Abstract
Fargesia Franchet emend. Yi is closely allied with Thamnocalamus Munro but differs in many major morphological characteristics. Based on traditional morphological characters, it is difficult to differentiate these two genera. The current study measured 19 species in these two genera to determine whether variations in 12 categories of major characters are continuous. In addition, a self-organizing map (SOM) and cluster analysis were used together to reveal whether the known species of Fargesia represent discontinuous sampling of Thamnocalamus. The results show that 46 morphological characteristics exhibited high variation at the generic and species levels. In addition, the cluster analysis showed that 32 morphological characteristics of Thamnocalamus and Fargesia were divided between two species and well separated from the outgroup. Additionally, significant differences (P < 0.01) were observed in the reproductive structures between these two genera. The unrooted dendrogram, which was based on the SOM neural network, shows the same results as the cluster analysis of morphological characteristics. These data indicate that Fargesia is not a result of discontinuous sampling of Thamnocalamus; thus, Fargesia should not be treated as a synonym for Thamnocalamus.
Introduction
Bambusoideae (bamboo) represents one of 12 recognized sub-families of perennial grasses (Poaceae, formerly known as Gramineae) and is widely found from sea level to high plateaus worldwide (ca. 4000 m.a.s.l. in the Himalayas), primarily in forest and grassland habitats, except for in Europe and Antarctica (see Figure S1)1–3. Bamboo is also one of the most valuable plants; it is used for many goods and services4, 5. However, information on the distribution, ecology and intraspecific variation of bamboo is inadequate, and its biodiversity in a broader sense remains largely unexplored3, 6. Additionally, the taxonomic characteristics that can be used to classify or group bamboo are confusing, and estimates of the total diversity have described 1,480 species in 115 genera (ca. 48 genera and nearly 500 species in China)2. Traditional taxonomy has concentrated principally on morphological features, such as the culm and culm-sheath, complex branching, and the rhizome/root system7, 8. However, because herbarium specimens are often collected from limited locations and represent a limited number of plants or only parts of a pant, the continuity of variation in characteristics within a species cannot be specifically quantified9. For example, numerical taxonomy10, one of the traditional taxonomy systems, addresses grouping using numerical methods with taxonomic units based on their character states. In recent years, many authors have treated numerical taxonomy and phenetics as being synonymous5, 11, 12 because phenetics, along with cladistics, originated from numerical taxonomy as a way to describe taxonomic relationships based on the patterns of overall similarities and the estimated evolutionary history of the taxa10. Although numerical taxonomy seems to be to be effective for species-level classification of woody bamboos, it is very difficult to apply at the genus level13, especially among closely related genera, e.g., Fargesia Franchet emend. Yi, Thamnocalamus Munro and Yushania Keng f., of woody bamboos14. Infrequent flowering events (flowering cycles of up to 120 years)1 or flowering only once before culm death, which is one of the special traits in bamboo, restricts the opportunity to gather fresh reproductive material8, 15, 16. In addition, inconsistent results among taxonomists usually stem from different interpretations of the characteristics, and confusion with regard to terminology17, 18 might result in a virtual species being defined as different species.
To address this issue, advances in modern taxonomic tools have been combined with traditional methods to achieve a more objective classification of bamboo. Recently, DNA sequence data have become available to help review classification systems, such as internal transcribed spacer (ITS) and partial granule-bound starch synthase I gene (GBSSI) sequences19–21, restriction site–associated DNA (RAD) and plastid DNA sequences (e.g., trnL-F)22–24, and random amplified polymorphic DNA (RAPD) and next generation sequencing (NGS) technologies11, 15. However, biological data are considered difficult to analyze because numerous biotic/abiotic components are involved in growth processes at all hierarchical levels of life5, 25. The components are related not only within but also between the hierarchical levels, eventually leading to trans-disciplinary holism. Therefore, exploring an approach that can convert complex non-linear statistical relationships between high-dimensional data items into simple geometric relationships would be helpful in accurately and effectively analyzing ecological data. In recent years, the development of biologically inspired machine intelligence (BIMI), including supervised and unsupervised learning methods and artificial neural networks (ANNs), has provided convenient tools to understand the ecological and physiological functions of living systems5, 26. Among these tools, a self-organizing map (SOM)27 is widely utilized because of its similarities to biological nervous systems, its simplicity, and the wide variety of problems to which it might be applied28. Lin & Chen29 demonstrated that using a SOM is a superior clustering technique and that its relative advantage over conventional techniques increases with higher levels of relative cluster dispersion in the data; it even performed better than seven other hierarchical clustering methods. SOMs have been used extensively for the extraction and clustering of various ecological data, including community classification evaluations30, water-quality assessments31, and population and community predictions26, 32. The Kohonen neural network (or SOM network) can project high-dimensional input space on low-dimensional topologies (Figure S2), thereby overcoming various traditional problems with conventional statistical and operational research techniques27, 28. Moreover, many researchers have demonstrated that using a SOM is a superior clustering technique that performs better than other hierarchical clustering methods25, 28, 32. These advantages, coupled with the unsupervised nature of the learning algorithm27, have made SOMs an attractive and interesting alternative for resolving confusion in plant taxonomy.
Bamboo is a versatile and important component of the ecology, culture, and economic livelihood of people in tropical countries, particularly in the Asia-Pacific region28, 33. Bamboo forests are significant for biodiversity, and they provide food and shelter for large animals and birds, soil organisms, insects, and the other plants that compose the bamboo forest ecosystem34. The most famous of these is certainly the critically endangered giant panda (Ailuropoda melanoleuca), whose distribution is mainly determined by the distribution of Fargesia species (e.g., Fargesia qinlingensis and Fargesia robusta)35, 36. Fargesia Franchet emend. Yi, as part of the Thamnocalamus group37, comprises approximately 90 species distributed in alpine areas in southwest China, Vietnam, and the adjacent Himalayas2, 38. In China, there are 78 species, and 61 of them were recognized and published without a description of their reproductive organs, and many of them are narrowly distributed species9, 38. Guo & Li19 and Li39 divided the Arundinariinae into two generic complexes (i.e., Arundinaria and Thamnocalamus groups) based on the differentiation of rhizome types and leaf anatomy. The Thamnocalamus group has pachymorph rhizomes and appears to have large microhairs and dumbbell-shaped silica bodies, although certain species lack fusoid cells7. This group consists of Thamnocalamus Munro, Fargesia Franchet (including Borinda Stapleton and Sinarundinaria Nakai)40, 41 and Yushania Keng f. and is closely related to Ampelocalamus Chen, Wen et Sheng, Chimonocalamus Hsueh et Yi, Drepanostachyum Keng f., Himalayacalamus Keng f. and Gaoligongshania Li, Hsueh et Xia19, 37, 42. Although the classification and delimitation of the genera in this group is highly controversial, it remains critical to the systematics of temperate bamboos. Some studies have suggested that Fargesia and Yushania should be treated as synonyms for the two genera Thamnocalamus (with bracteate racemiform synflorescences) and Sinarundinaria (with open panicles), respectively43. In addition, Soderstrom & Calderon7 were the first to propose merging Fargesia with Thamnocalamus based on their simple, open, semelauctant synflorescence without enclosing spathes, and this classification was also supported by other taxonomists such as Guo et al.37, Wang & Ye40 and Chao & Tang44. However, Li et al.45 believed that Fargesia and Yushania are comparatively large genera within Arundinarieae with ca. 90 and 80 species, respectively. Species in these two genera are mostly distributed above 2000 m, with some taxa even found at 4000 m, and have pachymorph rhizomes, many branches per node (one to five branches for some Yushania), semelauctant inflorescence and three stamens21. In addition, Fargesia species have short-necked rhizomes usually with unicaespitose culms and foliage-like bracts under the inflorescence, while Yushania species have long-necked rhizomes with diffuse culms and small and reduced bracts under the inflorescence13. Therefore, Li39 designated Fargesia (Borinda, Sinarundinaria), Thamnocalamus and Yushania as the Thamnocalamus group and Ampelocalamus, Chimonocalamus, Drepanostachyum (or Himalayacalamus) and Gaoligongshania as its close allies mainly based on their pachymorph rhizomes and features of their leaf anatomy. One explanation is that attention was paid only to the Thamnocalamus group and its allies and that these taxa were resolved as a monophyletic group based on GBSSI and ITS sequences19, 20. With more genera in Arundinarieae involved and an analysis of plastid regions46, the Thamnocalamus group and its allies were shown to be polyphyletic or paraphyletic. Interestingly, Yang et al.23 confirmed that alpine Bashania, Chimonocalamus, Thamnocalamus, and species currently placed in Fargesia and Yushania formed a clade based on LEAFY [which is a master regulator orchestrating the whole floral network, and exists across all land plants47] and combined their nuclear phylogenies. In addition, based on the original text descriptions and illustrations of a flowering branch with respect to the collections of Thamnocalamus species in general, Yi41 and Yi et al.48 noted that the Thamnocalamus inflorescences are lateral, whereas Fargesia inflorescences are terminal, thereby indicating that Fargesia should not be merged with Thamnocalamus. Thus, the establishment of a well-defined and unambiguous classification of bamboos requires the adoption of a novel approach.
To verify this issue, specimens of Thamnocalamus and Fargesia from a total of 19 species, which essentially cover the total distribution of the two genera, were collected. The 46 morphological characteristics for 66 specimens of the two genera were measured and analyzed to determine whether variations in these characteristics are continuous. Furthermore, to improve our understanding of the interspecific relationships of vegetative morphologic characteristics, a SOM, principal component analysis (PCA), and hierarchical cluster analysis (HCA) were used simultaneously for the first time. The SOM technique represents one of the most reliable tools in ecological systematic analyses26, 28, 32. Thus, in this work, we attempted to integrate morphological data with SOM-network data to assess differences in the patterns of variation in the characteristics between Thamnocalamus and Fargesia.
Results and Discussion
Morphological and taxonomic comparison of Thamnocalamus and Fargesia
In this study, the Kruskal–Wallis analysis showed that the values for WOL (presence/absence of leaf sheath), IGF (genuine/false inflorescence), PWOC (two cristae present/absent on palea), PAS (apex split into two in palea), LN (lodicule number), LMH (hairs on margins present/absent in lodicule), SUN (stamen number), and GWOH (hairs present/absent in gynoecium) did not differ among the species studied, which indicates that these characteristics cannot be used to differentiate the species belonging the Thamnocalamus and Fargesia at P < 0.05 (Tables 1 and 2). Among the significant characteristics, 16 morphological characteristics [i.e., TUS (type of underground stem), LSN (number of leaf sheaths), ES (extent of expansion around the spathe in leaf sheath), IB/IC (botryose/conical inflorescences), IC/IS (compact/squarrose inflorescences), IL/IT (terminal/lateral inflorescences), IWOB (bracts present/absent in inflorescences), SN (spikelet number), SFN (number of florets in spikelet), SFC (color of florets in spikelet), SPH (hairs on pedicel present/absent in spikelet), SPB (presence/absence of bracts at the base of pedicel in spikelet), LS (lemmashape), LNN (number of nerves in lemma), PBH (presence/absence of hairs between cristae in lemma), AAC (color of anther in lemma), and GSTN (number of stigmas in gynoecium)] could be differentiated at P < 0.01, and ILG (inflorescence length), SLG (spikelet length), LM (lemma texture), and LSSN (relative size of lodicule) could be differentiated at P < 0.05 (Tables 2 and 3). Any grouping based on vegetative characteristics should account for parallel trends in inflorescence structure37, and many agrostologists have indicated that inflorescences with 6 stamens, such as Bambusa Retz. corr. Schreber, are more derived, whereas those with 3 stamens, such Thamnocalamus or Fargesia, are ancestral49. Accordingly, most grasses having 3 stamens33, 50 and inflorescences with bracts originated via a reduction of bractless panicles to support vegetative bract-bearing axes. However, according to our field observations, the inflorescence characteristics of representatives of the genera in the Thamnocalamus group (see Table 4), including the trend of reduced bracts from Thamnocalamus to Fargesia and even Drepanostachyum, are distinct. This finding is accompanied by a reduction in the presence of bud-like structures in the axes of glumes, an overall expansion from compressed to open inflorescences, and the occurrence of specialized features, such as fasciculation and pulvini, which are more typical of non-bambusoid grass inflorescences36, 51. Additionally, in the specimen analysis, inflorescence characteristics (e.g., IB/IC, IC/IS, IL/IT, and IWOB in Table 2) were used to differentiate between the genera at a statistically significant level (P < 0.01), indicating that the main differences are the number of spathes and the growth point of the inflorescence. Consistent with these observations, Yi41 amended Fargesia greatly, adding species without enlarged spathe-like structures and bracteate inflorescences on the basis of shorter rhizome necks. Similarly, Guo et al.37 and Li et al.45 clearly discriminated two genera: Thamnocalamus, with inflorescence panicles consisting of racemes, with each subtended by a spathe; and Fargesia, with terminal inflorescences subtended by several enlarged or not enlarged spathes. We also believe that there is a clear evolutionary trend in spathe size from large to small among Fargesia and related genera, which provides strong evidence for the discrimination of species. The trend in spathe size is also correlated with other characteristics [e.g., nutritional (TUS) and reproductive (SN, SFN, and SFC) features], as shown in Table 2 and supported by Guo & Li19 and Stapleton50. In contrast, a totally different opinion was held by Wang & Ye40, who accepted Fargesia and Yushania as genera in addition to Thamnocalamus, with Sinarundinaria as a synonym for Fargesia, while Chao & Tang44 treated Fargesia as a synonym for Thamnocalamus based on whether the inflorescence was subtended by one to several enlarged spathes. Stapleton52 supported the opinion of Wang & Ye40 and created a new genus, Borinda, which was somewhat intermediate between Fargesia and Yushania. Thus, some taxonomists have suggested the “very widest” Arundinaria Michaux to accommodate the members of the Thamnocalamus group from Sri Lanka due to the confusion surrounding these genera.
Table 1.
Plant material used in the study.
| Species | Acronym | Location | No. of individuals | No. of samples | Altitude (m) | Longitude (E) | Latitude (N) |
|---|---|---|---|---|---|---|---|
| Fargesia adpressa T.P. Yi | F.a | Mianning, Liangshan, Sichuan, China | 3 | 16 | 2360–2700 | 102.17 | 28.57 |
| Fargesia brevissima T.P. Yi | F.ba | Hongchi, Wuxi, Chongqing, China | 3 | 24 | 2300–2750 | 109.07 | 31.54 |
| Fargesia denudata T.P. Yi | F.d | Beichuan, Mianyang, Sichuan, China | 3 | 18 | 3500–3600 | 104.81 | 31.70 |
| Fargesia dracocephala T.P. Yi | F.dr | Guandu, Wanyuan, Dazhou, Sichuan, China | 3 | 30 | 1500–2200 | 108.06 | 32.15 |
| Fargesia ferax (Keng f.) T.P. Yi | F.f | Kangding, Ganzi, Sichuan, China | 3 | 28 | 1700–2800 | 101.95 | 30.13 |
| Fargesia fungosa T.P. Yi | F.fu | Baiji, Weixi, Diqing, Yunnan, China | 3 | 31 | 1800–2700 | 99.08 | 27.36 |
| Fargesia grossa T.P. Yi | F.g | Cuona, Shannan, Tibet, China | 3 | 19 | 2300–3000 | 91.94 | 28.21 |
| Fargesia hsuchiana T.P. Yi | F.h | Shuiping, Jinping, Honghe, Yunnan, China | 3 | 32 | 1950–2100 | 103.24 | 22.85 |
| Fargesia lincangensis T.P. Yi | F.l | Daxueshan, Yongde, Lincang, Yunnan, China | 3 | 21 | 3300–3400 | 99.74 | 24.02 |
| Fargesia mairei (Hack. ex Hand.-Mazz) T.P. Yi | F.m | Baidiao, Muli, liangshan, Sichuan, China | 3 | 20 | 3100–3500 | 101.46 | 28.05 |
| Fargesia melanostachys (Hand.-Mazz) T.P. Yi | F.me | Baimang snow mountain, Deqin, Yunnan, China | 3 | 22 | 3300–3400 | 98.96 | 28.39 |
| Fargesia nitida (Mitford ex Stapf) Keng f. | F.n | Dalu, Jiuzhaigou, Aba, Sichuan, China | 3 | 27 | 2900–3000 | 103.67 | 33.56 |
| Fargesia ostrina T.P.Yi | F.b | Taiping,Fengdu, Chongqing, China | 3 | 23 | 2000–2150 | 107.73 | 29.92 |
| Fargesia pauciflora (Keng f.) T.P. Yi | F.p | Yaoshang, Qiaojia, Yunnan, China | 3 | 17 | 2100–2200 | 103.03 | 27.08 |
| Fargesia robusta T.P. Yi | F.r | Wolong, Wenchuan, Aba, Sichuan, China | 3 | 29 | 1700–2800 | 103.19 | 31.04 |
| Fargesia spathacea Franch. | F.sp | Yuanping, Chengkou, Chongqing, China | 3 | 25 | 1600–2400 | 108.34 | 31.92 |
| Fargesia scabrida T.P. Yi | F.sc | Beichuan, Mianyang, Sichuan, China | 3 | 26 | 1450–2520 | 104.81 | 31.70 |
| Thamnocalamus aristatus E.G. Camus | T.a | Eastern India | 1 | 15 | 2500–3000 | 81.51 | 24.45 |
| Thamnocalamus unispiculatus T.P. Yi & J.Y. Shi | T.u | Zhangmu, Nielamu, Shigatse, Tibet, China | 14 | 1~14 | 2650–3300 | 85.98 | 28.00 |
Table 2.
Coding of qualitative characteristics into binary or ordered multistate characteristics.
| Categories | Characteristics | Acronym | Encoding number | Standard deviation | Kruskal– | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | TM | FM | All | Wallis Test | |||
| Stem | Type of underground stem | TUS | Sympodial-clumping | Sympodial-scattering | — | — | 0.00 | 1.12 | 1 | 0.00** |
| Leaf sheath | Number | LSN | — | — | — | — | 0.00 | 0.00 | 1 | 0.00** |
| Length relative to inflorescences | CPI | Longer | Shorter | Equal | — | 0.49 | 1.17 | 1 | 0.05 | |
| Leaf present or absent | WOL | Absent | Present | — | — | 0.00 | 0.00 | 0 | — | |
| Extent of expansion around the spathe | ES | Not expanded | Expanded slightly | Expanded | — | 0.00 | 1.01 | 1 | 0.00** | |
| Inflorescences | Genuine or false | IGF | False | Genuine | — | — | 0.00 | 0.00 | 0 | — |
| Botryose or conical | IB/IC | Botryose | Conical | — | — | 0.00 | 0.94 | 1 | 0.00** | |
| Compact or squarrose | IC/IS | Compact | Squarrose | — | — | 0.54 | 1.06 | 1 | 0.00** | |
| Terminal or lateral | IL/IT | Terminal | Lateral | — | — | 0.00 | 0.00 | 1 | 0.00** | |
| Bracts present or absent | IWOB | Absent | Present | — | — | 0.00 | 0.00 | 1 | 0.00** | |
| Length (mm) | ILG | — | — | — | — | 0.90 | 0.99 | 1 | 0.02* | |
| Spikelet | Number | SN | — | — | — | — | 0.27 | 0.81 | 1 | 0.00** |
| Length (mm) | SLG | — | — | — | — | 0.43 | 1.18 | 1 | 0.04* | |
| Number of florets | SFN | — | — | — | — | 0.43 | 1.07 | 1 | 0.00** | |
| Color of florets | SFC | Light yellow | Light green | Green | — | 0.00 | 0.77 | 1 | 0.00** | |
| Length of rachilla (mm) | SRLG | — | — | — | — | 0.38 | 1.21 | 1 | 0.42 | |
| Length of pedicel (mm) | SPLG | — | — | — | — | 0.27 | 1.20 | 1 | 0.15 | |
| Hairs on pedicel present or absent | SPH | Absent | Present | — | — | 0.00 | 1.18 | 1 | 0.01** | |
| Bracts at the base of pedicel present or absent | SPB | Absent | Present | — | — | 0.00 | 0.98 | 1 | 0.00** | |
| Glume texture | Glume and texture | GM | Papery | Membranous | — | — | 1.14 | 0.92 | 1 | 0.76 |
| First glume | Shape | FS | Linear-lanceolate | Ovate-lanceolate | Long/narrow lanceolate | — | 0.00 | 1.29 | 1 | 0.20 |
| Length (mm) | FLG | — | — | — | — | 0.49 | 1.22 | 1 | 0.09 | |
| Number of nerves | FNN | — | — | — | — | 0.00 | 1.29 | 1 | 0.37 | |
| Second glume | Shape | SS | Linear-lanceolate | Ovate-lanceolate | Long/narrow lanceolate | — | 0.00 | 1.30 | 1 | 0.21 |
| Length (mm) | SLG | — | — | — | — | 0.45 | 1.25 | 1 | 0.60 | |
| Number of nerves | SNN | — | — | — | — | 0.00 | 1.28 | 1 | 0.10 | |
| Lemma | Shape | LS | Linear-lanceolate | Ovate-lanceolate | Long/narrow lanceolate | — | 0.00 | 0.85 | 1 | 0.00** |
| Length (mm) | LLG | — | — | — | — | 0.60 | 1.21 | 1 | 0.73 | |
| Number of nerves | LNN | — | — | — | — | 0.40 | 1.09 | 1 | 0.00** | |
| Hairs present or absent | LWOH | Absent | Present | — | — | 0.00 | 1.23 | 1 | 0.06 | |
| Texture | LM | Papery | Membranous | — | — | 0.00 | 1.2 | 1 | 0.02* | |
| Palea | Length (cm) | PLG | — | — | — | — | 0.64 | 1.15 | 1 | 0.15 |
| Two cristae present or absent | PWOC | Absent | Present | — | — | 0.00 | 0.00 | 0 | — | |
| Apex split into two | PAS | No | Yes | — | — | 0.00 | 0.00 | 0 | — | |
| Hairs on cristae present or absent | PCH | Absent | Present | — | — | 0.00 | 1.26 | 1 | 0.15 | |
| Hairs between cristae present or absent | PBH | Absent | Present | — | — | 0.54 | 1.06 | 1 | 0.00** | |
| Lodicule | Number | LN | — | — | — | — | 0.00 | 0.00 | 0 | — |
| Shape | LS | Lanceolate | Triangular | Ovate | — | 0.00 | 1.27 | 1 | 0.25 | |
| Relative size | LSSN | Equal | Not equal | — | — | 0.00 | 1.21 | 1 | 0.03* | |
| Hairs on margins present or absent | LMH | Absent | Present | — | — | 0.00 | 0.00 | 0 | — | |
| Stamen | Number | SUN | — | — | — | — | 0.00 | 0.00 | 0 | — |
| Color of anther | AAC | Yellow | Brownish yellow | Purple | — | 0.00 | 0.50 | 1 | 0.00** | |
| Gynoecium | Shape of ovary | GOS | Elliptic | Ovate | Oblong | — | 0.30 | 1.23 | 1 | 0.18 |
| Hairs present or absent | GWOH | Absent | Present | — | — | 0.00 | 0.00 | 0 | — | |
| Number of styles | GSN | — | — | — | — | 0.00 | 1.26 | 1 | 0.15 | |
| Number of stigmas | GSTN | — | — | — | — | 0.46 | 0.97 | 1 | 0.00** | |
The significance of differences and standard deviations between Fargesia and Thamnocalamus were calculated with the Kruskal–Wallis test. ** P < 0.01, * P < 0.05. TM, the mean standard deviation (SD) of Thamnocalamus species; FM, the mean SD of Fargesia species; All, the SD of Fargesia and Thamnocalamus species.
Table 3.
Pearson’s correlation coefficients between the significant discriminatory characteristics as indicated by the Kruskal–Wallis test results in Table 2.
| TUS | LSN | ES | IB/IC | IC/IS | IL/IT | IWOB | SN | SFN | SFC | SPH | SPB | LS | PBH | AAC | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LSN | 0.58 | ||||||||||||||
| ES | 0.01 | −0.62 | |||||||||||||
| IB/IC | −0.60 | −0.73 | 0.31 | ||||||||||||
| IC/IS | −0.56 | −0.55 | 0.08 | 0.66 | |||||||||||
| IL/IT | −0.58 | −1.00 | 0.62 | 0.73 | 0.55 | ||||||||||
| IWOB | 0.58 | 1.00 | −0.62 | −0.73 | −0.55 | −1.00 | |||||||||
| SN | 0.65 | 0.81 | −0.34 | −0.51 | −0.53 | −0.81 | 0.81 | ||||||||
| SFN | −0.53 | −0.59 | 0.14 | 0.49 | 0.74 | 0.59 | −0.59 | −0.44 | |||||||
| SFC | 0.34 | 0.78 | −0.52 | −0.47 | −0.43 | −0.78 | 0.78 | 0.74 | −0.39 | ||||||
| SPH | 0.13 | 0.50 | −0.18 | −0.21 | −0.25 | −0.50 | 0.50 | 0.31 | −0.55 | 0.39 | |||||
| SPB | 0.10 | 0.63 | −0.51 | −0.31 | −0.22 | −0.63 | 0.63 | 0.47 | −0.29 | 0.66 | 0.30 | ||||
| LS | −0.51 | −0.84 | 0.55 | 0.74 | 0.54 | 0.84 | −0.84 | −0.60 | 0.57 | −0.62 | −0.47 | −0.57 | |||
| PBH | −0.01 | −0.48 | 0.40 | 0.38 | 0.32 | 0.48 | −0.48 | −0.41 | 0.11 | −0.67 | −0.17 | −0.70 | 0.46 | ||
| AAC | −0.39 | −0.91 | 0.59 | 0.58 | 0.46 | 0.91 | −0.91 | −0.68 | 0.48 | −0.69 | −0.58 | −0.67 | 0.71 | 0.53 | |
| GSTN | 0.50 | 0.58 | −0.31 | −0.48 | −0.54 | −0.58 | 0.58 | 0.61 | −0.31 | 0.72 | 0.30 | 0.36 | −0.64 | −0.58 | −0.46 |
Values highlighted in bold are statistically significant at P < 0.01(**).
Table 4.
Feature comparison of the inflorescences of representatives of the genera in the Thamnocalamus group.
| Genera | Glume buds | Bracts | Fasciculation | Pulvini | Florets |
|---|---|---|---|---|---|
| Thamnocalamus Munro | Common | Usual | None | No | Many |
| Ampelocalamus Chen, Wen et Sheng | Common | Often | Yes | No | Many |
| Fargesia Franchet | Rare | Few | No | Few | Many |
| Yushania Keng f. | None | None | No | Many | Many |
| Drepanostachyum Keng f. | None | None | Yes | No | 2-several |
| Himalayacalamus Keng f. | None | None | Reduced | No | 1 |
In addition, approximately 60% of all studied characteristics showed highly significant correlations (P < 0.01; Table 3). Among them, the highest positive correlations were detected between LSN and IWOB (1.00**), ACC and LS (0.91**), IL/IT and LS (0.84**), LSN and SN (0.81**), and IWOB and SN (0.81**), indicating that the quantifiable characteristics of the leaf sheath and inflorescences are generally dependent on each other. In addition, leaf sheath (LSN) and spikelet (SFC, SPH, and SPB), gynoecium (GSTN) and spikelet (SN and SFC), and palea (PBH) and lemma (LS) characteristics are also positively correlated (P < 0.01) with each other. Importantly, differentiations among individuals (i.e., those highly significantly correlated at P < 0.01) are also indicated between leaf sheath (ES) and spikelet [SFC (−0.52**) and SPB (−0.51**)], inflorescence (IB/IC, IC/IS, and IL/IT) and spikelet (SN and SFC; see Table 3), and the stamen (AAC) and spikelet [SN (−0.68**), SFC (−0.69**), SPH (−0.58**), and SPB (−0.67**)] characteristics, suggesting that Fargesia is actually substantially different from Thamnocalamus and presents several more derived morphological characteristics19, 41, 48. Thus, recognizing Fargesia as a separate genus appears to be reasonable.
Cluster analysis of vegetative characteristics between Fargesia and Thamnocalamus
A PCA was performed to understand how the vegetative characteristics contribute to the definitions of these two genera. A factor analysis showed that the 32 accessions were divided into two distinct groups based on the scatterplot for the two principal component axes (Fig. 1). Evidently, the two groups of accessions comprise the separate genera Fargesia (No. 1~15 in Table 1) and Thamnocalamus (No. 16~32). Further analysis showed that the 14 specimens of T. unispiculatus (No. 1~14) were grouped especially closely along PC1, whereas T. aristatus (No. 15) was the most dissimilar specimen in the Thamnocalamus group but was separated from the T. unispiculatus specimen primarily along PC1. One possible explanation is that T. aristatus is a variant of T. unispiculatus and that the identification of T. aristatus 52 is the result of the discontinuous sampling of T. unispiculatus. Cases of discontinuous sampling that artificially isolate morphological characters have also been found for bamboo or other taxa9. Comparatively, the 17 Fargesia species (No. 16~32) showed a maximum distance between absolute values of approximately four units on PC1 and showed greater separation on PC2, with a maximum distance of approximately six units.
Figure 1.
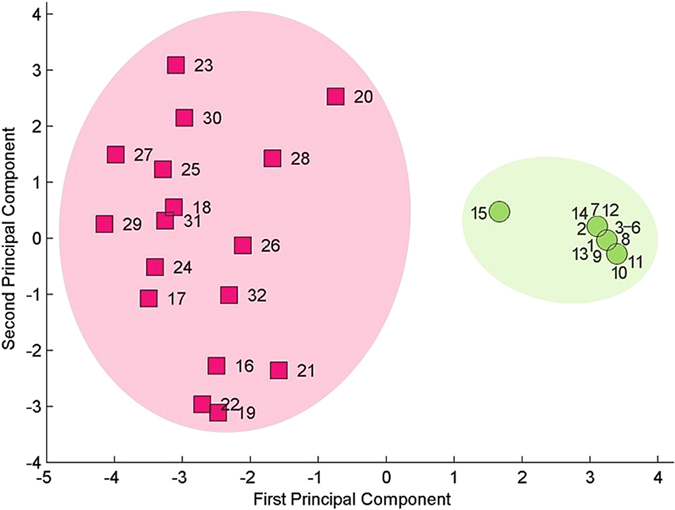
Scatterplot of the scores for the first (PC1) and second (PC2) principal components based on the principal component analysis (PCA). The data were subjected to z-score standardization prior to analysis. Red squares represent Fargesia species, and green circles represent Thamnocalamus species. Please see Table 1 for details.
However, in the HCA, the specimens were grouped on the basis of similarities without taking into account information on their class membership53. In our analysis, the two most used methods to calculate the distance between pairs of objects in a HCA were applied to analyze the data, and clustering using Euclidean distances grouped the 32 accessions into two main clusters that corresponded to the genera Fargesia and Thamnocalamus (see Fig. 2A). The maximum intra-cluster distances for the two genera were 14 and 9 Euclidean distance units, whereas a distance of almost 25 Euclidean distance units was observed between these clusters. As shown in Fig. 2, the specimens were obviously divided into two groups, with a large inter-group Euclidean distance. Correlation coefficients between each pair of specimens were calculated as distances and clustered based on a furthest-distance linkage model (Fig. 2B). Both methods had similar cluster results, especially with regard to the consistent division of the 32 accessions into two clusters as Thamnocalamus and Fargesia. Furthermore, dendrograms generated by the HCA using both Euclidean and city-block distances54, 55 and by the SOM neural network also clearly divided the accessions into two distinct clusters corresponding to the two genera, suggesting that no interaction between accessions of Thamnocalamus and Fargesia was observed and that morphological traits served as effective discriminators of the two genera (Figs 1–3). Therefore, the cluster results also confirmed the hypothesis that Thamnocalamus and Fargesia should be segregated as separate genera and should not be merged.
Figure 2.
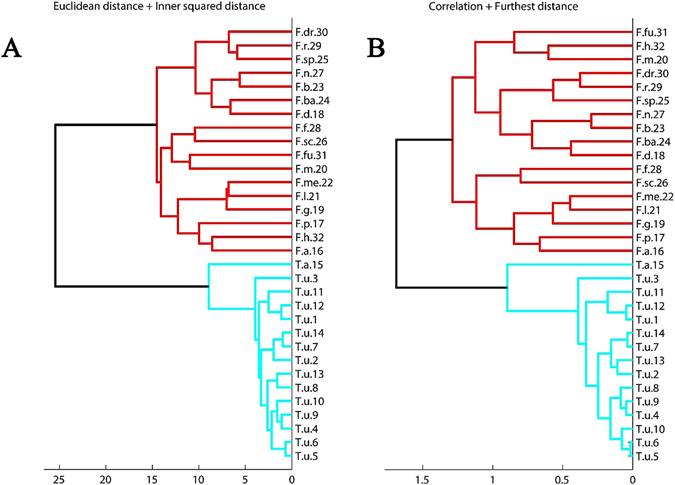
Hierarchical clustering (HC) dendrogram of 32 accessions derived from 46 morphological characteristics. (A) Distances between pairs of objects were calculated based on Euclidean distances, and the cluster linkage model utilized the inner squared distance; (B) distances between objects (one minus the sample correlation). The cluster linkage model had the greatest distance. In both figures, red lines represent Fargesia accessions, and green lines represent Thamnocalamus accessions. Please see Table 1 for details.
Figure 3.
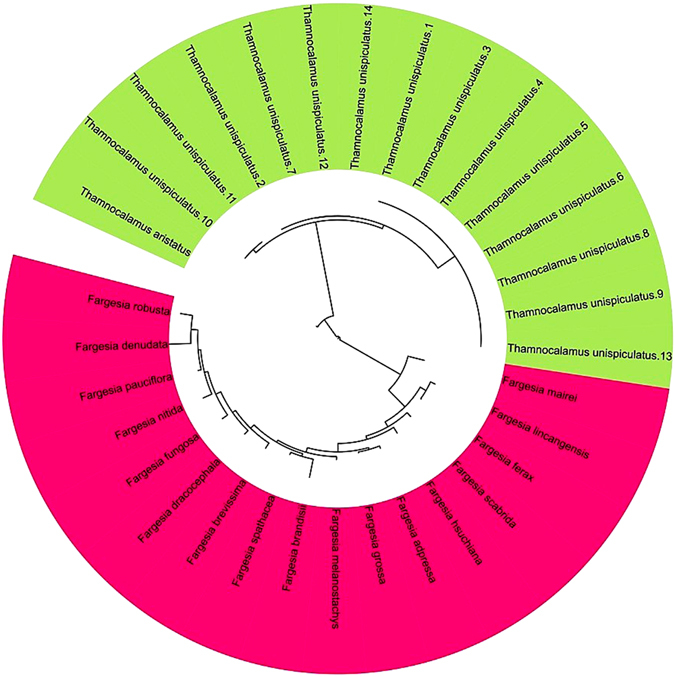
Unrooted dendrogram representing relationships among Fargesia and Thamnocalamus species estimated with a SOM neural network. For all characters, the mean value was used to construct the dendrogram. Tree distances were calculated with the SOM Neural Network Toolbox for MATLAB software (MathWorks Inc., Natick, MA, USA) and the online tool Interactive Tree Of Life (ITOL; http://itol.embl.de/).
Relationship and discrimination based on the Kohonen neural network
In recent decades, considerable research on bamboo classification has been published; however, systematic studies and critical assessments of taxa are rarely undertaken, which inevitably leads to an increasing number of taxa and little resolution of taxonomic confusion1, 17, 51. The issues related to bamboo taxonomy indicate that investigations of bamboo classification should focus on revising taxonomic groups to clarify uncertainties56. For example, Zhang et al.9 committed to integrating the data relating to morphology and ITS regions to assess the patterns of variation in characteristics between two closely allied species (F. decurvata and F. dracocephala), and the results indicated that F. dracocephala should be treated as a synonym of F. decurvata. At present, the neural-network approach to recognition and classification is widely used for the discrimination of indistinguishable species27, 28, and further investigations should be performed to determine the adaptability and effectiveness of this approach. In this work, the estimated dendrogram, which was based on measured distances57 for the morphological characteristics and calculated using a SOM/Kohonen neural network, provided an alternative representation of the relationships among the 32 accessions (Fig. 3). Among them, T. unispiculatus was shown to have a very consistent structure; eight specimens had identical phenotypes, whereas a longer branch separated the T. aristatus accession from the T. unispiculatus accessions. Moreover, the longer branch obviously separated the Fargesia and Thamnocalamus accessions, whereas the branches among the Fargesia species were relatively short, suggesting that Fargesia should not be treated as a synonym for Thamnocalamus. Similarly, the results from the SOM analysis of multiple vegetative characteristics, which has been successful in studies of other species25, 32, showed that ten populations were divided into two genera. Generally, a species is a collection of individuals with common characteristics, and defining a species represents the delimitation of the characteristics of the group8, 9, with each specimen representing only a reference point for naming. However, if a specimen can be considered to exhibit characteristics that define the species, then continuous variation of characteristics will be presented intermittently, causing difficulties and even errors in classification7. Thus, it is essential to investigate as many collections as possible from numerous herbaria, not only from restricted areas but also from entire areas where a taxon may occur.
Conclusion
As an important constituent of the Flora, bamboo is one of the most important plants for humans. However, knowledge of the taxonomic characteristics that can be used to classify or group bamboo species is still rudimentary and confusing. To the best of our knowledge, this is the first detailed report on the discrimination of Fargesia Franchet emend. Yi from Thamnocalamus Munro via an integration of morphological and SOM data to assess the differences in the patterns of variation of characteristics between these two genera. Our main conclusions are as follows: (1) 46 morphological characteristics exhibited considerable variation at the genus and species levels; (2) the PCA and HCA cluster analyses showed that 32 morphological characteristics of Thamnocalamus and Fargesia were divided among two species and indicated that significant differences (P < 0.01) occurred in the floral organs between the two genera; and (3) the unrooted dendrogram based on the SOM neural network representing the relationships among species from the two genera showed the same results as the cluster analysis of the morphological characteristics. These novel findings improve our understanding of the application and role of ANNs (e.g., SOM neural networks) in plant taxonomy. Further studies are required to determine how to merge molecular taxonomy and neural networks and review the classification systems based on morphological traits.
Materials and Methods
Plant materials
Specimens were collected for 19 species, including Fargesia Franchet emend. Yi (17 species) and Thamnocalamus Munro (2 species) (Table 1). Specimens of all the studied species (except for T. aristatus E.G., Camus) were collected from the Tibet, Sichuan, Chongqing, and Yunnan provinces in China by Prof. Tongpei Yi from 1975 to 2006. These accessions were deposited in the Campus of Dujiangyan at the Sichuan Agricultural University (SICAU), Sichuan, China. In addition, the accessions of T. aristatus were collected in 1960 from eastern India (2500~3000 m.a.s.l.) and obtained from the University of Tokyo (U-Tokyo), Japan. In this work, 15 specimens (15 individuals) from two species (one specimen of T. aristatus and 14 specimens of T. unispiculatus) in the genera Thamnocalamus were studied. Additionally, 17 specimens (54 individuals) in the Fargesia genera were studied (see Table 1 and Fig. 4).
Figure 4.
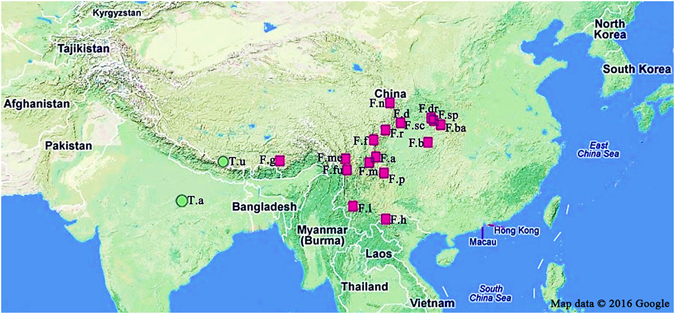
Geographic distribution of accessions of Fargesia and Thamnocalamus species used in this study. Red squares represent Fargesia species, and green circles indicate Thamnocalamus species. Please see Table 1 for details. The figure was generated with Adobe® Photoshop® CS3 extend 10.0.1 software (Adobe Systems Inc., San Jose, CA, USA; URL link: https://www.adobe.com/products/photoshop.html ? promoid = KLXLS) based on Google® maps (Google Inc., Mountain View, CA, USA; URL link: https://www.google.com/maps).
Quantification of morphological characteristics
Based on the Flora Reipublicae Popularis Sinicae (FRPS)38 and Flora of China 45, 12 major morphological characteristics and 46 expanded characteristics from a total 19 species were used to identify Thamnocalamus and Fargesia: (1) type of underground stem [acronym-TUS; see Table 2], (2) leaf sheath (length relative to inflorescences/CPI, LSN, WOL, ES), (3) inflorescences (IGF, IB/IC, IC/IS, IL/IT, IWOB, ILG), (4) spikelet (length of rachilla/SRLG, length of pedicel/SPLG, SN, SLG, SFN, SFC, SPH, SPB), (5) glume texture (GM), (6) first glume (shape/FS, length/FLG, number of nerves/FNN), (7) second glume (shape/SS, number of nerves/SNN, SLG), (8) lemma (LS, LLG, LNN, LWOH, LM), (9) palea (PLG, PWOC, PAS, PCH, PBH), (10) lodicule (LN, LS, LSSN, LMH), (11) stamen (SUN, AAC), and (12) gynoecium [shape of ovary/GOS, number of styles/GSN, GWOH, GSTN] (Tables 1 and 2 and Fig. 4). According to the rule for transforming qualitative data into quantitation indexes (Table S1), we obtained a coding matrix for the studied species (Table S2). In addition, the lengths of the inflorescences, spikelet, rachilla, pedicel, first/second glume, lemma, and palea were measured using a Micrometer Screw-Gauge (Mitutoyo Inc., Kanagawa, Japan).
For the quantitative analysis, all qualitative data for vegetative morphological characteristics were transformed via binary encoding with the following formula58:
| 1 |
where
| 2 |
To eliminate the impact of dimensional differences between different types of data and enable the use of multivariate analytical techniques, the average value of three individual measurements for each character was calculated and then standardized using the Z-score transformation algorithm59. Specifically, the raw intensity (I) data for each character were [log10] transformed and then used for the calculation of Z-scores, which were calculated by subtracting the overall average intensity from the raw intensity data for each character and dividing that result by the standard deviations (SDs) of all measured intensities according to the following formula60:
| 3 |
where C is any character of the species, C 1…C n represent the aggregate measure of all characteristics, and MI represents the mean intensity. The significance of differences in the mean values of each character between the Thamnocalamus and Fargesia species were analyzed via the Kruskal–Wallis test61.
Principal component and hierarchical cluster analysis
A principal component analysis (PCA) was applied to the data set after standardization (the mean of the values for each variable was subtracted from each variable value and the result was divided by the standard deviation of the values for each variable). The PCA was performed using the Unscrambler software package (Version 9.7; CAMO Software AS, Oslo, Norway), and it transforms the original, measured variables into new uncorrelated variables called principal components62. The first principal component covers as much of the variation in the data as possible. The second principal component is orthogonal to the first and covers as much of the remaining variation as possible (CAMO Software AS, Oslo, Norway). Euclidean distances and city-block distances between accessions were estimated from all recorded characteristics according to Ward Jr.’s method54. A HCA was also applied to the standardized data to investigate similarities between different specimens and specimen types (MINITAB, 15.1.1.0, 2007). The HCA calculates the distances (or correlations) between all specimens using a defined metric, such as Euclidean distance or Manhattan distance18, 53.
Self-organizing map (SOM) algorithm
A SOM is a neural-network algorithm that implements a characteristic nonlinear projection from a high-dimensional space of input signals onto a low-dimensional array of weights27, 32. Forty-six morphological characteristics were converted into normalized vectors of codon usage x(t) (32 accessions were classified by characteristic factors; see Fig. 5). Each component of input vectors was scaled with the following formula so that its mean became 0 and its variance became one:
| 4 |
where x i old is the original value of component i of the data vector x, Oi is the mean of values of x i, and s i is their SDs. The scaling is used to ensure that no component has excessive influence on the learning results due to a greater variance or larger absolute value63 (see Supplemental Methods S1).
Figure 5.
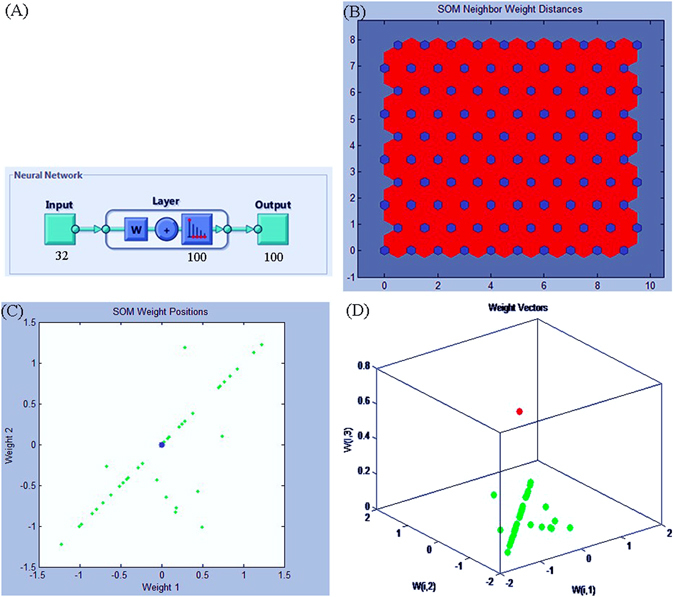
Trained classification structure model (A) and weight structure (B,C and D) of the SOM neural network. We converted the 46 morphological characters into normalized vectors of codon usage x(t), and 32 accessions were classified by character factors. Symmetrical effects and differences among the samples are more obvious and significant, although off-diagonal weight points are observed.
To avoid dead neurons that might be caused by the random generation of an initial network by the neural network algorithm, the number of training specimens was increased as the network was re-initialized 1000 times. For our analysis, we used the SOM Toolbox (http://www.cis.hut.fi/projects/somtoolbox), a MATLAB-based SOM (MathWorks Inc., Natick, MA, USA), and the interactive Tree Of Life (iTOL; http://itol.embl.de), and a new web-based tool, for the display, manipulation, and annotation of phylogenetic trees64.
Electronic supplementary material
Acknowledgements
This work was partially supported by the National Natural Science Foundation of China (NSFC, No. 31470636 and No. 31570700). Special thanks are given to associate Prof. S.R. Pu (SICAU, Dujiangyan, China) and Dr. H.Z. Tang (SICAU, Chengdu, China) for their technical assistance in the data analysis and to the University of Tokyo (U-Tokyo) for providing the accessions of Thamnocalamus aristatus. We thank the professional editing service company (American Journal Experts) for their assistance in reviewing the English language of the manuscript. The authors are also grateful to the Editors and two Reviewers for their comments and suggestions on an earlier draft of the manuscript.
Author Contributions
S.L.L., M.D.M., T.P.Y. and Q.B.C. designed the research; S.L.L., R.J.Y. and J.Y. performed the research; S.L.L., R.J.Y., J.Y., M.Y.J. and T.P.Y. analyzed the data; S.L.L., R.J.Y. and J.Y. wrote the first version of the manuscript; S.L.L., H.X.S., D.K.T., M.Y.J., M.D.M. and Q.B.C. contributed substantially to the revisions.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-04613-9
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mingdong Ma, Email: 610245498@qq.com.
Qibing Chen, Email: cqb@sicau.edu.cn.
References
- 1.Bamboo Phylogeny Group [BPG]. An updated tribal and subtribal classification of the bamboos (Poaceae: Bambusoideae). In: Gielis J, Potters G. (Eds) Proceedings of the 9th World Bamboo Congress, 10–15 April 2012 World Bamboo Organization, Antwerp, Belgium, 3–27 (2012).
- 2.Clark, L. G., Londoño, X. & Ruiz-Sanchez, E. Bamboo taxonomy and habitat. In: Liese, W., Köhl, M. (eds) Bamboo, tropical forestry 10. Springer International Publishing, Switzerland, 1–30 (2015).
- 3.Wysocki WP, Clark LG, Attigala L, Ruiz-Sanchez E, Duvall MR. Evolution of the bamboos (Bambusoideae; Poaceae): a full plastome phylogenomic analysis. BMC Evol. Bio. 2015;15:e50. doi: 10.1186/s12862-015-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baskin JM. Death of bamboo triggers regeneration of overstory tree in a southern beech forest. New Phytol. 2009;181:749–751. doi: 10.1111/j.1469-8137.2009.02757.x. [DOI] [PubMed] [Google Scholar]
- 5.Wu M, et al. Genome-wide identification and expression analysis of the IQD gene family in moso bamboo (Phyllostachys edulis) Sci. Rep. 2016;6:e24520. doi: 10.1038/srep24520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yeasmin L, Ali MN, Gantait S, Chakraborty S. Bamboo: an overview on its genetic diversity and characterization. 3 Biotech. 2015;5:1–11. doi: 10.1007/s13205-014-0201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soderstrom TR, Calderon CE. A commentary on the bamboos (Poaceae: Bambusoideae) Biotropica. 1979;11:161–172. doi: 10.2307/2388036. [DOI] [Google Scholar]
- 8.Giordano CV, Sánchez RA, Austin AT. Gregarious bamboo flowering opens a window of opportunity for regeneration in a temperate forest of Patagonia. New Phytol. 2009;181:880–889. doi: 10.1111/j.1469-8137.2008.02708.x. [DOI] [PubMed] [Google Scholar]
- 9.Zhang YQ, Wang XM, Wu AL, Ren Y. Merging Fargesia dracocephala into Fargesia decurvata (Bambusoideae, Poaceae): implications from morphological and ITS sequence analysis. PLoS ONE. 2014;9:e101362. doi: 10.1371/journal.pone.0101362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sneath, P. H. A., & Sokal, R. R. Numerical taxonomy. W. H. Freeman Press, San Francisco, 315–320 (1973).
- 11.Das M, Bhattacharya S, Basak J, Pal A. Phylogenetic relationships among the bamboo species as revealed by morphological characters and polymorphism analyses. Biol. Plant. 2007;51:667–672. doi: 10.1007/s10535-007-0140-7. [DOI] [Google Scholar]
- 12.Desai P, et al. Comparative assessment of genetic diversity among Indian bamboo genotypes using RAPD and ISSR markers. Mol. Biol. Rep. 2015;42:1265–1273. doi: 10.1007/s11033-015-3867-9. [DOI] [PubMed] [Google Scholar]
- 13.Li DZ. On some problems of methodology of bamboo classification with special reference to the circumscription of Dendrocalamus. Acta Phytotax. Sin. 1994;31:283–289. [Google Scholar]
- 14.Hodkinson TR, et al. Phylogenetic analyses of plastid and nuclear DNA sequences indicate a rapid late Miocene radiation of the temperate bamboo tribe Arundinarieae (Poaceae, Bambusoideae) Plant Ecol. Divers. 2010;3:109–120. doi: 10.1080/17550874.2010.521524. [DOI] [Google Scholar]
- 15.Zhao HS, et al. Developing genome-wide microsatellite markers of bamboo and their applications on molecular marker assisted taxonomy for accessions in the genus Phyllostachys. Sci. Rep. 2015;5:e8018. doi: 10.1038/srep08018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W, Franklin SB, Lu ZJ, Rude BJ. Delayed flowering in bamboo: evidence from Fargesia qinlingensis in the Qinling Mountains of China. Front. Plant Sci. 2016;7:e151. doi: 10.3389/fpls.2016.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malcomber ST, Kellogg EA. Evolution of unisexual flowers in grasses (Poaceae) and the putative sex-determination gene. TASSELSEED2 (TS2). New Phytol. 2006;170:885–899. doi: 10.1111/j.1469-8137.2006.01726.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhou YY, Wang YQ, Zhang L, Hu ZM. Mathematic classification of 46 species in Rhododendron with the morphologic characteristics. Sci. Sil. Sin. 2009;45:67–75. [Google Scholar]
- 19.Guo ZH, Li DZ. Phylogenetics of the Thamnocalamus group and its allies (Gramineae: Bambusoideae): inference from the sequences of GBSSI gene and ITS spacer. Mol. Phylogenet. Evol. 2004;30:1–12. doi: 10.1016/S1055-7903(03)00161-1. [DOI] [PubMed] [Google Scholar]
- 20.Yang HQ, et al. A molecular phylogenetic and fruit evolutionary analysis of the major groups of the paleotropical woody bamboos (Gramineae: Bambusoideae) based on nuclear ITS, GBSSI gene and plastid trnL-F DNA sequences. Mol. Phylo. Evol. 2008;48:809–824. doi: 10.1016/j.ympev.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Zhang YX, Zeng CX, Li DZ. Complex evolution in Arundinarieae (Poaceae: Bambusoideae): Incongruence between plastid and nuclear GBSSI gene phylogenies. Mol. Phylo. Evol. 2012;63:777–797. doi: 10.1016/j.ympev.2012.02.023. [DOI] [PubMed] [Google Scholar]
- 22.Wang XQ, Zhao L, Eaton DAR, Li DZ, Guo ZH. Identification of SNP markers for inferring phylogeny in temperate bamboos (Poaceae: Bambusoideae) using RAD sequencing. Mol. Ecol. Resour. 2013;13:938–945. doi: 10.1111/1755-0998.12136. [DOI] [PubMed] [Google Scholar]
- 23.Yang HM, Zhang YX, Yang JB, Li DZ. The monophyly of Chimonocalamus and conflicting gene trees in Arundinarieae (Poaceae: Bambusoideae) inferred from four plastid and two nuclear markers. Mol. Phylo. Evol. 2013;68:340–356. doi: 10.1016/j.ympev.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Attigala L, Kathriarachchi HS, Clark LG. Taxonomic revision of the temperate woody bamboo genus kuruna (Poaceae: Bambusoideae: Arundinarieae) Syst. Bot. 2016;23:174–196. doi: 10.1600/036364416X690570. [DOI] [Google Scholar]
- 25.Lin GF, Chen LH. Identification of homogeneous regions for regional frequency analysis using the self-organizing map. J. Hydrol. 2006;324:1–9. doi: 10.1016/j.jhydrol.2005.09.009. [DOI] [Google Scholar]
- 26.Céréghino R, Santoul F, Compin A, Mastrorillo S. Using self-organizing maps to investigate spatial patterns of non-native species. Biol. Conserv. 2005;125:459–465. doi: 10.1016/j.biocon.2005.04.018. [DOI] [Google Scholar]
- 27.Kohonen T. Self-organized formation of topologically correct feature maps. Biol. Cybern. 1982;43:59–69. doi: 10.1007/BF00337288. [DOI] [Google Scholar]
- 28.Kohonen T. Essentials of the self-organizing map. Neural Networks. 2013;37:52–65. doi: 10.1016/j.neunet.2012.09.018. [DOI] [PubMed] [Google Scholar]
- 29.Lin GF, Chen LH. Time series forecasting by combining the radial basis function network and the self-organizing map. Hydrol. Process. 2005;19:1925–1937. doi: 10.1002/hyp.5637. [DOI] [Google Scholar]
- 30.Tison J, et al. Typology of diatom communities and the influence of hydro-ecoregions: a study on the French hydrosystem scale. Water Res. 2005;39:3177–3188. doi: 10.1016/j.watres.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 31.Ley R, Casper MC, Hellebrand H, Merz R. Catchment classification by runoff behavior with self-organizing maps (SOM) Hydrol. Earth Syst. Sci. 2011;15:2947–2962. doi: 10.5194/hess-15-2947-2011. [DOI] [Google Scholar]
- 32.Ghaseminezhad MH, Karami A. A novel self-organizing map (SOM) neural network for discrete groups of data clustering. Appl. Soft Comput. 2011;11:3771–3778. doi: 10.1016/j.asoc.2011.02.009. [DOI] [Google Scholar]
- 33.Grass Phylogeny Working Group II [GPWG II] New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytol. 2012;193:304–312. doi: 10.1111/j.1469-8137.2011.03972.x. [DOI] [PubMed] [Google Scholar]
- 34.Bai SB, et al. Can native clonal moso bamboo encroach on adjacent natural forest without human intervention? Sci. Rep. 2016;6:e31504. doi: 10.1038/srep31504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gielis J, Goetghebeur P, Debergh P. Physiological aspects and experimental reversion of flowering in Fargesia murieliae (Poaceae, Bambusoideae) Syst. Geogr. Plants. 1999;68:147–158. doi: 10.2307/3668597. [DOI] [Google Scholar]
- 36.Wang W, Franklin SB, Ouellette JR. Clonal regeneration of an arrow bamboo, Fargesia qinlingensis, following giant panda herbivory. Plant Ecol. 2007;192:97–106. doi: 10.1007/s11258-006-9229-x. [DOI] [Google Scholar]
- 37.Guo ZH, Chen YY, Li DZ. Phylogenetic studies on the Thamnocalamus group and its allies (Gramineae: Bambusoideae) based on ITS sequence data. Mol. Phylogenet. Evol. 2002;22:20–30. doi: 10.1006/mpev.2001.1039. [DOI] [PubMed] [Google Scholar]
- 38.Yi, T. P. Fargesia. Flora reipublicae popularis sinicae. Science Press, Beijing, 387–378 (in Chinese with English abstract) (1996).
- 39.Li, D.Z. The flora of China Bambusoideae project—Problems and current understanding of bamboo taxonomy in China. Academic Press, London, 215–216 (1997).
- 40.Wang, C.P., & Ye, G.H. On the problems of the classification of Chinese bamboos with creeping rhizomes. Acta Phytotax. Sin. 18, 292–307 (in Chinese with English abstract) (1980).
- 41.Yi, T.P. A study on the genus Fargesia Fr. from China. J. Bamboo Res. 7, 111–119 (in Chinese with English abstract) (1988).
- 42.Sun MS, Guo XJ, Dong YR, Li DZ, Yang HQ. Fargesia microauriculata (Poaceae, Bambusoideae), a new species from northwest Yunnan, China. Ann. Bot. Fenn. 2016;53:280–284. doi: 10.5735/085.053.0603. [DOI] [Google Scholar]
- 43.Soderstrom TR. Some evolutionary trends in the Bambusoideae (Poaceae) Ann. Misso. Bot. Gard. 1981;68:15–47. doi: 10.2307/2398809. [DOI] [Google Scholar]
- 44.Chao CS, Tang GG. The present status and problems of bamboo classification in China. J. Nanjing Forestry Univ. (Natural Science Edition) 1993;17:1–8. [Google Scholar]
- 45.Li, D. Z. et al. Bambuseae (Poaceae). In: Wu, Z. Y., Raven, P. H., & Hong, D. Y. (Eds.), Flora of China. Science Press and Missouri Botanical Garden Press, Beijing and St. Louis, 7–180 (2006).
- 46.Zhang, et al. Multi-locus plastid phylogenetic biogeography supports the Asian hypothesis of the temperate woody bamboos (Poaceae: Bambusoideae) Mol. Phylo. Evol. 2016;96:118–129. doi: 10.1016/j.ympev.2015.11.025. [DOI] [PubMed] [Google Scholar]
- 47.Moyroud E, Kusters E, Monniaux M, Koes R, Parcy F. LEAFY blossoms. Trends Plant Sci. 2010;15:346–352. doi: 10.1016/j.tplants.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 48.Yi, T. P., Shi, J. Y., Ma, L. S., Wang, H. T. & Yang, L. Pictorial Record of Chinese Bamboo. Science Press, Beijing, 236–237 (in Chinese with English abstract) (2008).
- 49.Tzvelev NN. The system of grasses (Poaceae) and their evolution. Bot. Rev. 1989;55:141–203. doi: 10.1007/BF02858328. [DOI] [Google Scholar]
- 50.Stapleton CMA. Bergbambos and Oldeania, new genera of African bamboos (Poaceae, Bambusoideae) PhytoKeys. 2013;25:87–103. doi: 10.3897/phytokeys.25.6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kelchner SA. Higher level phylogenetic relationships within the bamboos (Poaceae: Bambusoideae) based on five plastid markers. Mol. Phylogenet Evol. 2013;67:404–413. doi: 10.1016/j.ympev.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 52.Stapleton CMA. The bamboos of the Nepal and Bhutan, Part II: Arundinaria, Thamnocalamus, Borinda and Yushania (Gramineae: Poaceae, Bambusoideae) Edinb. J. Bot. 1994;51:275–295. doi: 10.1017/S0960428600000883. [DOI] [Google Scholar]
- 53.Jasińska AK, et al. Relationships among Cedrus libani, C brevifolia and C atlantica as revealed by the morphological and anatomical needle characteristics. Plant Syst. Evol. 2013;299:35–48. doi: 10.1007/s00606-012-0700-y. [DOI] [Google Scholar]
- 54.Ward JH., Jr. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963;58:236–244. doi: 10.1080/01621459.1963.10500845. [DOI] [Google Scholar]
- 55.Sahigara F, et al. Comparison of different approaches to define the applicability domain of QSAR models. Molecules. 2012;17:4791–4810. doi: 10.3390/molecules17054791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng, W. J. Editorial Committee of the Records of Chinese Trees. Records of Chinese Trees. China Forestry Publishing House, Beijing, 236–237 (in Chinese with English abstract) (2004).
- 57.Ceyhan E, et al. Quantization and analysis of hippocampal morphometric changes due to dementia of Alzheimer type using metric distances based on large deformation diffeomorphic metric mapping. Comput. Med. Imag. Graph. 2011;35:275–293. doi: 10.1016/j.compmedimag.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen SL, Xu KX, Sheng GY. On the numerical classification and determination of taxa of Chinese bamboos with leptomorph rhizomes. J. Grad. School Chinese Acad. Sci. 1983;21:113–120. [Google Scholar]
- 59.Larsen, R. J. & Marx, M. L. An introduction to mathematical statistics and its applications (Third Edition). Pearson Education Press, San Antonio, 257–258 (2000).
- 60.Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z-score transformation. J. Mol. Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sokal, R. R. & Rohlf, T. J. Biometry: principles and practice of statistics in biological research. W.H. Freeman Press, San Francisco, 125–126 (1997).
- 62.Ҫam M, Yaşar H, Durmaz G. Classification of eight pomegranate juices based on antioxidant activity measured by four methods. Food Chem. 2009;112:721–726. doi: 10.1016/j.foodchem.2008.06.009. [DOI] [Google Scholar]
- 63.Vesanto J. SOM-based data visualization methods. Intelligent Data Anal. 1999;3:111–126. doi: 10.1016/S1088-467X(99)00013-X. [DOI] [Google Scholar]
- 64.Ciccarelli FD, et al. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


