoming and outgoing signaling pathways that broadly influence cell function. Furthermore, Golgi secretory trafficking itself is under the control of signals that converge to regulate the GOLPH3 complex. The role of the GOLPH3 complex in cancer highlights the significance of Golgi secretory function in cell physiology and human disease.
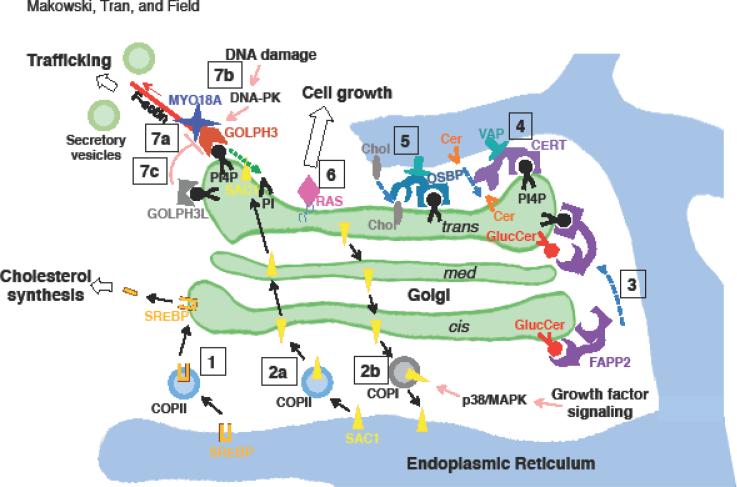
Figure.
Summary of some of the regulatory events that occur at the Golgi, as described in the main text.
[1] When cholesterol levels are low, SREBP is transported by COPII vesicles from the ER to the Golgi. This allows SREBP to be cleaved by Golgi-specific proteases, releasing an N-terminal fragment that enters the nucleus to activate the transcription of genes involved in cholesterol uptake and synthesis.
[2a] In the absence of growth factors, oligomerized SAC1 is transported by COPII vesicles and 14-3-3 from the ER to the Golgi, where it dephosphorylates PtdIns(4)P (PI4P) to produce PtdIns (PI), resulting in reduced secretory trafficking.
[2b] Growth factor signaling activates the p38/MAPK pathway, which stimulates dissociation of SAC1 oligomers and COPI-mediated trafficking of SAC1 from the Golgi to the ER. This increases PtdIns(4)P levels at the Golgi, which drives increased anterograde trafficking.
[3] FAPP2, which localizes to the trans-Golgi through binding to both PtdIns(4)P and ARF, transports glucosylceramide (GlucCer) from the cis- to trans-Golgi to facilitate globotriaosylceramide production.
[4] CERT binds to PtdIns(4)P at the Golgi and VAP at the ER, transporting ceramide (Cer) from the ER to the trans-Golgi at ER-Golgi contact sites.
[5] OSBP binding to PtdIns(4)P at the Golgi and VAP at the ER allows transport of cholesterol (Chol) from the ER to the trans-Golgi at ER-Golgi contact sites.
[6] While RAS signaling is typically thought to occur at the PM, a growing body of literature reports RAS localization to the Golgi, where it may trigger signaling that promotes cell growth.
[7a] The GOLPH3 complex, involving PtdIns(4)P/GOLPH3/MYO18A/F-actin, links the Golgi to the actin cytoskeleton and applies a tensile force to the Golgi membrane which aids in vesicle budding for forward trafficking to the PM.
[7b] DNA damage triggers DNA-PK activation, which phosphorylates GOLPH3 at Thr 143 and enhances its interaction with MYO18A, resulting in Golgi vesiculation and fragmentation. The resulting change in trafficking is required for normal cell survival following DNA damage.
[7c] GOLPH3L asserts a dominant-negative effect on the GOLPH3 complex. GOLPH3L binds to PtdIns(4)P but is unable to bind to MYO18A.
Highlights.
The Golgi hosts signaling events independent of its secretory function.
Golgi PtdIns(4)P effectors regulate vesicular and non-vesicular Golgi transport.
Signals converge on the GOLPH3 complex to regulate Golgi secretory function.
Oncogenic GOLPH3 and MYO18A implicate Golgi secretory function in cancer.
Acknowledgements
We apologize to colleagues whose work could not be referenced due to length restrictions. We thank members of the Field lab for critical reading of the manuscript. SLM acknowledges support from National Institutes of Health 2T32DK7044-36A1. SJF acknowledges support from the Breast Cancer Research Program Era of Hope Scholar Award (W81XWH-10-1-0822), National Institutes of Health (R01 CA201303, R01 GM120055), and the Burroughs Wellcome Fund Career Award in Biomedical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 2.DeBose-Boyd RA, Brown MS, Li WP, Nohturfft A, Goldstein JL, Espenshade PJ. Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell. 1999;99:703–712. doi: 10.1016/s0092-8674(00)81668-2. [DOI] [PubMed] [Google Scholar]
- 3.Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 4.Sakai J, Duncan EA, Rawson RB, Hua X, Brown MS, Goldstein JL. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 1996;85:1037–1046. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- 5.Sakai J, Rawson RB, Espenshade PJ, Cheng D, Seegmiller AC, Goldstein JL, Brown MS. Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol. Cell. 1998;2:505–514. doi: 10.1016/s1097-2765(00)80150-1. [DOI] [PubMed] [Google Scholar]
- 6.Rawson RB, Zelenski NG, Nijhawan D, YE J, Sakai J, Hasan MT, Chang TY, Brown MS, Goldstein JL. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol. Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 7•.Cheng C, Ru P, Geng F, Liu J, Yoo JY, Wu X, Cheng X, Euthine V, Hu P, Guo JY, et al. Glucose-Mediated N-glycosylation of SCAP Is Essential for SREBP-1 Activation and Tumor Growth. Cancer Cell. 2015;28:569–581. doi: 10.1016/j.ccell.2015.09.021. [This paper identifies a mechanistic link between elevated glucose supply and lipogenesis in tumor growth. EGFR signaling promotes glucose uptake and N-glycosylation of SCAP, stimulating SREBP pathway activation and promoting tumorigenesis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8•.Ricoult SJH, Yecies JL, Ben-Sahra I, Manning BD. Oncogenic PI3K and K-Ras stimulate de novo lipid synthesis through mTORC1 and SREBP. Oncogene. 2016;35:1250–1260. doi: 10.1038/onc.2015.179. [This paper highlights an essential role for mTORC1 activation of SREBP in mediating oncogenic PI3K- and K-RAS-induced de novo lipogenesis in cancer.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blagoveshchenskaya A, Cheong FY, Rohde HM, Glover G, Knödler A, Nicolson T, Boehmelt G, Mayinger P. Integration of Golgi trafficking and growth factor signaling by the lipid phosphatase SAC1. The Journal of Cell Biology. 2008;180:803–812. doi: 10.1083/jcb.200708109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10•.Bajaj Pahuja K, Wang J, Blagoveshchenskaya A, Lim L, Madhusudhan MS, Mayinger P, Schekman R. Phosphoregulatory protein 14-3-3 facilitates SAC1 transport from the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E3199–206. doi: 10.1073/pnas.1509119112. [This paper details the mechanism by which SAC1 is trafficked from the ER to the Golgi, involving 14-3-3 proteins and COPII vesicles, and further elaborates on the role of growth factor signaling in regulating SAC1 ER-Golgi localization.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godi A, Pertile P, Meyers R, Marra P, Di Tullio G, Iurisci C, Luini A, Corda D, De Matteis MA. ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat Cell Biol. 1999;1:280–287. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- 12.Godi A, Campli AD, Konstantakopoulos A, Tullio GD, Alessi DR, Kular GS, Daniele T, Marra P, Lucocq JM, De Matteis MA. FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat Cell Biol. 2004;6:393–404. doi: 10.1038/ncb1119. [DOI] [PubMed] [Google Scholar]
- 13.Walch-Solimena C, Novick P. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- 14.Hama H, Schnieders EA, Thorner J, Takemoto JY, DeWald DB. Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1999;274:34294–34300. doi: 10.1074/jbc.274.48.34294. [DOI] [PubMed] [Google Scholar]
- 15.Audhya A, Foti M, Emr SD. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang YJ, Wang J, Sun H-Q, Martinez M, Sun YX, Macia E, Kirchhausen T, Albanesi JP, Roth MG, Yin HL. Phosphatidylinositol 4 phosphate regulates targeting of clathrin adaptor AP-1 complexes to the Golgi. Cell. 2003;114:299–310. doi: 10.1016/s0092-8674(03)00603-2. [DOI] [PubMed] [Google Scholar]
- 17.Lenoir M, Overduin M. PtdIns(4)P signalling and recognition systems. Adv. Exp. Med. Biol. 2013;991:59–83. doi: 10.1007/978-94-007-6331-9_5. [DOI] [PubMed] [Google Scholar]
- 18.De Matteis MA, Wilson C, D'Angelo G. Phosphatidylinositol-4-phosphate: the Golgi and beyond. Bioessays. 2013;35:612–622. doi: 10.1002/bies.201200180. [DOI] [PubMed] [Google Scholar]
- 19.Olkkonen VM. OSBP-Related Protein Family in Lipid Transport Over Membrane Contact Sites. Lipid Insights. 2015;8:1–9. doi: 10.4137/LPI.S31726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- 21.Levine TP, Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr Biol. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- 22.Hanada K, Kumagai K, Tomishige N, Yamaji T. CERT-mediated trafficking of ceramide. Biochim. Biophys. Acta. 2009;1791:684–691. doi: 10.1016/j.bbalip.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 23.Kawano M, Kumagai K, Nishijima M, Hanada K. Efficient trafficking of ceramide from the endoplasmic reticulum to the Golgi apparatus requires a VAMP-associated protein-interacting FFAT motif of CERT. J. Biol. Chem. 2006;281:30279–30288. doi: 10.1074/jbc.M605032200. [DOI] [PubMed] [Google Scholar]
- 24.Wyles JP, McMaster CR, Ridgway ND. Vesicle-associated Membrane Protein-associated Protein-A (VAP-A) Interacts with the Oxysterol-binding Protein to Modify Export from the Endoplasmic Reticulum. Journal of Biological Chemistry. 2002;277:29908–29918. doi: 10.1074/jbc.M201191200. [DOI] [PubMed] [Google Scholar]
- 25.Beh CT, Rine J. A role for yeast oxysterol-binding protein homologs in endocytosis and in the maintenance of intracellular sterol-lipid distribution. Journal of Cell Science. 2004;117:2983–2996. doi: 10.1242/jcs.01157. [DOI] [PubMed] [Google Scholar]
- 26.Mesmin B, Bigay J, Moser von Filseck J, Lacas-Gervais S, Drin G, Antonny B. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 2013;155:830–843. doi: 10.1016/j.cell.2013.09.056. [DOI] [PubMed] [Google Scholar]
- 27.Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D'Angelo G, Polishchuk E, Tullio GD, Santoro M, Campli AD, Godi A, West G, Bielawski J, Chuang C-C, van der Spoel AC, et al. Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature. 2007;449:62–67. doi: 10.1038/nature06097. [DOI] [PubMed] [Google Scholar]
- 29.D'Angelo G, Uemura T, Chuang C-C, Polishchuk E, Santoro M, Ohvo-Rekilä H, Sato T, Di Tullio G, Varriale A, D'Auria S, et al. Vesicular and non-vesicular transport feed distinct glycosylation pathways in the Golgi. Nature. 2013;501:116–120. doi: 10.1038/nature12423. [DOI] [PubMed] [Google Scholar]
- 30.Halter D, Neumann S, van Dijk SM, Wolthoorn J, de Mazière AM, Vieira OV, Mattjus P, Klumperman J, van Meer G, Sprong H. Pre- and post-Golgi translocation of glucosylceramide in glycosphingolipid synthesis. The Journal of Cell Biology. 2007;179:101–115. doi: 10.1083/jcb.200704091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munro S. The Golgi apparatus: defining the identity of Golgi membranes. Curr. Opin. Cell Biol. 2005;17:395–401. doi: 10.1016/j.ceb.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 32•.Yamaji T, Hanada K. Sphingolipid metabolism and interorganellar transport: localization of sphingolipid enzymes and lipid transfer proteins. Traffic. 2015;16:101–122. doi: 10.1111/tra.12239. [This paper provides a detailed review of sphingolipid metabolism and interorganelle sphingolipid transport.] [DOI] [PubMed] [Google Scholar]
- 33.Bell AW, Ward MA, Blackstock WP, Freeman HN, Choudhary JS, Lewis AP, Chotai D, Fazel A, Gushue JN, Paiement J, et al. Proteomics characterization of abundant Golgi membrane proteins. J. Biol. Chem. 2001;276:5152–5165. doi: 10.1074/jbc.M006143200. [DOI] [PubMed] [Google Scholar]
- 34.Wu CC, Taylor RS, Lane DR, Ladinsky MS, Weisz JA, Howell KE. GMx33: a novel family of trans-Golgi proteins identified by proteomics. Traffic. 2000;1:963–975. [PubMed] [Google Scholar]
- 35.Dippold HC, Ng MM, Farber-Katz SE, Lee S-K, Kerr ML, Peterman MC, Sim R, Wiharto PA, Galbraith KA, Madhavarapu S, et al. GOLPH3 bridges phosphatidylinositol-4- phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009;139:337–351. doi: 10.1016/j.cell.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snyder CM, Mardones GA, Ladinsky MS, Howell KE. GMx33 associates with the trans-Golgi matrix in a dynamic manner and sorts within tubules exiting the Golgi. Mol. Biol. Cell. 2006;17:511–524. doi: 10.1091/mbc.E05-07-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng MM, Dippold HC, Buschman MD, Noakes CJ, Field SJ. GOLPH3L antagonizes GOLPH3 to determine Golgi morphology. Mol. Biol. Cell. 2013;24:796–808. doi: 10.1091/mbc.E12-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taft MH, Behrmann E, Munske-Weidemann L-C, Thiel C, Raunser S, Manstein DJ. Functional characterization of human myosin-18A and its interaction with F-actin and GOLPH3. J. Biol. Chem. 2013;288:30029–30041. doi: 10.1074/jbc.M113.497180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bishé B, Syed GH, Field SJ, Siddiqui A. Role of phosphatidylinositol 4-phosphate (PI4P) and its binding protein GOLPH3 in hepatitis C virus secretion. J. Biol. Chem. 2012;287:27637–27647. doi: 10.1074/jbc.M112.346569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40•.Buschman MD, Xing M, Field SJ. The GOLPH3 pathway regulates Golgi shape and function and is activated by DNA damage. Front Neurosci. 2015;9:362. doi: 10.3389/fnins.2015.00362. [This paper reviews the relationship between Golgi morphology and secretory function that was discovered through study of the GOLPH3 complex. The relationship between Golgi morphology, function, and human disease states is discussed, as illuminated by the GOLPH3 complex.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Boukhelifa M, Tribble E, Morin-Kensicki E, Uetrecht A, Bear JE, Bankaitis VA. The Sac1 phosphoinositide phosphatase regulates Golgi membrane morphology and mitotic spindle organization in mammals. Mol. Biol. Cell. 2008;19:3080–3096. doi: 10.1091/mbc.E07-12-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tenorio MJ, Ross BH, Luchsinger C, Rivera-Dictter A, Arriagada C, Acuña D, Aguilar M, Cavieres V, Burgos PV, Ehrenfeld P, et al. Distinct Biochemical Pools of Golgi Phosphoprotein 3 in the Human Breast Cancer Cell Lines MCF7 and MDA-MB-231. PLoS ONE. 2016;11:e0154719. doi: 10.1371/journal.pone.0154719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farber-Katz SE, Dippold HC, Buschman MD, Peterman MC, Xing M, Noakes CJ, Tat J, Ng MM, Rahajeng J, Cowan DM, et al. DNA damage triggers Golgi dispersal via DNA-PK and GOLPH3. Cell. 2014;156:413–427. doi: 10.1016/j.cell.2013.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44••.Buschman MD, Rahajeng J, Field SJ. GOLPH3 links the Golgi, DNA damage, and cancer. Cancer Res. 2015;75:624–627. doi: 10.1158/0008-5472.CAN-14-3081. [This paper examines the role of the GOLPH3 complex in Golgi trafficking, its role in the Golgi DNA damage response, and implications for understanding GOLPH3's function as an oncogene.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 46.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 47.Zhou B, Der CJ, Cox AD. The role of wild type RAS isoforms in cancer. Seminars in Cell & Developmental Biology. 2016;58:60–69. doi: 10.1016/j.semcdb.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kano Y, Cook JD, Lee JE, Ohh M. New structural and functional insight into the regulation of Ras. Seminars in Cell & Developmental Biology. 2016;58:70–78. doi: 10.1016/j.semcdb.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 49.Lowy DR, Willumsen BM. Function and regulation of ras. Annu Rev Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 50.Prior IA, Hancock JF. Compartmentalization of Ras proteins. Journal of Cell Science. 2001;114:1603–1608. doi: 10.1242/jcs.114.9.1603. [DOI] [PubMed] [Google Scholar]
- 51.Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 2011;75:50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 53.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 54.Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, Ivanov IE, Philips MR. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- 55.Lynch SJ, Snitkin H, Gumper I, Philips MR, Sabatini D, Pellicer A. The Differential Palmitoylation States of N-Ras and H-Ras Determine Their Distinct Golgi Subcompartment Localizations. J. Cell. Physiol. 2014;230:610–619. doi: 10.1002/jcp.24779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prior IA, Hancock JF. Ras trafficking, localization and compartmentalized signalling. Seminars in Cell & Developmental Biology. 2012;23:145–153. doi: 10.1016/j.semcdb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chiu VK, Bivona T, Hach A, Sajous JB, Silletti J, Wiener H, Johnson RL, Cox AD, Philips MR. Ras signalling on the endoplasmic reticulum and the Golgi. Nat Cell Biol. 2002;4:343–350. doi: 10.1038/ncb783. [DOI] [PubMed] [Google Scholar]
- 58.Mor A, Campi G, Du G, Zheng Y, Foster DA, Dustin ML, Philips MR. The lymphocyte function-associated antigen-1 receptor costimulates plasma membrane Ras via phospholipase D2. Nat Cell Biol. 2007;9:713–719. doi: 10.1038/ncb1592. [DOI] [PubMed] [Google Scholar]
- 59•.Herrero A, Casar B, Colón-Bolea P, Agudo-Ibáñez L, Crespo P. Defined spatiotemporal features of RAS-ERK signals dictate cell fate in MCF-7 mammary epithelial cells. Mol. Biol. Cell. 201627:1958–1968. doi: 10.1091/mbc.E15-02-0118. [This study identifies differences in RAS signaling at the PM versus the Golgi by comparing MCF-7 breast cancer cell responses to EGF versus heregulin.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bivona TG, Pérez De Castro I, Ahearn IM, Grana TM, Chiu VK, Lockyer PJ, Cullen PJ, Pellicer A, Cox AD, Philips MR. Phospholipase Cgamma activates Ras on the Golgi apparatus by means of RasGRP1. Nature. 2003;424:694–698. doi: 10.1038/nature01806. [DOI] [PubMed] [Google Scholar]
- 61.Caloca MJ, Zugaza JL, Bustelo XR. Exchange factors of the RasGRP family mediate Ras activation in the Golgi. J. Biol. Chem. 2003;278:33465–33473. doi: 10.1074/jbc.M302807200. [DOI] [PubMed] [Google Scholar]
- 62.Roose JP, Mollenauer M, Gupta VA, Stone J, Weiss A. A diacylglycerol-protein kinase C-RasGRP1 pathway directs Ras activation upon antigen receptor stimulation of T cells. Molecular and Cellular Biology. 2005;25:4426–4441. doi: 10.1128/MCB.25.11.4426-4441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aran V, Prior IA. Compartmentalized Ras signaling differentially contributes to phenotypic outputs. Cell. Signal. 2013;25:1748–1753. doi: 10.1016/j.cellsig.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64••.Hernandez-Valladares M, Prior IA. Comparative proteomic analysis of compartmentalised Ras signalling. Sci Rep. 2015;5:17307. doi: 10.1038/srep17307. [In this study, the authors perform proteomic analysis to detect differences in signaling between forms of K-RAS that have been tethered to different subcellular sites, including the Golgi.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scott KL, Kabbarah O, Liang M-C, Ivanova E, Anagnostou V, Wu J, Dhakal S, Wu M, Chen S, Feinberg T, et al. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature. 2009;459:1085–1090. doi: 10.1038/nature08109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sanchez-Garcia F, Villagrasa P, Matsui J, Kotliar D, Castro V, Akavia U-D, Chen B-J, Saucedo-Cuevas L, Rodriguez Barrueco R, Llobet-Navas D, et al. Integration of genomic data enables selective discovery of breast cancer drivers. Cell. 2014;159:1461–1475. doi: 10.1016/j.cell.2014.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BVSK, Varambally S, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nature Medicine. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eirew P, Steif A, Khattra J, Ha G, Yap D, Farahani H, Gelmon K, Chia S, Mar C, Wan A, et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature. 2015;518:422–426. doi: 10.1038/nature13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martelotto LG, De Filippo MR, Ng CKY, Natrajan R, Fuhrmann L, Cyrta J, Piscuoglio S, Wen H-C, Lim RS, Shen R, et al. Genomic landscape of adenoid cystic carcinoma of the breast. J. Pathol. 2015;237:179–189. doi: 10.1002/path.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin W-C, Mansour J, Mollaee M, Wagner K-U, Koduru P, Yopp A, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cancer Genome Atlas Research Network Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zeng Z, Lin H, Zhao X, Liu G, Wang X, Xu R, Chen K, Li J, Song L. Overexpression of GOLPH3 promotes proliferation and tumorigenicity in breast cancer via suppression of the FOXO1 transcription factor. Clin. Cancer Res. 2012;18:4059–4069. doi: 10.1158/1078-0432.CCR-11-3156. [DOI] [PubMed] [Google Scholar]
- 75.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roos WP, Thomas AD, Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer. 2016;16:20–33. doi: 10.1038/nrc.2015.2. [DOI] [PubMed] [Google Scholar]
- 78.Lord CJ, Ashworth A. The DNA damage response and cancer therapy. Nature. 2012;481:287–294. doi: 10.1038/nature10760. [DOI] [PubMed] [Google Scholar]
- 79.Isaji T, Im S, Gu W, Wang Y, Hang Q, Lu J, Fukuda T, Hashii N, Takakura D, Kawasaki N, et al. An Oncogenic Protein Golgi Phosphoprotein 3 Up-regulates Cell Migration via Sialylation. J. Biol. Chem. 2014;289:20694–20705. doi: 10.1074/jbc.M113.542688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tokuda E, Itoh T, Hasegawa J, Ijuin T, Takeuchi Y, Irino Y, Fukumoto M, Takenawa T. Phosphatidylinositol 4-Phosphate in the Golgi Apparatus Regulates Cell-Cell Adhesion and Invasive Cell Migration in Human Breast Cancer. Cancer Res. 2014;74:3054–3066. doi: 10.1158/0008-5472.CAN-13-2441. [DOI] [PubMed] [Google Scholar]
- 81••.Xing M, Peterman MC, Davis RL, Oegema K, Shiau AK, Field SJ. GOLPH3 drives cell migration by promoting Golgi reorientation and directional trafficking to the leading edge. Mol. Biol. Cell. 2016 doi: 10.1091/mbc.E16-01-0005. doi:10.1091/mbc.E16-01-0005. [This paper demonstrates that GOLPH3 drives cell migration by acting through the PtdIns(4)P/GOLPH3/MYO18A/F-actin complex to promote Golgi reorientation and directional trafficking.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82••.Halberg N, Sengelaub CA, Navrazhina K, Molina H, Uryu K, Tavazoie SF. PITPNC1 Recruits RAB1B to the Golgi Network to Drive Malignant Secretion. Cancer Cell. 2016;29:339–353. doi: 10.1016/j.ccell.2016.02.013. [This paper identifies PITPNC1 as a novel oncogene that acts to promote secretion of factors that are pro-invasive and pro-angiogenic. The authors find that PITPNC1 drives secretion by increasing the activity of GOLPH3 at the Golgi.] [DOI] [PMC free article] [PubMed] [Google Scholar]