Abstract
Aging is accompanied by a decline in tissue function, regeneration, and repair. A large part of this decline is due to the deterioration of tissue stem cell function. Understanding the mechanisms that drive stem cell aging and how to counteract them is a critical step for enhancing tissue repair and maintenance during aging. Emerging evidence indicates that epigenetic modifiers and metabolism regulators interact to impact lifespan, suggesting that this mechanism may also affect stem cell function with age. This review focuses on the interaction between chromatin and metabolism in the regulation of tissue stem cells during aging. We also discuss how these mechanisms integrate environmental stimuli such as nutrient stress to regulate stem cell function. Finally, this review examines new perspectives for regeneration, rejuvenation, and treatment of age-related decline of stem cell function.
Tissue-specific stem cells are present in virtually every adult tissue in mammals and are essential for tissue homeostasis and repair after injury (Box 1). The striking decline in stem cell function during aging, coupled with a bias in the type of differentiated cells they generate, could drive the deterioration in tissue function and diminished capacity for tissue repair in older individuals [1]. Stem cell function is regulated in response to exposure of individuals to a variety of external stimuli, including nutrient stress (e.g. starvation or caloric restriction, or high fat diets). Knowledge of the mechanisms by which stem cells integrate signals from the environment will be critical to identify strategies to preserve or reactivate their function in old age.
Box 1.
In many stem cell compartments, the lineage consists of a quiescent stem cell that can activate (proliferate) and generate more committed progenitors and differentiated progeny [66]. Adult stem cells can be unipotent (i.e. generate one differentiated cell type) or multipotent (i.e. generate several differentiated cell types). For example, muscle stem cells (MuSCs) are unipotent and give rise to one cell type – muscle fibers. In contrast, hematopoietic stem cells (HSCs) and neural stem cells (NSCs) are multipotent and can give rise to several types of differentiated cells (NCS, for example, give rise to neurons, astrocytes and oligodendrocytes). Some stem cells are very important for tissue homeostasis (HSCs, intestinal stem cells [ISCs]). Other stem cells are activated in response to injury (MuSCs and to a lesser extent NSCs), although they can also contribute to some aspect of homeostasis (NSCs).
During aging, two key aspects of stem cell function are primarily affected in multiple stem cell lineages: the transition from quiescent stem cells to activated stem cells and the bias in generated differentiated cell types. For example, HSCs exhibit a myeloid bias during aging whereas NSCs exhibit an astrocytic bias. Single cells studies have recently revealed additional cellular transitions in stem cell lineages, and such transitions could also be affected during aging [67-69].
Stem cells are present within complex “niches” that are often in tight connection with blood vessels, thereby providing an interface with the systemic environment and factors in the blood (metabolites, hormones, growth factors, etc.) [70,71]. In the brain, neural stem cells are also in contact with the cerebral spinal fluid [72], providing an additional source for external stimuli, such as metabolites.
Understanding the interaction between cellular metabolism and chromatin features in stem cells is confounded by the complexity of stem cell fates compared to many other somatic cells. Stem cells, by their very nature, give rise to progeny that are vastly different in terms of virtually every biological parameter. For example, quiescent stem cells exhibit minimal metabolic activity, have few mitochondria and other organelles, and have a minuscule cytoplasmic volume. In contrast, proliferating progeny manifest dramatic energetic shifts, increases in biosynthetic activity, and cell growth. As these progeny differentiate into mature, tissue-specific cells, there are again dramatic structural and functional changes. The dynamic interaction between cellular metabolism and the epigenome is emerging as pivotal to the control of stem cell transitions and function. This review will discuss recent work connecting metabolism and chromatin regulators in mammalian tissue-specific stem cells, focusing primarily on mechanisms that are relevant to the process of aging and that could be used to restore function to old stem cells.
Interaction between epigenetic and metabolic pathways in organismal aging
The importance of epigenetic mechanisms in controlling aging has been extensively reviewed [2-5]. In this section, we will present recent studies that have uncovered intriguing connections between key chromatin regulators and metabolic pathways in the regulation of lifespan in yeast, worms, and flies. These studies help illustrate the importance of the interactions between chromatin and metabolism in the regulation of processes that may be important in cell and tissue aging. For example, deletion of the chromatin remodeler SWI/SNF (ISW2) extends replicative lifespan in yeast, in a manner that mimics caloric restriction [6]. Consistent with these findings, genome-wide analysis indicates that in ISW2-deficient yeast, changes in nucleosome positioning partially replicate those of calorie-restricted cells [6]. As nucleosome positioning is critical for gene expression regulation, these results suggest that a common mechanism, regulated by both pathways, impacts many target genes in a coordinated manner. Chromatin remodelers of the SWI/SNF family also play a key role in modulating lifespan in C. elegans [6,7], notably in partnership with FOXO/DAF-16 transcription factor downstream of the insulin-signaling pathway [7]. Furthermore, global chromatin structure changes were observed in conditions (mitochondrial deficiency) that extend lifespan in C. elegans [8]. These studies in yeast and worms, coupled with the observed changes in nucleosome positioning during aging in mouse tissues [9], highlight the importance of chromatin structure and nucleosome positioning in integrating metabolic changes to regulate lifespan [10].
In addition to chromatin remodelers, several histone modifiers, including Sirtuin deacetylases, have been shown to link metabolic state to organismal lifespan regulation [11]. Recently, in Drosophila, key metabolites including acetylCoA were found to be increased during aging. This increase is accompanied by an increase in global histone acetylation, notably acetylated lysine 12 on histone H4 (H4K12ac). Consistent with these observations, mutation in the H4K12 acetyltransferase Chameau extends the lifespan of male flies [12]. Histone methylation regulators, including H3K4me3, H3K36me3, and H3K27me3 regulators, have also been found to regulate lifespan in C. elegans [13-17]. Interestingly, the conserved H3K27me3 demethylase Jmjd-3.1/JMJD3 and the H3K27me2 demethylase Jmjd-1.2/PHF8 were recently found to specifically mediate longevity due to mitochondrial deficiency in C. elegans [18]. Mitochondrial deficiency triggers the specific upregulation of nuclear-encoded genes involved in the response to mitochondrial unfolded protein response (UPR) [18]; this could mediate long-lasting transcriptional responses in response to deficit in mitochondrial function. In mammals, PHF8 and JMJD3 expression is correlated with the longevity of outbred mice [18]. Thus, histone modifiers that are responsive to cellular metabolic state are key determinants of organismal longevity.
These recent examples highlight the interaction between chromatin state and cellular metabolism in influencing longevity, at least in model organisms. The interrelated processes of chromatin regulation and metabolism are increasingly being studied in the areas of stem cell aging and general stem cell biology in mammals. In the following sections, we discuss recent findings that reveal key epigenetic changes that are associated with stem cell aging and that, in some cases, reflect the potential impact of cellular metabolism on the epigenetic state.
Epigenetics and stem cell aging
Epigenetic changes associated with somatic stem cell aging have been reported for multiple stem cell populations, notably hematopoietic stem cells (HSCs) and muscle stem cells (MuSCs)/satellite cells [19,20]. In both HSCs and MuSCs, there is an age-dependent increase in the repressive histone modification H3K27me3, whereas H3K4me3, a mark associated with active genes, shows increase in breadth in HSCs but decreases slightly in intensity in quiescent MuSCs with age. In each case, the increase in H3K27me3 is associated with a down-regulation of a limited number of genes associated with stem cell function. These results are interesting in light of the recent finding that the H3K27me3 demethylase UTX is important for MuSC-mediated muscle regeneration [21]. However, the specific effects of these chromatin changes on stem cell function remain to be tested, particularly given that different regulators of the same H3K27me3 mark (for example, the H3K27me3 demethylases UTX and JMJD3) can either extend or shorten lifespan in model organisms [14,18,22].
Changes in a number of other chromatin features, most of which are also altered with age, have been shown to regulate stem cell function [23]. For example, changes in DNA methylation affect the differentiation potential of HSCs [24,25]. Furthermore, H3K9 methylation, a mediator of heterochromatin formation, is important for HSC differentiation [26]. H4K20 methylation controls MuSC quiescence by promoting the formation of facultative heterochromatin [27]. Finally, exceptionally broad H3K4me3 domains form a signature for cell function/identity, and these extended domains mark genes that are critical for the ability of neural stem cells (NSCs) to self-renew and to differentiate into neurons [28]. How these different epigenetic marks are affected in aging stem cells is not yet known. It will be important to develop methods to modify the epigenetic state of specific genetic loci to test for causal effects on stem cell function [29,30] to determine if the epigenome of old stem cells can be “reset” to a youthful state [31].
Direct modulation of epigenetic regulator activity in stem cells by metabolites
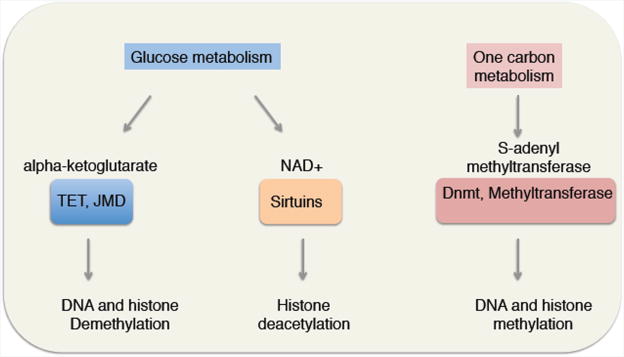
While epigenetic states are clearly under multiple levels of control, their direct regulation by metabolites in a variety of cell types, including stem cells, is an area that has recently been of great interest. It is noteworthy that the co-factors important for enzymes that regulate chromatin modifications (e.g. DNA methylation/demethylation, histone acetylation/deacetylation, and histone methylation/demethylation) are metabolites whose levels are determined by the metabolic status of the cell [32]. This direct link raises the intriguing possibility that changes in metabolism could have global effects on the stem cell epigenome and stem cell function (Figure 2).
Figure 2. Metabolites are co-factors of many chromatin regulators, thereby directly connecting metabolic state with chromatin state.
It was recently shown that decreases in levels of cellular NAD(+), a co-substrate for Sirtuin deacetylases and other enzymes [33], lead to elevated H4K16 acetylation and the induction of expression of a myogenic program in MuSCs during activation out of quiescence [34]. NAD(+) has been shown to become limiting during aging and these changes could drive the global changes in histone acetylation levels [35]. Other metabolites could play important roles in modifying chromatin to impact stem cell function or transitions. For example, changes in alpha-ketoglutarate levels in response to knockdown of phosphoserine aminotransferase 1 (PSAT1) affects embryonic stem cell (ESC) differentiation, especially in the ectoderm (neural) lineage [36]. Alpha-ketoglutarate is a co-factor not only for TET enzymes (implicated in DNA demethylation) but also for Jumonji family proteins (histone demethylases), and could thereby exert widespread effects on chromatin in many stem cells. Likewise, S-adenosyl-methionine (SAM) is a cofactor for histone methyltransferases and has been shown to connect H3K4 methylation to one carbon metabolism [37] and threonine metabolism [38]. How SAM levels change with age in different cell types, including stem cells, is not known, but changes in SAM levels could play an important role in orchestrating age-dependent changes in histone methylation marks in response to variations in one-carbon metabolism. These examples illustrate a diverse set of connections between metabolites and specific chromatin regulators. Thus, select metabolites could promote long-lasting changes in gene expression and stem cell state changes in response to environmental stimuli. Given the epigenetic changes associated with stem cell aging, it will be of interest to understand how changes in metabolic regulation associated with aging underlie these age-related changes in chromatin.
Indirect regulation of stem cell function by metabolism: Effects of nutrient sensing pathways
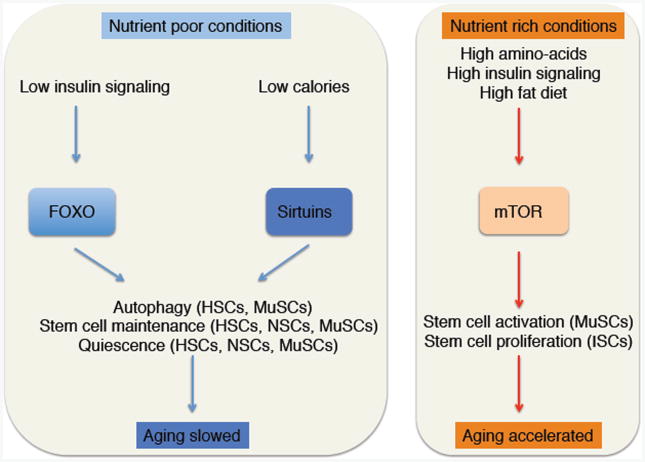
While some key regulators of lifespan and healthspan, such as Sirtuins, are directly regulated by metabolites, other crucial players, including the insulin-FOXO and the mTOR signaling pathways, are globally integrators of cellular nutrient and metabolic status (Figure 3). The role of these pathways in the control of stem cell fate has been extensively reviewed [39]. Here, we will focus in how these pathways affect stem cell function and their changes with age, at least in part through modulation of chromatin/transcriptional states.
Figure 3. Dietary control of regulators of stem cell function and consequences for tissue aging.
Sirtuins play important roles in the function of several somatic stem cells. Inactivation of the Sirtuin SIRT1 in mouse NSCs leads to the expansion of oligodendrocyte progenitors and leads to the misregulation of genes involved in metabolism, notably amino-acid metabolism [40]. In HSCs, the Sirtuin SIRT7 is important for the modulation of the mitochondrial unfolded protein response (mtUPR), downstream of mitochondrial stressors. SIRT7 expression is downregulated during aging in HSCs, and its expression maintains HSC function in part by repressing NRF1 genomic targets [41]. Finally, in MuSCs, as described above, SIRT1 responds to the cellular energetic state via NAD(+) to act as an epigenetic regulator that influences stem cell fate [34]. Interestingly, SIRT1 has been implicated in the regulation of autophagy in MuSCs [42], and the induction of autophagy is necessary for MuSCs activation out of the quiescent state by providing essential building blocks for macromolecular synthesis associated with the rapid growth of stem cells during this transition [42].
The transcription factor FOXO3 is critical for the maintenance of NSC and MuSC quiescence [43-46]. FOXO3 enhances HSC survival in response to complete starvation by poising the cells for the rapid induction of autophagy [47]. Indeed, several of FOXO3 targets are implicated in autophagy [48]. In contrast, in nutrient rich environments, mTOR signaling, a key nutrient sensing pathway that influences lifespan and suppresses autophagy [49], is essential for the induction of quiescent cells MuSCs and HSCs into a poised state (GAlert) for more rapid activation [50]. Thus, autophagic processes that are catabolic but also support cellular anabolic activities may be important as determinants of stem cell aging by their roles in linking metabolism to the production of co-factors essential for epigenetic changes.
The role of external stimuli: effects of starvation/refeeding and high fat diets on stem cell function
The impact of metabolism on stem cell aging is suggested by the profound effect that caloric restriction has in extending lifespan. In addition, caloric excess (for example high fat diets) often has negative healthspan and lifespan effects [51]. Dietary changes that mimic fasting and that delay the onset of age-related pathologies are able to enhance the function of stem cells in multiple tissues [52]. Prolonged fasting itself promotes HSC-based regeneration of the hematopoietic system [53]. Caloric restriction also enhances intestinal stem cell (ISC) function and intestinal regeneration [54]. The expansion of ISCs by caloric restriction depends upon the key nutrient sensors, mTOR and SIRT1 [55]. In contrast, high fat diet promotes dysregulation of ISCs and their progeny, resulting in an increased incidence of intestinal tumors [56].
Whether changes in stem cell function in response to nutrient availability are mediated by epigenetic changes is still largely unclear, but a few examples are emerging. For instance, the regulation of NSC proliferation in response to decreased glucose availability is governed by the nutrient sensors CREB and SIRT1, and the effects of caloric restriction are accompanied by an increase in H3K9Ac [57]. Changes in dietary intake are accompanied by massive changes in chromatin states in several species [58,59]. Although these studies were not done in stem cells, it is likely that extensive changes in the epigenome also accompany the altered stem cell activity in response to changes in nutrient availability. In the future, it will be interesting to explore the extent to which those epigenetic changes mediate stem cell aging.
Conclusion and future outlook
While exciting progress has been made in understanding how metabolism and chromatin factors interact in stem cells, it is still unclear how this interaction affects stem cell function, how it is impacted by environmental stimuli, and how it changes during aging. A deeper knowledge of the interaction between chromatin regulators and metabolic processes will be critical for understanding stem cell responses to other physiological processes that impact metabolism such as circadian rhythm, exercise, or sleep. As chromatin changes can have a long-lasting impact, it will be important to determine the kinetics and window of action of the key regulators of metabolism and chromatin states. This interaction could explain how changes in nutrition early in life could have long-lasting consequences on tissue function and regeneration in adulthood (and even possibly in the subsequent generations).
It is important to note that most studies so far have been conducted in bulk populations of cells, thereby obscuring potential cell heterogeneity and cell-to-cell variability in chromatin states and metabolic responses. Initial studies on limited numbers of genes have reported increased in cell-to-cell transcriptional variability during aging of cardiomyocytes [60], but not of HSCs [61]. Recent technological developments have allowed single cell transcriptomic and chromatin accessibility studies. Thus, an interesting future area of study will be to uncover how chromatin and metabolites are modulated at the single cell level and whether there is an increase in cell-to-cell variability during aging. Recent evidence has provided some link between chromatin regulators and cell-to-cell variability. For example, H3K4me3 regulators have recently found to be associated with reduced cell-to-cell transcriptional variability [28]. Furthermore, the linker histone H1.0 has been shown to exhibit cell-to-cell heterogeneity in cancer stem cells [62]. Cell-to-cell variability within a stem cell niche during aging could be particularly deleterious for the coordination of tissue regeneration or repair upon injury.
It will also be interesting to determine the role that the interaction between metabolism and epigenetic factors plays in the rejuvenation of old stem cell function in response to interventions that revert some aspects of aging. For example, heterochronic parabiosis, the fusion of an old and young organism by the blood circulation, can revert hallmarks of aging in several stem cells [63,64]. The mechanisms of rejuvenation of old stem cells could be mediated at least in part by the interaction between metabolites and epigenetic regulators. Similarly, cellular reprogramming has also been shown to reset several age-dependent phenotypes including metabolic (mitochondria) and epigenetic features [65]. Thus, the manipulation of specific metabolic and epigenetic pathways, especially those that occur early during the reprogramming process, could be sufficient to promote rejuvenation without inducing de-differentiation.
Collectively, the knowledge of the interaction between metabolism and epigenetics will be critical to identify new strategies for preserving youthful cellular function as organisms age or to counteract age-related dysfunction of stem cells in old tissues.
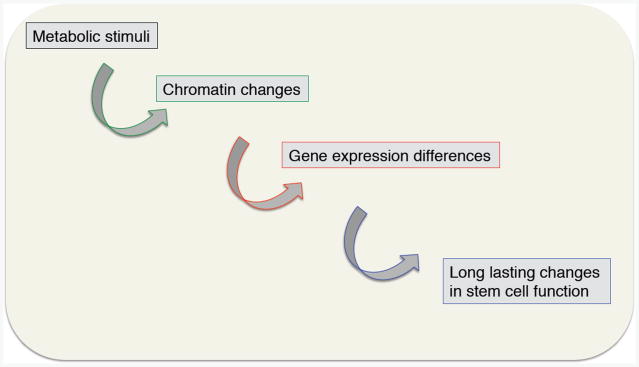
Figure 1. Changes in metabolism could have long-lasting impact on stem cell function.
Acknowledgments
We apologize to the authors whose work we could not cite due to space limitation. We thank Bérénice Benayoun for feedback on the manuscript. Supported by P01 AG036695 (A.B and T.A.R.) and by the Glenn Center for the Biology of Aging (A.B. and T.A.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goodell MA, Rando TA. Stem cells and healthy aging. Science. 2015;350:1199–1204. doi: 10.1126/science.aab3388. [DOI] [PubMed] [Google Scholar]
- 2.Benayoun BA, Pollina EA, Brunet A. Epigenetic regulation of ageing: linking environmental inputs to genomic stability. Nat Rev Mol Cell Biol. 2015;16:593–610. doi: 10.1038/nrm4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sen P, Shah PP, Nativio R, Berger SL. Epigenetic Mechanisms of Longevity and Aging. Cell. 2016;166:822–839. doi: 10.1016/j.cell.2016.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Booth LN, Brunet A. The Aging Epigenome. Mol Cell. 2016;62:728–744. doi: 10.1016/j.molcel.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pal S, Tyler JK. Epigenetics and aging. Sci Adv. 2016;2:e1600584. doi: 10.1126/sciadv.1600584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dang W, Sutphin GL, Dorsey JA, Otte GL, Cao K, Perry RM, Wanat JJ, Saviolaki D, Murakami CJ, Tsuchiyama S, et al. Inactivation of yeast Isw2 chromatin remodeling enzyme mimics longevity effect of calorie restriction via induction of genotoxic stress response. Cell Metab. 2014;19:952–966. doi: 10.1016/j.cmet.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riedel CG, Dowen RH, Lourenco GF, Kirienko NV, Heimbucher T, West JA, Bowman SK, Kingston RE, Dillin A, Asara JM, et al. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat Cell Biol. 2013;15:491–501. doi: 10.1038/ncb2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian Y, Garcia G, Bian Q, Steffen KK, Joe L, Wolff S, Meyer BJ, Dillin A. Mitochondrial Stress Induces Chromatin Reorganization to Promote Longevity and UPR(mt) Cell. 2016;165:1197–1208. doi: 10.1016/j.cell.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bochkis IM, Przybylski D, Chen J, Regev A. Changes in nucleosome occupancy associated with metabolic alterations in aged mammalian liver. Cell Rep. 2014;9:996–1006. doi: 10.1016/j.celrep.2014.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyler JK. Nucleosomes Find Their Place in Life. Trends Genet. 2016;32:689–690. doi: 10.1016/j.tig.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 201213:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12•.Peleg S, Feller C, Forne I, Schiller E, Sevin DC, Schauer T, Regnard C, Straub T, Prestel M, Klima C, et al. Life span extension by targeting a link between metabolism and histone acetylation in Drosophila. EMBO Rep. 2016;17:455–469. doi: 10.15252/embr.201541132. In this report, the authors show that middle age in Drosophila is accompanied by a concomitant increase in the acetyl-CoA metabolite and the histone mark H4K12ac. Reducing the levels of H4K12ac by mutating the acetyltransferase, called Chameau in Drosophila (MYST-type acetyltransferase in mammals), extends the lifespan of Drosophila. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greer EL, Maures TJ, Hauswirth AG, Green EM, Leeman DS, Maro GS, Han S, Banko MR, Gozani O, Brunet A. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–387. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maures TJ, Greer EL, Hauswirth AG, Brunet A. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell. 2011;10:980–990. doi: 10.1111/j.1474-9726.2011.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pu M, Ni Z, Wang M, Wang X, Wood JG, Helfand SL, Yu H, Lee SS. Trimethylation of Lys36 on H3 restricts gene expression change during aging and impacts life span. Genes Dev. 2015;29:718–731. doi: 10.1101/gad.254144.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ni Z, Ebata A, Alipanahiramandi E, Lee SS. Two SET domain containing genes link epigenetic changes and aging in Caenorhabditis elegans. Aging Cell. 2012;11:315–325. doi: 10.1111/j.1474-9726.2011.00785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sen P, Dang W, Donahue G, Dai J, Dorsey J, Cao X, Liu W, Cao K, Perry R, Lee JY, et al. H3K36 methylation promotes longevity by enhancing transcriptional fidelity. Genes Dev. 2015;29:1362–1376. doi: 10.1101/gad.263707.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18••.Merkwirth C, Jovaisaite V, Durieux J, Matilainen O, Jordan SD, Quiros PM, Steffen KK, Williams EG, Mouchiroud L, Tronnes SU, et al. Two Conserved Histone Demethylases Regulate Mitochondrial Stress-Induced Longevity. Cell. 2016;165:1209–1223. doi: 10.1016/j.cell.2016.04.012. This study, together with Tian et al 2016, show that deficiency in mitochondria early during development, triggers profound changes in chromatin regulators, including the H3K27me2 demethylase JMJD-1.2 (PHF8) and the H3K27me3 demethylases JMJD-3.1 (JMJD3) (Merkwirth et al) and H3K9me2 regulators (Tian et al). Together, these chromatin changes lead to global alterations in chromatin structure and the expression of mitochondrial stress response genes (encoded by the nucleus). Interestingly, increased expression of PHF8 and JMJD3 is also associated with increased lifespan in outbred mouse strains, suggesting conservation in mammals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chambers SM, Shaw CA, Gatza C, Fisk CJ, Donehower LA, Goodell MA. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 2007;5:e201. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu L, Cheung TH, Charville GW, Hurgo BM, Leavitt T, Shih J, Brunet A, Rando TA. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 2013;4:189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faralli H, Wang C, Nakka K, Benyoucef A, Sebastian S, Zhuang L, Chu A, Palii CG, Liu C, Camellato B, et al. UTX demethylase activity is required for satellite cell-mediated muscle regeneration. J Clin Invest. 2016;126:1555–1565. doi: 10.1172/JCI83239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labbadia J, Morimoto RI. Repression of the Heat Shock Response Is a Programmed Event at the Onset of Reproduction. Mol Cell. 2015;59:639–650. doi: 10.1016/j.molcel.2015.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avgustinova A, Benitah SA. Epigenetic control of adult stem cell function. Nat Rev Mol Cell Biol. 2016;17:643–658. doi: 10.1038/nrm.2016.76. [DOI] [PubMed] [Google Scholar]
- 24.Mayle A, Yang L, Rodriguez B, Zhou T, Chang E, Curry CV, Challen GA, Li W, Wheeler D, Rebel VI, et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood. 2015;125:629–638. doi: 10.1182/blood-2014-08-594648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Challen GA, Sun D, Mayle A, Jeong M, Luo M, Rodriguez B, Mallaney C, Celik H, Yang L, Xia Z, et al. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell. 2014;15:350–364. doi: 10.1016/j.stem.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ugarte F, Sousae R, Cinquin B, Martin EW, Krietsch J, Sanchez G, Inman M, Tsang H, Warr M, Passegue E, et al. Progressive Chromatin Condensation and H3K9 Methylation Regulate the Differentiation of Embryonic and Hematopoietic Stem Cells. Stem Cell Reports. 2015;5:728–740. doi: 10.1016/j.stemcr.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Boonsanay V, Zhang T, Georgieva A, Kostin S, Qi H, Yuan X, Zhou Y, Braun T. Regulation of Skeletal Muscle Stem Cell Quiescence by Suv4-20h1-Dependent Facultative Heterochromatin Formation. Cell Stem Cell. 2016;18:229–242. doi: 10.1016/j.stem.2015.11.002. This study shows that a chromatin modifying enzyme, Suv4-20H1, which is a histone methyltrasferase, helps to maintain the quiescent state of MuSCs by promoting heterochromatin formation and the suppression of transcription of a master regulator of the myogenic lineage program, MyoD. [DOI] [PubMed] [Google Scholar]
- 28.Benayoun BA, Pollina EA, Ucar D, Mahmoudi S, Karra K, Wong ED, Devarajan K, Daugherty AC, Kundaje AB, Mancini E, et al. H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell. 2014;158:673–688. doi: 10.1016/j.cell.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vora S, Tuttle M, Cheng J, Church G. Next stop for the CRISPR revolution: RNA-guided epigenetic regulators. FEBS J. 2016;283:3181–3193. doi: 10.1111/febs.13768. [DOI] [PubMed] [Google Scholar]
- 30.Hathaway NA, Bell O, Hodges C, Miller EL, Neel DS, Crabtree GR. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149:1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rando TA, Chang HY. Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell. 2012;148:46–57. doi: 10.1016/j.cell.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berger SL, Sassone-Corsi P. Metabolic Signaling to Chromatin. Cold Spring Harb Perspect Biol. 2016;8 doi: 10.1101/cshperspect.a019463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verdin E. NAD(+) in aging, metabolism, and neurodegeneration. Science. 2015;350:1208–1213. doi: 10.1126/science.aac4854. [DOI] [PubMed] [Google Scholar]
- 34••.Ryall JG, Dell'Orso S, Derfoul A, Juan A, Zare H, Feng X, Clermont D, Koulnis M, Gutierrez-Cruz G, Fulco M, et al. The NAD(+)-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell Stem Cell. 2015;16:171–183. doi: 10.1016/j.stem.2014.12.004. The authors of this study show that the activation of MuSCs out of quiescence exhibit a metabolic switch with a reduction in NAD(+) levels. This leads to a reduction in SIRT1 activity, and increase in H4K16 acetylation, and an increase in expression of myogenic genes associated with the activation and ultimate differentiation of MuSCs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imai S, Guarente L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014;24:464–471. doi: 10.1016/j.tcb.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang IY, Kwak S, Lee S, Kim H, Lee SE, Kim JH, Kim YA, Jeon YK, Chung DH, Jin X, et al. Psat1-Dependent Fluctuations in alpha-Ketoglutarate Affect the Timing of ESC Differentiation. Cell Metab. 2016;24:494–501. doi: 10.1016/j.cmet.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 37.Mentch SJ, Mehrmohamadi M, Huang L, Liu X, Gupta D, Mattocks D, Gomez Padilla P, Ables G, Bamman MM, Thalacker-Mercer AE, et al. Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab. 2015;22:861–873. doi: 10.1016/j.cmet.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chandel NS, Jasper H, Ho TT, Passegue E. Metabolic regulation of stem cell function in tissue homeostasis and organismal ageing. Nat Cell Biol. 2016;18:823–832. doi: 10.1038/ncb3385. [DOI] [PubMed] [Google Scholar]
- 40.Rafalski VA, Ho PP, Brett JO, Ucar D, Dugas JC, Pollina EA, Chow LM, Ibrahim A, Baker SJ, Barres BA, et al. Expansion of oligodendrocyte progenitor cells following SIRT1 inactivation in the adult brain. Nat Cell Biol. 2013;15:614–624. doi: 10.1038/ncb2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41••.Mohrin M, Shin J, Liu Y, Brown K, Luo H, Xi Y, Haynes CM, Chen D. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science. 2015;347:1374–1377. doi: 10.1126/science.aaa2361. This work shows that the deacetylase SIRT7 responds to mitochondrial stress (unfolded protein response in the mitochondria) and regulates genome-wide the transcription factor NRF1. Interestingly, SIRT7 expression is reduced during aging in mouse HSCs and expression of SIRT7 preserves old HSCs, in part by maintaining their reversible quiescence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang AH, Rando TA. Induction of autophagy supports the bioenergetic demands of quiescent muscle stem cell activation. EMBO J. 2014;33:2782–2797. doi: 10.15252/embj.201488278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Renault VM, Rafalski VA, Morgan AA, Salih DA, Brett JO, Webb AE, Villeda SA, Thekkat PU, Guillerey C, Denko NC, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5:527–539. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gopinath SD, Webb AE, Brunet A, Rando TA. FOXO3 promotes quiescence in adult muscle stem cells during the process of self-renewal. Stem Cell Reports. 2014;2:414–426. doi: 10.1016/j.stemcr.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Webb AE, Pollina EA, Vierbuchen T, Urban N, Ucar D, Leeman DS, Martynoga B, Sewak M, Rando TA, Guillemot F, et al. FOXO3 shares common targets with ASCL1 genome-wide and inhibits ASCL1-dependent neurogenesis. Cell Rep. 2013;4:477–491. doi: 10.1016/j.celrep.2013.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paik JH, Ding Z, Narurkar R, Ramkissoon S, Muller F, Kamoun WS, Chae SS, Zheng H, Ying H, Mahoney J, et al. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell. 2009;5:540–553. doi: 10.1016/j.stem.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warr MR, Binnewies M, Flach J, Reynaud D, Garg T, Malhotra R, Debnath J, Passegue E. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494:323–327. doi: 10.1038/nature11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webb AE, Kundaje A, Brunet A. Characterization of the direct targets of FOXO transcription factors throughout evolution. Aging Cell. 2016;15:673–685. doi: 10.1111/acel.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kennedy BK, Lamming DW. The Mechanistic Target of Rapamycin: The Grand ConducTOR of Metabolism and Aging. Cell Metab. 2016;23:990–1003. doi: 10.1016/j.cmet.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodgers JT, King KY, Brett JO, Cromie MJ, Charville GW, Maguire KK, Brunson C, Mastey N, Liu L, Tsai CR, et al. mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert) Nature. 2014;510:393–396. doi: 10.1038/nature13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lopez-Otin C, Galluzzi L, Freije JM, Madeo F, Kroemer G. Metabolic Control of Longevity. Cell. 2016;166:802–821. doi: 10.1016/j.cell.2016.07.031. [DOI] [PubMed] [Google Scholar]
- 52••.Brandhorst S, Choi IY, Wei M, Cheng CW, Sedrakyan S, Navarrete G, Dubeau L, Yap LP, Park R, Vinciguerra M, et al. A Periodic Diet that Mimics Fasting Promotes Multi-System Regeneration, Enhanced Cognitive Performance, and Healthspan. Cell Metab. 2015;22:86–99. doi: 10.1016/j.cmet.2015.05.012. In this study, the authors report that a fasting-mimicking diet (FMD), which alternates between periods of complete fasting and periods of refeeding, has beneficial effect on stem cells in multiple tissues from old mice. FMD cycles also promote rejuvenation of HSCs, and mesenchymal stem cells (MSCs) as well as the generation of new hippocampal neurons from NSCs. FMD cycles, when initiated at mid-age, extends median mouse lifespan (but not maximal lifespan) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng CW, Adams GB, Perin L, Wei M, Zhou X, Lam BS, Da Sacco S, Mirisola M, Quinn DI, Dorff TB, et al. Prolonged fasting reduces IGF-1/PKA to promote hematopoietic-stem-cell-based regeneration and reverse immunosuppression. Cell Stem Cell. 2014;14:810–823. doi: 10.1016/j.stem.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yilmaz OH, Katajisto P, Lamming DW, Gultekin Y, Bauer-Rowe KE, Sengupta S, Birsoy K, Dursun A, Yilmaz VO, Selig M, et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486:490–495. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Igarashi M, Guarente L. mTORC1 and SIRT1 Cooperate to Foster Expansion of Gut Adult Stem Cells during Calorie Restriction. Cell. 2016;166:436–450. doi: 10.1016/j.cell.2016.05.044. [DOI] [PubMed] [Google Scholar]
- 56.Beyaz S, Mana MD, Roper J, Kedrin D, Saadatpour A, Hong SJ, Bauer-Rowe KE, Xifaras ME, Akkad A, Arias E, et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature. 2016531:53–58. doi: 10.1038/nature17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57•.Fusco S, Leone L, Barbati SA, Samengo D, Piacentini R, Maulucci G, Toietta G, Spinelli M, McBurney M, Pani G, et al. A CREB-Sirt1-Hes1 Circuitry Mediates Neural Stem Cell Response to Glucose Availability. Cell Rep. 2016;14:1195–1205. doi: 10.1016/j.celrep.2015.12.092. In this report, the authors show the metabolic state resulting from excess glucose inhibits the self-renewal so stem cells by a molecular pathway that involves the transcription factor CREB and the NAD(+)-dependent histone demethylase SIRT1. By contrast, low glucose promotes NSC proliferation and self-renewal, and caloric restriction triggers the CREB-SIRT1 metabolic switch in the mouse hippocampus. [DOI] [PubMed] [Google Scholar]
- 58.Horvath S, Erhart W, Brosch M, Ammerpohl O, von Schonfels W, Ahrens M, Heits N, Bell JT, Tsai PC, Spector TD, et al. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci U S A. 2014;111:15538–15543. doi: 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang N, Du G, Tobias E, Wood JG, Whitaker R, Neretti N, Helfand SL. Dietary and genetic effects on age-related loss of gene silencing reveal epigenetic plasticity of chromatin repression during aging. Aging (Albany NY) 2013;5:813–824. doi: 10.18632/aging.100614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bahar R, Hartmann CH, Rodriguez KA, Denny AD, Busuttil RA, Dolle ME, Calder RB, Chisholm GB, Pollock BH, Klein CA, et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- 61.Warren LA, Rossi DJ, Schiebinger GR, Weissman IL, Kim SK, Quake SR. Transcriptional instability is not a universal attribute of aging. Aging Cell. 2007;6:775–782. doi: 10.1111/j.1474-9726.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- 62.Torres CM, Biran A, Burney MJ, Patel H, Henser-Brownhill T, Cohen AS, Li Y, Ben-Hamo R, Nye E, Spencer-Dene B, et al. The linker histone H1.0 generates epigenetic and functional intratumor heterogeneity. Science. 2016;353 doi: 10.1126/science.aaf1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Conboy IM, Conboy MJ, Wagers AJ, Girma ER, Weissman IL, Rando TA. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433:760–764. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- 64.Villeda SA, Plambeck KE, Middeldorp J, Castellano JM, Mosher KI, Luo J, Smith LK, Bieri G, Lin K, Berdnik D, et al. Young blood reverses age-related impairments in cognitive function and synaptic plasticity in mice. Nat Med. 2014 doi: 10.1038/nm.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu J, Ocampo A, Izpisua Belmonte JC. Cellular Metabolism and Induced Pluripotency. Cell. 2016;166:1371–1385. doi: 10.1016/j.cell.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 66.Rando TA. Stem cells, ageing and the quest for immortality. Nature. 2006;441:1080–1086. doi: 10.1038/nature04958. [DOI] [PubMed] [Google Scholar]
- 67.Kowalczyk MS, Tirosh I, Heckl D, Rao TN, Dixit A, Haas BJ, Schneider RK, Wagers AJ, Ebert BL, Regev A. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015;25:1860–1872. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Llorens-Bobadilla E, Zhao S, Baser A, Saiz-Castro G, Zwadlo K, Martin-Villalba A. Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells that Become Activated upon Brain Injury. Cell Stem Cell. 2015;17:329–340. doi: 10.1016/j.stem.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 69.Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming GL, et al. Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell. 2015;17:360–372. doi: 10.1016/j.stem.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Silva-Vargas V, Crouch EE, Doetsch F. Adult neural stem cells and their niche: a dynamic duo during homeostasis, regeneration, and aging. Curr Opin Neurobiol. 2013;23:935–942. doi: 10.1016/j.conb.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 71.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 72.Silva-Vargas V, Maldonado-Soto AR, Mizrak D, Codega P, Doetsch F. Age-Dependent Niche Signals from the Choroid Plexus Regulate Adult Neural Stem Cells. Cell Stem Cell. 2016;19:643–652. doi: 10.1016/j.stem.2016.06.013. [DOI] [PubMed] [Google Scholar]