Abstract
In Bacillus subtilis, Listeria monocytogenes and in two Mycobacteria, it was previously shown that yvcK is a gene required for normal cell shape, for optimal carbon source utilization and for virulence of pathogenic bacteria. Here we report that the B. subtilis protein YvcK binds to Uridine diphosphate-sugars like Uridine diphosphate-Glucose (UDP-Glc) and Uridine diphosphate-N-acetylglucosamine (UDP-GlcNAc) in vitro. Using the crystal structure of Bacillus halodurans YvcK, we identified residues involved in this interaction. We tested the effect of point mutations affecting the ability of YvcK to bind UDP-sugars on B. subtilis physiology and on cell size. Indeed, it was shown that UDP-Glc serves as a metabolic signal to regulate B. subtilis cell size. Interestingly, we observed that, whereas a yvcK deletion results in the formation of unusually large cells, inactivation of YvcK UDP-sugar binding site does not affect cell length. However, these point mutations result in an increased sensitivity to bacitracin, an antibiotic which targets peptidoglycan synthesis. We thus propose that UDP-GlcNAc, a precursor of peptidoglycan, could be a good physiological ligand candidate of YvcK.
Introduction
In bacteria, several cellular processes like cell division, cell size and morphogenesis, are tightly coordinated with metabolism1. For example, during growth in a poor medium, bacterial cell size can be reduced to half the length in comparison to growth in a rich medium2. The Bacillus subtilis YvcK protein might play a role in the maintenance of bacterial shape in a nutrient dependent manner3–5.
Indeed, in B. subtilis, YvcK was previously shown to be unnecessary when bacteria were grown in glycolytic carbon sources but essential for growth in Krebs cycle intermediates and in carbon sources metabolized via the pentose phosphate pathway3. In these carbon sources, yvcK mutant cells display morphologic abnormalities, including bulging cells before lysis. The cell wall, with its major structural component the peptidoglycan (PG), forms the protective barrier that maintains the integrity and the shape of the cell. Its expansion is orchestrated by cytoskeletal proteins6. By deconvolution fluorescence microscopy, it was observed that YvcK has a helix-like cellular localization as it was previously described for MreB, the key actin-like cytoskeletal component in rod-shaped bacteria4, 7. The use of high resolution fluorescence microscopy has revealed that MreB forms motile patches whose movements are dependent on the cell wall synthesis machinery8, 9. The mreB mutant exhibits a characteristic bulging phenotype and PBP1, the major penicillin-binding protein implicated in PG synthesis, is mislocalized10. Amazingly, overproduction of YvcK restores proper PBP1 localization and rescues the morphology defects of cells lacking MreB. Moreover, PBP1 is also delocalized in a yvcK mutant grown in minimal medium supplemented with gluconate and the deletion of the PBP1 encoding gene restores the growth of a yvcK mutant in this restrictive medium4. Thus, all these results suggest that, though its mechanism of action is still unknown, YvcK is required for proper PBP1 localization and normal cell wall biosynthesis or integrity in B. subtilis. Also, a yvcK mutant is more sensitive than the WT strain to bacitracin, an antibiotic that inhibits cell wall synthesis by binding to undecaprenyl-pyrophosphate, the phosphorylated form of the carrier molecule for PG precursors11, 12.
The YvcK protein is present in a wide variety of bacteria and its role has also been investigated in pathogens. In two mycobacterial species, M. smegmatis and M. tuberculosis, the deletion of cuvA, the gene encoding the YvcK orthologue, leads to changes in nutrient uptake and/or metabolism that affect cell wall structure, morphology and bacterial virulence5. Strains lacking cuvA display growth defects in several conditions using different carbon sources. These strains also have morphological defects and a hypersensitivity to several β-lactam antibiotics that inhibit synthesis of PG. In both mycobacterial species, CuvA localizes only to the growing cell pole, the site of PG synthesis5. In Listeria monocytogenes, it has recently been shown that YvcK is also required for cell wall stress responses, growth in glycerol medium, cytosolic survival and virulence13.
To gain information on YvcK cellular function(s), a transposon mutagenesis approach was carried out in order to identify suppressors that rescue the B. subtilis ΔyvcK mutant and the M. tuberculosis ΔcuvA mutant3, 5. This approach was not very informative for B. subtilis despite the isolation of revertants with insertion in yfnI 3, a gene encoding a lipoteichoic acid synthase involved in cell wall synthesis14. However, for M. tuberculosis, the isolation of transductants with insertions in pbpA and rodA, two genes involved in PG synthesis and cell shape control, indicates a link between CuvA and cell wall synthesis and morphology5. This result is reminiscent of the rescue of the B. subtilis yvcK mutant by deletion of the gene encoding PBP14. In addition CuvA specifically localizes to the growing cell pole, where the peptidoglycan synthesis occurs in mycobacteria5, whereas YvcK is localized as a helical-like pattern along the length of the cell4, where the PG synthesis machinery is evenly dispersed allowing a cylindrical elongation of the rod-shaped Bacillus cells8, 9, 15, 16.
This YvcK/CuvA protein belongs to the UPF0052 uncharacterized protein family and its biochemical properties are unknown. It harbors a Rossmann fold, a structural motif that is commonly observed in enzymes regulated by dinucleotide coenzymes such as FAD and NAD(P)17. No binding of mononucleotides like GTP, GDP or ATP, ADP to B. subtilis YvcK was detected but the crystal structure of Bacillus halodurans YvcK was obtained when associated with NAD18. In this paper, we have undertaken the biochemical characterization of YvcK from B. subtilis and we have shown that this protein is able to bind UDP-sugars like UDP-GlcNAc and UDP-Glc in vitro. We have also identified the residues involved in UDP-sugar binding. We observe that inactivation of the UDP-sugar binding site does not affect cell size and growth on non-permissive medium. By contrast, mutants where the ability of YvcK to bind UDP-sugar is affected, have an increased sensitivity to bacitracin suggesting that UDP-GlcNAc, a key precursor of PG, could be the physiological ligand of YvcK.
Results and Discussion
YvcK is a UDP-sugar binding protein
To determine its biochemical properties, the YvcK protein was over-expressed, purified and its ability to bind dinucleotides was analyzed. However, we did not detect any binding with micromolar concentrations of FAD or NAD(P) (reduced and oxidized forms) with all the techniques tested. In an analysis of Lactobacillus rhamnosus genome predicting novel glycosyltransferases, YvcK was proposed as a putative one19. Glycosyltransferases are enzymes which transfer sugar moieties from an activated donor to a specific substrate. Since a yvcK mutant has an increased sensitivity to bacitracin12, we tested the binding of YvcK with two UDP-sugars, UDP-Glc and UDP-GlcNAc, that are cell wall precursor components.
For this purpose, we assessed YvcK for sensitivity to limited protease digestion in vitro. As shown in Fig. 1A, the presence of UDP-GlcNAc weakly modifies YvcK trypsin sensitivity. Indeed, after 20 min of digestion, the band at around 35 kDa corresponding to YvcK is partially degraded to four main bands. In the presence of UDP-GlcNAc, YvcK is still partially degraded to four main bands but the band of 30 kDa is more intense and thus seems to be more resistant to the trypsin cleavage. This result suggests a direct binding of UDP-GlcNAc to YvcK. This binding probably induces conformational changes of YvcK that result in the protection from trypsin cleavage. A similar result was obtained with UDP-Glc (data not shown). To confirm and quantify these interactions we carried out Thermal Shift Assay (TSA); this approach was successfully used to investigate protein-ligand interactions20 and to determine apparent K D 21. By monitoring SYPRO® Orange dye fluorescence in microplates using a thermal cycler, we have quantified the effects of ligands on YvcK melting temperature (Fig. 1B). We found that UDP-GlcNAc and UDP-Glc raise the melting temperature (Tm) of YvcK in a concentration-dependent manner (Fig. 1C). Moreover, the observed increases of Tm (of up to 8 °C) indicate that binding of UDP-sugars significantly stabilizes the structure of YvcK (Fig. 1B). In addition, we observed that YvcK possesses a five-fold higher affinity for the UDP-GlcNAc (apparent K D = 0.41 ± 0.24 mM) than UDP-Glc (apparent K D = 2.11 ± 0.65 mM). We also analyzed the effect of the two moieties of UDP-GlcNAc separately; namely UDP and GlcNAc on the Tm of YvcK. Whereas UDP was able to bind to YvcK, no binding of GlcNAc was detected (Fig. 1C). This observation suggests that the presence of the UDP moiety in the ligand is necessary for the interaction with YvcK. Moreover, UDP alone binds YvcK with an affinity lower than that of the two UDP-sugars tested (apparent K D = 6.21 ± 0.62 mM). This result indicates that the presence of a sugar to the UDP moiety increases the affinity (Fig. 1C) and suggests that YvcK may probably bind other UDP-sugars.
Figure 1.
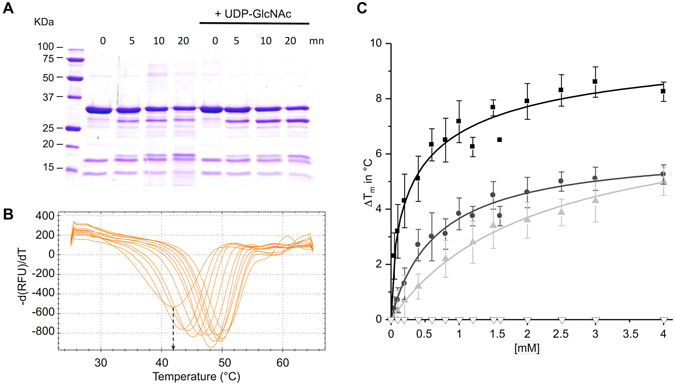
YvcK binds to UDP, UDP-Glc and UDP-GlcNAc. (A) Coomassie-stained SDS-PAGE of YvcK partial proteolysis profile. YvcK was incubated with Trypsin (Promega) in the absence or in the presence of 1 mM UDP-GlcNAc for 0, 5, 10 or 20 min at 37 °C. The digestion profiles were assessed by electrophoresis in 12.5% SDS-PAGE. Full-length gel is presented in Supplementary data. (B) Thermal Shift Assay (TSA) in the presence of increasing concentrations of UDP-GlcNAc. YvcK melting profiles were monitored in the presence of increasing concentration of UDP-GlcNAc (0 to 4 mM). One curve corresponds to data obtained for one concentration of UDP-GlcNAc. The melting temperature of the protein (Tm) is obtained at the midpoint of the each melting curve and corresponds to the minimum of the negative derivative curves (see the arrow that indicates the Tm of YvcK in the absence of UDP-GlcNAc). The Tm is an indicator of protein stability and is increased by the addition of UDP-GlcNAc (Tm = 42 °C in the absence of UDP-GlcNAc and Tm = 50.5 °C in the presence of 4 mM UDP-GlcNAc). (C) TSA results for the binding of UDP, UDP-Glc, UDP-GlcNAc and Glc-NAc. Assays were performed in the presence of increasing concentrations of ligands (0 to 4 mM). The difference of temperature (the shift of Tm induced by the presence of ligand) was plotted against the concentration of UDP (black circle), UDP-Glc (grey triangle), UDP-GlcNAc (black square) and Glc-NAc (white triangle). All the curves correspond to the average of data from at least 3 independent experiments and the standard deviations are representated by the error bars. Curve fitting was performed by using Microcal Origin 5.0 software.
YvcK binds UDP-Glc and UDP-GlcNAc with relatively low apparent affinities and we can wonder if the binding of these two UDP-sugars has any significance in vivo. Up to now, their intracellular concentrations were not determined in B. subtilis but we can suppose that they are likely high in actively growing cells because bacteria use large amounts of these metabolites in their peptidoglycan, teichoic acids and lipopolysaccharides biosynthesis. For example in Escherichia coli, the intracellular concentration of UDP-GlcNAc was determined and, in the conditions tested i.e. exponential phase growth in LB medium, the value obtained was approximately 0.43 mM22. This value belongs to the same scale to that of the apparent affinity of B. subtilis YvcK for this metabolite. Consequently, if the concentration of UDP-GlcNAc in B. subtilis cells is similar to that in E. coli cells, it is conceivable that UDP-GlcNAc could bind to YvcK in vivo.
A yvcK deletion affects cell length
In B. subtilis, UDP-Glc serves as a metabolic signal to regulate cell size in response to enhanced carbon availability by a well-characterized mechanism mediated by UgtP, an enzyme involved in the synthesis of glycolipids and the anchoring of lipoteichoic acids23–25. Under rich conditions, UgtP binds to UDP-Glc and is concentrated at midcell and inhibits FtsZ in rapidly growing cells, causing a delay of cell division. Under poor growth conditions, when the availability of UDP-Glc is low, UgtP is free and appears to be sequestered in randomly distributed foci where it is presumably unable to modulate FtsZ assembly. This change of UgtP localization seems to be controlled by the availability of UDP-Glc, a substrate of UgtP; a deletion of ugtP gene results in reduced cell size, in particular under rich growth conditions25, 26. Because YvcK function is in relation with cell wall synthesis (or integrity) and it also binds UDP-Glc in vitro, we wonder if it participates in cell size regulation. For this purpose, we constructed a non-polar markerless yvcK deletion mutant and checked that the genes downstream of yvcK were correctly expressed. Indeed it was previously observed that cells deleted for yvcL, the gene just downstream of yvcK, are longer than WT cells27.
For these experiments concerning the analysis of the ΔyvcK cell size, we chose experimental conditions in which yvcK mutant strain has a growth rate comparable to that of the WT strain; we thus excluded media where the sole carbon source is a Krebs cycle intermediate or a substrate of pentose phosphate pathway3. As it was previously done for the studies of ugtP mutant25, 26, we used two growth media. In the first one, we used minimal medium supplemented with sorbitol (glucitol) as sole carbon source; in these poor growth conditions the UDP-Glc intracellular concentration is supposed to be low and the cells short. The second medium used is LB; in these rich growth conditions the UDP-Glc intracellular concentration is supposed to be high, the cells large and the effect of a ugtP deletion was amplified. As expected, the yvcK mutant strain growth in minimal medium containing sorbitol is similar to that of the WT strain indicating that the growth rate is not impaired (Fig. 2A) and ref. 3. However, the yvcK mutant cells (average size about 2.9 μm) seem larger than the WT cells (average size about 2.4 μm) (Fig. 2B). This size increase is weak but, in this poor growth condition, cells are short25, 26. To confirm that this increase of cell length is really due to the absence of YvcK, we constructed a new strain by introducing a copy of yvcK allele fused to gfp under the control of P xyl promoter in the yvcK mutant strain. Then we analyzed the length of cells grown in the presence of xylose, in a LB rich medium where the bacterial cell size is large25, 26. We showed that, as observed in minimal medium, in LB medium the yvcK mutant cells are larger than the WT cells (average sizes about 6.3 μm for ΔyvcK cells vs 4.7 μm for WT cells) and this cell size increase is more obvious than in poor medium (Fig. 2C). Furthermore, in presence of xylose, the yvcK mutant cells overexpressing yvcK-gfp gene fusion under the control of P xyl, are smaller than the WT cells (about 0.6 μm of difference; average size about 4.1 μm for cells overproducing YvcK-GFP) (Fig. 2C and D). We conclude that YvcK regulates cell length and its gene deletion induces larger cells.
Figure 2.
YvcK regulates cell length. (A) Growth curve of WT strain (168) and ΔyvcK mutant strain (SG471) on minimal medium with sorbitol as sole carbon source. (B) Histogram of cell lengths for 168 and SG471 strains collected at OD600 = 0.4: WT (n = 306), ΔyvcK strain (n = 335). (C) Histogram of cell lengths for WT strain (168), ΔyvcK strain (SG471) and ΔyvcK strain with P xyl yvcK at amyE locus (SG517). These three strains were grown in LB medium in the presence of xylose and collected at OD600 = 0.4: WT (n = 319), ΔyvcK strain (n = 256), ΔyvcK, P xyl yvcK strain (n = 301). (D) Analysis of YvcK production from crude extracts of WT strain (168), ΔyvcK strain (SG471) and ΔyvcK strain with P xyl yvcK at amyE locus (SG517) by Western blotting. Bacteria were collected at OD600 = 0.4, lyzed and 10 μl of cell extracts were run on a 12.5% SDS-PAGE and transferred to nitrocellulose membrane by electroblotting. The blot membrane was exposed for each time set in interval time and accumulates images. Full-length blot is presented in Supplementary data.
It was recently shown that the availability of PG precursors plays an essential role in determining cellular dimensions across diverse species of bacteria28. Furthermore, another study showed that knockdowns of genes encoding PG biosynthesis enzymes also produced elongated B. subtilis cells29. The cell size increase of a yvcK mutant could be due to an alteration of PG synthesis but it can also have other causes and could also be related to the UDP-Glc and UgtP pathway.
A yvcK deletion enhances the effect of deletions of ugtP and gtaB genes
The two proteins YvcK and UgtP are both able to bind UDP-Glc but, contrarily to a ugtP deletion that results in the formation of unusually small cells25, we showed here that a deletion of yvcK produces large daughter cells. To study these antagonist effects, we decided to construct an ugtP yvcK double mutant in order to measure its cell length and compare it to that of a WT, a ugtP mutant and a yvcK mutant in a rich LB medium (Fig. 3A and B). Remarkably, the cell length distribution of the ugtP yvcK double mutant resembles that of the ugtP mutant but cells of the double mutant are even smaller than that of the single mutant (about 3.1 μm vs 3.5 μm) as if a yvcK deletion enhances the effect of a ugtP deletion. This result is unexpected because it is the opposite effect of that obtained with the single deletion yvcK mutant that causes elongated cells.
Figure 3.

A yvcK deletion enhances the effect of deletions of ugtP and gtaB genes. Strains were grown on LB medium until OD600 = 0.4. (A) Micrographs of WT strain (168) and mutant strains ΔyvcK strain (SG471), ΔugtP strain (SG571), ΔugtP ΔyvcK strain (SG561). Membranes were stained with FM1-43. (B) Histogram of cell lengths for WT strain (n = 1032), ΔyvcK strain (n = 611), ΔugtP strain (n = 970) and ΔugtP ΔyvcK strain (n = 626). (C) Simplified scheme of the first steps of the glycolipid metabolism. Enzymes involved in this pathways are PgcA: Phosphoglucomutase, GtaB: UTP-glucose-1-phosphate uridylyltransferase, UgtP: Processive diacylglycerol beta-glucosyltransferase. (D) Micrographs of WT strain (168) and mutant strains ΔyvcK strain (SG471), ΔgtaB strain (DKE35670), ΔgtaB ΔyvcK strain (SG562). Membranes were stained with FM1-43. E- Histogram of cell lengths for WT strain (n = 390), ΔyvcK strain (n = 183), ΔgtaB strain (n = 355) and ΔgtaB ΔyvcK strain (n = 270).
UgtP is not only an FtsZ inhibitor; it is also the third and the last enzyme of the glycolipid biosynthesis pathway. In this pathway, UDP-Glc is generated from Glc-6-P by two reversible steps, catalyzed by the two upstream enzymes PgcA or GtaB (Fig. 3C). A deletion of ugtP, gtaB or pgcA genes causes a similar cell length phenotype, i.e. reduced cell size when bacteria were grown in rich medium25. In order, to test whether the effect of a yvcK deletion associated with a deletion of a gene encoding another enzyme of the glycolipid biosynthesis pathway (in conditions where UDP-Glc synthesis is altered), we constructed a gtaB yvcK double mutant and analyzed its cell size in LB rich conditions (Fig. 3D and E). We observed that, as expected the gtaB simple mutant cells are smaller than WT cells but larger than that of the ugtP mutant. In addition, the cell length distribution showed that the gtaB yvcK mutant cells are smaller than the gtaB mutant cells and similar to that of the ugtP yvcK mutant. Indeed, for both double mutants, the average cell size is about 1.9 or 2 μm shorter than that of the WT cells (Fig. 3B and E). These data suggest that a yvcK deletion enhances the effect of deletions of genes encoding an enzyme of the glycolipid biosynthesis pathway, ugtP or gtaB, to obtain critically short cells.
Characterization of the YvcK residues involved in the binding of UDP-sugars
To gain more information on the mysterious role of YvcK on cell size, we analyzed mutants of yvcK encoding YvcK protein affected in its ability to UDP-sugar. YvcK from B. haludurans possesses 63% identity and 86% similarity with YvcK from B. subtilis (Fig. 4A). The structure of B. haludurans YvcK was previously obtained in complex with NAD (Fig. 4B). The residues found to interact with the dinucleotide in the crystal structure are conserved in B. subtilis YvcK (Fig. 4A). The structures of the three molecules NAD, UDP-Glc and UDP-GlcNAc possess similarities; the 2 phosphates and a D-ribose can be overlaid (Fig. 4C). We can thus speculate that some residues found to interact with NAD in the YvcK structure, Thr13, Asn217, Tyr264 and Arg300 (numbering from B. halodurans), could interact with UDP-Glc (+/−NAc). To test this hypothesis, we replaced these four amino acids by Ala in B. subtilis YvcK protein. The corresponding proteins, YvcK-T14A, YvcK-N218A, YvcK-Y265A and YvcK-R301A, were overexpressed and purified as for the wild-type enzyme. Then, their proteolysis pattern was checked; for each mutant protein, it was similar to that of the WT protein in the absence of UDP-GlcNAc suggesting that the overall structure seemed to be unaffected by the introduced mutations (data not shown). Hence, their ability to bind UDP-GlcNAc was tested by TSA. In the absence of nucleotide, the four mutant proteins have a Tm around 40+/− 2 °C (Tm of the WT protein is about 42 °C see Fig. 1B) confirming that the global structure is not affected by the mutations. We found that the ability to bind UDP-GlcNAc is affected at different levels in the four modified proteins (Fig. 4D and E). In particular, whereas the replacement of Thr14 by an Ala, and at a less extent that of Asn218 by an Ala, slightly affects the binding of UDP-GlcNAc, the replacement of Tyr265 or Arg301 by Ala strongly affects this binding. These results show that some residues found to interact with NAD in the structure interact with UDP-sugars in vitro.
Figure 4.
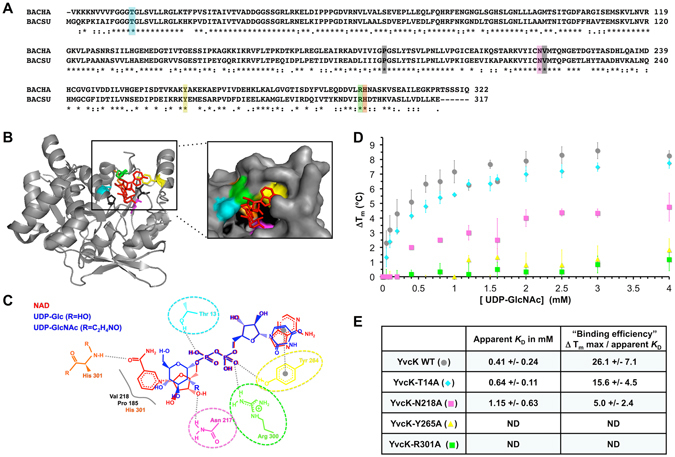
Identification of residues involved in UDP-sugars binding. (A) Sequence alignment of YvcK from B. subtilis and from B. halodurans. The highlighted amino acids are those found to interact with NAD in the crystal structure. (B) Crystal structure of a monomer of B. halodurans YvcK in complex with NAD (PDB ID: 2O2Z). The colored residues of YvcK were found to interact with NAD. The figure was realized with PyMOL software. (C) Identification of YvcK residues susceptible to interact with UDP-sugars. This scheme generated by PoseView software represents the residues of B. halodurans YvcK that interact with NAD. A molecule of UDP-Glc or UDP-GlcNAc was superposed to NAD and residues of YvcK that could interact with UDP-Glc or UDP-GlcNAc are encircled. (D) TSA results for the binding of UDP-GlcNAc to B. subtilis YvcK modified proteins. Four YvcK residues found to interact with NAD in the crystal structure were replaced by Ala to generate YvcK-T14A, YvcK-N218A, YvcK-Y265A and YvcK-R301A. The proteins were purified and TSA was performed in the presence of increasing concentrations of UDP-GlcNAc (0 to 4 mM). The difference of temperature was plotted against the concentration of UDP-GlcNAc for YvcK (grey circle) and each modified protein: YvcK-T14A (cyan diamond), YvcK-N218A (pink square), YvcK-Y265A (yellow triangle) and YvcK-R301A (green square). All the values correspond to the average of data from at least 3 independent experiments and the standard deviations are representated by the error bars. (E) Kinetic parameters for the binding of UDP-GlcNAc to B. subtilis YvcK modified proteins from TSA experiments. The apparent K D, the Δ Tm max and the standard deviations were calculated using Microcal Origin 5.0 software. ND indicates that the binding of UDP-GlcNAc to YvcK and the Δ Tm values are too weak to calculate an apparent K D.
Point mutations of the UDP-sugar binding site do not affect neither cell size in rich medium nor growth and morphology in minimal medium supplemented with gluconate
Since YvcK regulates cell length, we investigated whether yvcK point mutant cells are larger than WT cells when its ability to bind UDP-Glc is affected by point mutations. For this purpose, we constructed non-polar markerless strains expressing the mutated alleles of yvcK at a physiological level. After growth of these strains in LB medium, we carried out western blot analysis in order to verify the expression level of each mutant protein (YvcK-T14A, YvcK-N218A, YvcK-Y265A and YvcK-R301A). We observed that, as expected, the growth of the mutant strains was normal and similar to WT strain (data not shown) and that each mutant protein was expressed (Fig. 5A). Then, we analyzed the cell size of these strains grown in LB medium (Fig. 5B). However, surprisingly, when we measured the cell size and in particular the average length of each strain, we did not find any obvious correlation between the increase of cell length and the inability of the corresponding mutant YvcK protein to bind UDP-sugars (Fig. 5B vs Fig. 4D and E). Indeed cells expressing the proteins YvcK-Y265A or YvcK-R301A, whose ability to bind UDP-sugars is almost lost, have an average length similar to that of the strains expressing WT YvcK or YvcK-N218A, whose ability to bind UDP-sugars is moderately affected. Cells expressing the YvcK-T14A protein, whose ability to bind UDP-sugars is slightly affected, have an average cell length between that WT and yvcK mutant strains (about 5.4 μm vs 5.0 μm and 6.0 μm, respectively).
Figure 5.
Mutations of residues interacting with UDP-sugars do not affect cell length and bacterial growth on minimal medium containing gluconate as sole carbon source. (A) Analysis of YvcK production by Western blotting. WT strain (168), ΔyvcK strain (SG471) and strains producing YvcK-T14A (SG520), YvcK-N218A (SG521), YvcK-Y265A (SG522) and YvcK-R301A (SG523) were grown at 37 °C in 20 ml of LB medium and collected at OD600 = 1. 16 μl of cell extracts were run on a 12.5% SDS-PAGE and transferred to nitrocellulose membrane by electroblotting. The blot membrane was read by the LAS4000 mini Imager. Full-length blot is presented in Supplementary data. (B) Bar graph of cell length averages for WT strain (168), ΔyvcK strain (SG471) and strains producing YvcK-T14A (SG520), YvcK-N218A (SG521), YvcK-Y265A (SG522) and YvcK-R301A (SG523). Bacteria were grown on LB medium until OD600 = 0.4 and analyzed by microscopy. The data were obtained from four independent experiments and the standard deviations are representated by the error bars. (C) Effect of yvcK point mutations on B. subtilis growth on gluconate as sole carbon source. WT strain (168; grey circle), ΔyvcK strain (SG471; empty grey diamond) and strains producing YvcK-T14A (SG520; cyan diamond), YvcK-N218A (SG521; YvcK pink square), YvcK-Y265A (SG522; yellow triangle) and YvcK-R301A (SG523; green square) were grown overnight on LB medium with 1% glucose. After centrifugation, cells were grown in CE-gluconate liquid medium at 37 °C supplemented with 0.5% xylose. (D) Effect of yvcK point mutations on cell morphology during growth on gluconate as sole carbon source. Cells were grown in minimal medium supplemented with gluconate at 37 °C and collected after 300 mn of growth. Their morphology was analyzed by microscopy: see micrographs of WT strain (168; grey circle), ΔyvcK strain (SG471; empty grey diamond) and strains producing YvcK-T14A (SG520; cyan diamond), YvcK-N218A (SG521; pink square), YvcK-Y265A (SG522; yellow triangle) and YvcK-R301A (SG523; green square).
We also tested the effect of these four substitutions on bacterial growth in minimal medium containing gluconate as source of carbon (Fig. 4C). In these growth conditions, a strain deleted for yvcK has a growth defect3, 4. All the strains expressing the YvcK modified proteins have a normal growth and a normal cell shape, even for the strains expressing the proteins YvcK-Y265A or YvcK-R301A, whose ability to bind UDP-sugars is strongly disturbed (Fig. 4C and D). By contrast, as expected, the strain deleted for yvcK cannot grow normally in this medium. These data indicate that the ability of YvcK to bind UDP-sugars is not crucial for B. subtilis growth and morphology in minimal medium containing gluconate.
Point mutations of the UDP-sugar binding site affect bacitracin sensitivity
We previously observed that a yvcK mutant has an increased bacitracin sensitivity in comparison with the WT strain12. This antibiotic inhibits cell wall biosynthesis by interfering with the dephosphorylation of C55-isoprenyl pyrophosphate, a membrane carrier molecule that transports the building-blocks of the PG11, 12. Because UDP-sugars and particularly UDP-GlcNAc are key precursors of PG, we tested the effect of point mutations of the YvcK UDP-sugar binding site on bacitracin sensitivity. Thus, for each mutant strain, we determined the bacitracin concentration needed to inhibit half of the bacterial growth (Table 1). We observed that strains expressing the YvcK-Y265A or YvcK-R301A protein, whose ability to bind UDP-GlcNAc is almost lost in vitro (Fig. 4D), have IC50 values similar to the yvcK mutant strain (about 135 μg/ml and 172 μg/ml for strains producing YvcK-Y265A or YvcK-R301A protein respectively and 139 μg/ml for the yvcK mutant strain). By contrast, strains expressing the YvcK-T14A or YvcK-N218A protein, whose ability to bind UDP-sugars is more weakly affected (Fig. 4D and E), have IC50 values that tend towards that of the WT strain (about 282 μg/ml and 261 μg/ml for strains producing YvcK-T14A or YvcK-N218A protein respectively and 368 μg/ml for the WT strain). These data show a correlation between the increase of bacitracin sensitivity and the alteration of the corresponding mutant YvcK protein to bind UDP-GlcNAc and demonstrate a physiological role for the residues involved in UDP-sugars binding. UDP-GlcNAc being a key precursor of PG and bacitracin targeting the synthesis of PG, we propose that this UDP-sugar could be a good candidate to be the physiological ligand of YvcK.
Table 1.
Effect of yvcK point mutations on antibiotic resistance.
| Strains | Genotypes | Bacitracin IC50 in μg/ml |
|---|---|---|
| 168 | trpC2 | 368 ± 27 |
| SG471 | trpC2, yvcKΔ1 | 139 ± 18 |
| SG520 | trpC2, yvcK T14A | 282 ± 22 |
| SG521 | trpC2, yvcK N218A | 261 ± 63 |
| SG522 | trpC2, yvcK Y265A | 135 ± 30 |
| SG523 | trpC2, yvcK R301A | 172 ± 39 |
Bacitracin resistance of WT strain (168) and ΔyvcK strain (SG471) used as controls and of strains producing YvcK-T14A (SG520), YvcK-N218A (SG521), YvcK-Y265A (SG522) and YvcK-R301A (SG523) was determined in 96-well microplates. At the end of the incubation period, OD600 nm was monitored using a Tecan microplate reader. Results are expressed as average of IC50 values (i.e. bacitracin concentrations inhibiting 50% of bacterial growth) obtained in at least 3 independent experiments ± standard deviations.
Furthermore, because alteration of UDP-sugar binding site does not have any influence on growth in gluconate medium and no obvious effect on cell size but affects sensitivity to bacitracin, this implies that YvcK has at least two cellular roles. One, independent of its ability to bind UDP-sugars, is involved in the regulation of cell size and of bacterial growth and morphology where the sole carbon source available is a Krebs cycle intermediate or a substrate of the pentose phosphate pathway. Another one, dependent on its ability to bind UDP-sugars, is related to bacitracin sensitivity and potentially to PG synthesis. This multifunctional aspect of YvcK was already mentioned in previous studies. Indeed, YvcK was found to be phosphorylated in B. subtilis, in M. tuberculosis and in L. monocytogenes 5, 12, 13, 30. The role of this phosphorylation was analyzed by mutations of the phosphorylated residue(s) in YvcK and inactivation of the corresponding kinase. In B. subtilis, it was shown that YvcK phosphorylation is irrelevant to growth in restrictive medium but is involved in bacitracin sensitivity12. In L. monocytogenes, YvcK phosphorylation is unrelated to metabolism and cell wall stress responses, but has a key role in cytosolic survival and virulence13. Phosphorylated residues are not conserved across the bacterial species but, in these three bacteria, they are all located in the C-terminus part of YvcK. In B. subtilis, we can imagine that the phosphorylation of Thr304, that is located near the Arg301 in the sequence and in the structure, modifies YvcK ability to bind UDP-sugars and thus the bacitracin sensitivity of Bacillus cells.
Conclusion
In this paper we showed that YvcK is a UDP-sugar binding protein whose gene deletion affects cell size. The molecular mechanism by which a yvcK deletion influences cell size is unknown but it will continue to be explored in the future. Altogether, our data suggest that YvcK’s intracellular role is complex. It is a protein with at least two cellular roles that can be dissociated by inactivation of the UDP-sugar binding site or of the phosphorylation site. One of YvcK’s function is independent of its ability to bind UDP-sugars (and of its level of phosphorylation) and is involved in the control of cell size and in the regulation of growth and morphology in Krebs cycle intermediates and substrates of the pentose phosphate pathway. The other one is dependent on its ability to bind UDP-sugars (and of its level of phosphorylation) and is associated with bacitracin sensitivity and potentially with PG synthesis.
Experimental Procedures
Plasmid and strain constructions
Standard procedures for molecular cloning and cell transformation of B. subtilis or E. coli were used.
To generate a non-polar markerless yvcK deletion mutant, we first replaced the yvcK gene by a chloramphenicol gene cassette, carrying out isothermal assembly31. For that, the upstream and downstream regions of yvcK were amplified by PCR and mixed with a DNA fragment encoding chloramphenicol resistance. The three DNA fragments were assembled into a single big fragment, which was used to transform 168 WT strain. Then, the antibiotic resistance cassette was removed using the plasmid pDR244, available in the Bacillus Genetic Stock Center (http://www.bgsc.org/). The expression of the genes downstream yvcK was checked by qRT PCR.
To introduce point mutations in yvcK gene in B. subtilis chromosome, first we amplified the yvcK mutant alleles from pEFK15 to pEFK18, the upstream and downstream regions of yvcK, and mixed with the DNA fragment encoding chloramphenicol resistance. For each point mutation, the four DNA fragments were isothermally assembled into a single big fragment, used to transform 168 WT strain. Then, the antibiotic gene cassette was removed using the plasmid pDR244.
In B. subtilis strains, yvcKgfp gene fusion was expressed from the P xyl promoter in the presence of 0.5% xylose. All the strains and plasmids used in this study are listed in Tables 2 and 3, respectively. Primer sequences are available upon request.
Table 2.
B. subtilis strains used in this study.
| Strains | Relevant structures or Genotypes | References |
|---|---|---|
| 168 | trpC2 | Laboratory stock |
| SG470 | trpC2, yvcK::cat | This study |
| SG471 | trpC2, yvcKΔ1 | pDR244 → SG470 |
| SG517 | trpC2, yvcKΔ1, amyE::(P xyl yvcKgfp), spec | pEFK6 → SG471 |
| BTM218 | ugtP::erm | Laboratory stock |
| SG571 | ugtPΔ1 | pDR244 → BTM218 |
| BKE35670 | gtaB::erm | (Koo et al., unpublished) BGSC* |
| SG567 | trpC2, yvcK::cm, ugtP::erm | BTM218 → SG470 |
| SG561 | trpC2, yvcKΔ1, ugtPΔ1 | pDR244 → SG567 |
| SG562 | trpC2, yvcKΔ1, gtaB::erm | BKE35670 → SG471 |
| SG520 | trpC2, yvcK T14A | This study |
| SG521 | trpC2, yvcK N218A | This study |
| SG522 | trpC2, yvcK Y265A | This study |
| SG523 | trpC2, yvcK R301A | This study |
Table 3.
Plasmids used in this study.
Site-directed Mutagenesis
Point mutations were introduced into the gene by site-directed mutagenesis by PCR amplification of the whole pEFK6 plasmid. The resulting constructs were verified by DNA sequencing.
Growth tests
B. subtilis strains were grown in LB and CE-minimal medium as previously described3.
Expression and Purification of YvcK Proteins
E. coli AD494 was transformed with PQE30-YvcK (wild type or mutated allele) and the resulting strains were used for expression and purification of His6-tagged YvcK with NTA resin (Qiagen) as described in refs 12, 32.
Limited proteolysis
For each 20 μl sample, 5 μg of YvcK were pre-incubated for 10 mn at 37 °C with 40 mM NaCl, 1 mM MgCl2, 10 mM Tris/HCl, pH 8.0 in the absence or in the presence of UDP-GlcNAc (1 mM). After addition of 0.005 μg of Trypsin (Promega), the reaction mixture was incubated for 0, 5, 10 or 20 min at 37 °C. The digestion was stopped by adding an equal volume of electrophoresis loading buffer to the assay mixtures and by heating 5 min at 100 °C before applying the samples onto a 12.5% sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Thermal Shift Assay (TSA)
In thin-walled 96-well PCR plates, each well (20 μl) contained 10 μM of YvcK protein and 2 μl of the fluorescent SYPRO® Orange dye solution (Molecular Probes, 5000x, diluted to 100x in water), in 40 mM NaCl, 1 mM MgCl2, 10 mM Tris/HCl, pH 8.0 and was heated from 25 °C to 65 °C in 0.5 °C steps. The fluorescence intensity (Ex/Em = 470/570 nm) of SYPRO® Orange was monitored using a real-time PCR apparatus CFX96 (Bio-Rad). The fluorescence of SYPRO® Orange dye changes when it interacts with the protein undergoing thermal unfolding. The denaturation temperature (Tm) was analyzed from the melt peak using CFX Manager software (Bio-Rad). The shift of Tm (ΔTm) induced by the presence of ligand was plotted against the concentration of ligand. Curve fitting was performed by using Microcal Origin 5.0 software using the following equation y = ΔTm max * xn/(apparent K D n + xn), were n is the cooperative binding site.
Western blot
The cells were grown at 37 °C in LB medium to OD600 = 1 then 1 ml of culture was centrifuged for 1 min at 14000 rpm. Cell pellets were resuspended in 1/10 volume of lysis buffer containing 50 mM HEPES pH 8.0, 200 mM NaCl, 1 mM DTT, 1 mM MgCl2, 1 mM CaCl2, 1 mM PMSF, 25 U/ml benzonase and 0.6 mg/ml lyzozyme. Extracts were incubated for 30 min at 37 °C then 0.5% SDS were added at room temperature for 30 min. Samples were afterwards heated at 100 °C for 10 min. Samples were run on a 12.5% SDS-PAGE and transferred to nitrocellulose membrane by electroblotting. The membrane was blocked with phosphate buffer saline (PBS) solution containing 5% milk powder (w/v), for 2 h at room temperature with shaking. Then, the membrane was incubated either with anti-GFP antibody (AgroBio) used at 1/10000 dilution and the secondary antibody, a goat anti-rabbit IgG-HRP (Thermo Fisher) used at 1/2000 or with anti-YvcK antibody used at 1/2500 dilution and the secondary antibody, a goat anti-rabbit IgG-HRP (Thermo Fisher) used at 1/2000. After three washes, the Blot membrane was reading by the LAS4000 mini Imager (by chemiluminescence).
Microscopy
Strains were grown in LB medium or in minimal medium at 37 °C. Live cells were analyzed by microscopy on a Zeiss Upright Axio Imager M2 microscope as described previously12 and stained for membrane using FM1-43 (Invitrogen) where necessary. Cell length was calculated as the distance between adjacent septa.
Measurement of bacitracin resistance
The antibiotic concentration giving 50% growth inhibition (IC50) was determined using the microplate assay described previously33.
Electronic supplementary material
Acknowledgements
This research was supported by the CNRS, the ANR and Aix-Marseille University. We thank Y. Denis for his help with the thermal cycler apparatus and software at the IMM Transcriptomic facility. We thank also F. Pompeo for stimulating discussion and critical reading of the manuscript and B. Khadaroo for toning down our French touch.
Author Contributions
E.F. designed, performed and analyzed the experiments and A.G. conceived the study and wrote the paper. All the authors analyzed the results and approved the final version of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-04064-2
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wang JD, Levin PA. Metabolism, cell growth and the bacterial cell cycle. Nat Rev Microbiol. 2009;7:822–827. doi: 10.1038/nrmicro2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schaechter M, Maaloe O, Kjeldgaard NO. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 3.Görke, B., Foulquier, E. & Galinier, A. YvcK of Bacillus subtilis is required for a normal cell shape and for growth on Krebs cycle intermediates and substrates of the pentose phosphate pathway. Microbiology151, 3777–3791, doi:151/11/3777 [pii]10.1099/mic.0.28172-0 (2005). [DOI] [PubMed]
- 4.Foulquier E, Pompeo F, Bernadac A, Espinosa L, Galinier A. The YvcK protein is required for morphogenesis via localization of PBP1 under gluconeogenic growth conditions in Bacillus subtilis. Mol Microbiol. 2011;80:309–318. doi: 10.1111/j.1365-2958.2011.07587.x. [DOI] [PubMed] [Google Scholar]
- 5.Mir M, et al. Mycobacterial Gene cuvA Is Required for Optimal Nutrient Utilization and Virulence. Infect Immun. 2014;82:4104–4117. doi: 10.1128/IAI.02207-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenton AK, Gerdes K. Direct interaction of FtsZ and MreB is required for septum synthesis and cell division in Escherichia coli. EMBO J. 2013;32:1953–1965. doi: 10.1038/emboj.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones LJ, Carballido-López R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/S0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 8.Domínguez-Escobar J, et al. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 9.Garner EC, et al. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawai Y, Daniel RA, Errington J. Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol Microbiol. 2009;71:1131–1144. doi: 10.1111/j.1365-2958.2009.06601.x. [DOI] [PubMed] [Google Scholar]
- 11.Storm DR, Strominger JL. Complex formation between bacitracin peptides and isoprenyl pyrophosphates. The specificity of lipid-peptide interactions. J Biol Chem. 1973;248:3940–3945. [PubMed] [Google Scholar]
- 12.Foulquier E, et al. PrkC-mediated Phosphorylation of Overexpressed YvcK Protein Regulates PBP1 Protein Localization in Bacillus subtilis mreB Mutant Cells. J Biol Chem. 2014;289:23662–23669. doi: 10.1074/jbc.M114.562496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pensinger DA, et al. The Listeria monocytogenes PASTA Kinase PrkA and Its Substrate YvcK Are Required for Cell Wall Homeostasis, Metabolism, and Virulence. PLoS Pathog. 2016;12:e1006001. doi: 10.1371/journal.ppat.1006001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wörmann ME, Corrigan RM, Simpson PJ, Matthews SJ, Gründling A. Enzymatic activities and functional interdependencies of Bacillus subtilis lipoteichoic acid synthesis enzymes. Mol Microbiol. 2011;79:566–583. doi: 10.1111/j.1365-2958.2010.07472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Errington J. Bacterial morphogenesis and the enigmatic MreB helix. Nat Rev Microbiol. 2015;13:241–248. doi: 10.1038/nrmicro3398. [DOI] [PubMed] [Google Scholar]
- 16.Randich AM, Brun YV. Molecular mechanisms for the evolution of bacterial morphologies and growth modes. Front Microbiol. 2015;6:580. doi: 10.3389/fmicb.2015.00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rossmann MG, Argos P. Protein folding. Annu Rev Biochem. 1981;50:497–532. doi: 10.1146/annurev.bi.50.070181.002433. [DOI] [PubMed] [Google Scholar]
- 18.Forouhar F, et al. Molecular insights into the biosynthesis of the F420 coenzyme. J Biol Chem. 2008;283:11832–11840. doi: 10.1074/jbc.M710352200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sánchez-Rodríguez A, et al. A network-based approach to identify substrate classes of bacterial glycosyltransferases. BMC Genomics. 2014;15:349. doi: 10.1186/1471-2164-15-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rupesh KR, Smith A, Boehmer PE. Ligand induced stabilization of the melting temperature of the HSV-1 single-strand DNA binding protein using the thermal shift assay. Biochem Biophys Res Commun. 2014;454:604–608. doi: 10.1016/j.bbrc.2014.10.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grøftehauge MK, Hajizadeh NR, Swann MJ, Pohl E. Protein-ligand interactions investigated by thermal shift assays (TSA) and dual polarization interferometry (DPI) Acta Crystallogr D Biol Crystallogr. 2015;71:36–44. doi: 10.1107/S1399004714016617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Namboori SC, Graham DE. Enzymatic analysis of uridine diphosphate N-acetyl-D-glucosamine. Anal Biochem. 2008;381:94–100. doi: 10.1016/j.ab.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levin PA, Angert ER. Small but Mighty: Cell Size and Bacteria. Cold Spring Harb Perspect Biol. 2015;7:a019216. doi: 10.1101/cshperspect.a019216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vadia S, Levin PA. Growth rate and cell size: a re-examination of the growth law. Curr Opin Microbiol. 2015;24:96–103. doi: 10.1016/j.mib.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weart RB, et al. A metabolic sensor governing cell size in bacteria. Cell. 2007;130:335–347. doi: 10.1016/j.cell.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chien AC, Zareh SK, Wang YM, Levin PA. Changes in the oligomerization potential of the division inhibitor UgtP co-ordinate Bacillus subtilis cell size with nutrient availability. Mol Microbiol. 2012;86:594–610. doi: 10.1111/mmi.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Surdova K, et al. The conserved DNA-binding protein WhiA is involved in cell division in Bacillus subtilis. J Bacteriol. 2013;195:5450–5460. doi: 10.1128/JB.00507-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris LK, Theriot JA. Relative Rates of Surface and Volume Synthesis Set Bacterial Cell Size. Cell. 2016;165:1479–1492. doi: 10.1016/j.cell.2016.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters JM, et al. A Comprehensive, CRISPR-based Functional Analysis of Essential Genes in Bacteria. Cell. 2016;165:1493–1506. doi: 10.1016/j.cell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang CM, et al. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 2005;19:1692–1704. doi: 10.1101/gad.1311105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 32.Galinier A, et al. The Bacillus subtilis crh gene encodes a HPr-like protein involved in carbon catabolite repression. Proc Natl Acad Sci USA. 1997;94:8439–8444. doi: 10.1073/pnas.94.16.8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohki R, et al. The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol Microbiol. 2003;49:1135–1144. doi: 10.1046/j.1365-2958.2003.03653.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


