ABSTRACT
Vertebrate proteins that fulfill multiple and seemingly disparate functions are increasingly recognized as vital solutions to maintaining homeostasis in the face of the complex cell and tissue physiology of higher metazoans. However, the molecular adaptations that underpin this increased functionality remain elusive. In this Commentary, we review the PACS proteins – which first appeared in lower metazoans as protein traffic modulators and evolved in vertebrates to integrate cytoplasmic protein traffic and interorganellar communication with nuclear gene expression – as examples of protein adaptation ‘caught in the act’. Vertebrate PACS-1 and PACS-2 increased their functional density and roles as metabolic switches by acquiring phosphorylation sites and nuclear trafficking signals within disordered regions of the proteins. These findings illustrate one mechanism by which vertebrates accommodate their complex cell physiology with a limited set of proteins. We will also highlight how pathogenic viruses exploit the PACS sorting pathways as well as recent studies on PACS genes with mutations or altered expression that result in diverse diseases. These discoveries suggest that investigation of the evolving PACS protein family provides a rich opportunity for insight into vertebrate cell and organ homeostasis.
Summary: We summarize how the vertebrate PACS proteins acquired specific motifs that enabled them to function as metabolic switches that integrate cytoplasmic membrane trafficking and interorganellar communication with nuclear gene expression.
Introduction
At the turn of the 20th century, Archibald Garrod, Walter Sutton and Thomas Hunt Morgan ushered in a new era of biological research by replacing observational studies with mechanistic analyses (Kelves and Hood, 1992). Their discoveries that chromosomes encode heritable traits set the foundation for understanding genetic inheritance. More than 30 years later, through inactivation and mutational studies of the common bread mold Neurospora, George Beadle and Edward Tatum ultimately determined that genes encode enzymes and laid a cornerstone of modern molecular biology through the ‘one gene – one protein’ hypothesis (Beadle and Tatum, 1941). This provocative model inevitably collapsed under the weight of subsequent discoveries, which revealed that genes frequently encode multiple proteins or collections of peptides that drive complex physiology and contribute to molecular promiscuity through mechanisms ranging from alternative RNA splicing to the proteolytic processing of polyproteins (‘one gene – many proteins’; Yang et al., 2016). The essence of these studies led to the present-day ‘one protein – multiple functions’ mantra, which recognizes that individual proteins often have multiple and seemingly disparate functions – a phenomenon known as ‘moonlighting’ (Jeffery, 1999; Henderson and Martin, 2014). The glycolytic enzymes glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and enolase are widely cited examples of moonlighting proteins. Human GAPDH participates in as many as 20 different cellular functions ranging from the regulation of membrane traffic to the maintenance of genome integrity (Sirover, 2011). Human enolase can be present on the cell surface of activated monocytes where it serves as a plasminogen receptor that promotes recruitment of inflammatory cells to sites of tissue injury (Wygrecka et al., 2009).
Protein moonlighting, which by definition is limited to proteins that use a single primary sequence in the absence of posttranslational modifications to mediate multiple functions (Jeffery, 1999), is inherently limited for solving evolutionary challenges. Eventually, moonlighting gives way to gene duplication, which enables the resulting paralogs to acquire mutations that expand the molecular functions initiated by the ancestral protein. Inevitably, such a hit-and-miss approach to mutational diversification will hit a roadblock in the form of rigidly folded domains, such as SH2 or PDZ domains found in many proteins (Tompa et al., 2014). The dedicated structure of these ∼100-amino-acid protein domains, which are typically involved in high-affinity binding to discrete ligands, limits amino acid substitutions, thereby reducing their ability to increase the functional density of associated cellular proteins. Therefore, protein adaptation represents an additional and vital solution to increase the array of protein functions to an extent that is not possible by moonlighting and gene duplication alone (Tompa et al., 2014). Rather than coaxing rigid protein domains to accept new roles, such proteins – notably vertebrate proteins – have acquired intrinsically disordered regions (IDRs) to complement the limited utility of folded domains (Dunker et al., 2005; Pancsa and Tompa, 2016). IDRs do not fold into an autonomous structure, but contain flexible 5–15-amino-acid small linear motifs (SLiMs) that can assume an ensemble of ‘structures’, which in turn interact with a panoply of binding partners through low-affinity but high-specificity interactions (Davey et al., 2015). SLiMs are frequently sites of posttranslational modifications and undergo rapid evolution, greatly increasing the functional density of IDR-containing proteins (Tompa et al., 2014; Pancsa and Tompa, 2016). This plasticity enabled SLiMs from disparate proteins across a large taxonomic range to acquire similar amino acid mutations by convergent evolution so that they may bind to a common globular protein domain (Schlessinger et al., 2011; Davey et al., 2012, 2015; Van Roey et al., 2012; Jonas and Izaurralde, 2013). However, examples tracing how a single protein adapted to evolutionary pressure by acquiring specific SLiM mutations in order to fulfill new functions that are critical for vertebrate homeostasis are surprisingly limited. Studies on the phosphofurin acidic cluster sorting (PACS) proteins provide a powerful illustration of such evolutionary protein adaptations ‘caught in the act’ of connecting the increasingly complex organellar landscape that underpins vertebrate cell function.
Here, we discuss how PACS proteins first appeared in metazoans as membrane traffic regulators, and then expanded their functional repertoire in vertebrates by acquiring molecular switches, notably phosphorylation sites and nuclear localization signals (NLSs) within their SLiMs. These gain-of-function adaptations enabled the vertebrate PACS proteins to integrate cytoplasmic functions with nuclear gene expression. In particular, we focus on the acquisition of an Akt phosphorylation site and NLS in vertebrate PACS-2, which enable this protein to function as a metabolic switch that integrates cytoplasmic membrane traffic and interorganellar communication with nuclear gene expression in response to anabolic or catabolic cues.
PACS proteins direct membrane trafficking in worms to humans
The PACS proteins were discovered in a genetic screen for regulators of the secretory pathway proteinase furin (Wan et al., 1998; Thomas, 2002). Human PACS1 is located at 11q13.1-q13.2 and contains 24 exons and at least 12 alternatively spliced variants. Human PACS-2, which is located near the telomere at 14q32.33, also contains 24 exons and at least 11 alternatively spliced variants. Genome-wide association studies (GWAS) identified the human PACS1 locus as a susceptibility gene in severe early-onset obesity (Wheeler et al., 2013) and developmental delay (Deciphering Developmental Disorders Study, 2017), and mutations in PACS1 underlie ‘PACS-1 syndrome’ (Schuurs-Hoeijmakers et al., 2012, 2016; Stern et al., 2017), characterized by epileptic seizures, heart defects, hypotonia and craniofacial malformations. The PACS2 locus is highly susceptible to loss of heterozygosity in colorectal cancer, leading to a reduction or loss of PACS-2 protein expression (Anderson et al., 2001; Aslan et al., 2009). Similar to PACS1, mutations in PACS2 are associated with neonatal onset epilepsy, global developmental delay and intellectual disability (C. Thauvin and H. Olson, personal communication).
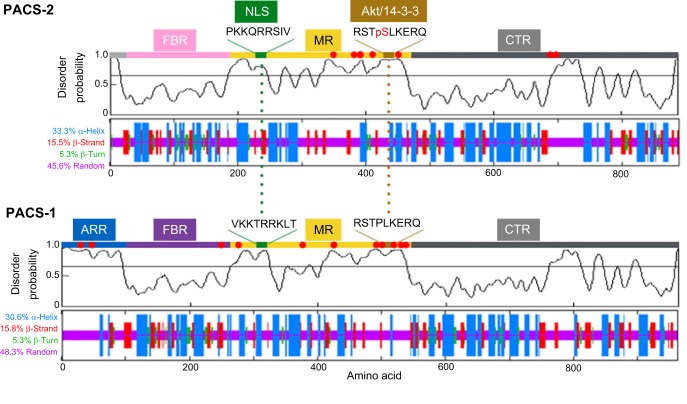
The canonical 963-amino-acid PACS-1 and 889-amino-acid PACS-2 proteins share 54% sequence identity. Bioinformatics analyses suggest both proteins lack folded globular domains and are instead nearly 50% disordered (Fig. 1). The ∼150-amino-acid cargo (furin)-binding regions (FBRs) in PACS-1 and PACS-2, which are predicted to be structured (Fig. 1), share nearly 80% sequence identity and bind client proteins, as well as the cytoplasmic membrane trafficking machinery as described below. PACS-1 and PACS-2 also share a disordered middle region (MR), which contains an autoregulatory domain as well as NLSs and, specifically in PACS-2, a critical Akt phosphorylation site that binds to 14-3-3 proteins. The function of the shared C-terminal region (CTR) is unknown. PACS-1 additionally contains an N-terminal extension called the atrophin-1-related region (ARR), which has homology to this nuclear transcriptional co-repressor (Zhang et al., 2006).
Fig. 1.
Disorder prediction for PACS-1 and PACS-2. The PrDOS server (http://prdos.hgc.jp/cgi-bin/top.cgi; Ishida and Kinoshita, 2007) was used to predict natively disordered regions from the amino acid sequences of the human PACS-1 (UniProt Q6VY07) and PACS-2 (UniProt Q86VP3) proteins [false discovery rate (FDR)=2%]. The disorder probabilities for each residue were plotted as a function of length and the graphical profiles were juxtaposed with the predicted secondary structures, which were obtained using an improved self-optimized prediction method (SOPMA) on a set of aligned members of the PACS-1 or PACS-2 protein families (lower plots). The PACS-2 nuclear localization signal (NLS) and Akt site, which binds 14-3-3 proteins, together with the corresponding sequences in PACS-1 are shown and predicted to reside in disordered regions. ARR, atrophin-1-related region; FBR, furin (cargo)-binding region; MR, middle region; CTR, C-terminal region. Red dots, phosphorylation sites [as predicted by PhosphoSitePlus (http://www.phosphosite.org/; Hornbeck et al., 2015)].
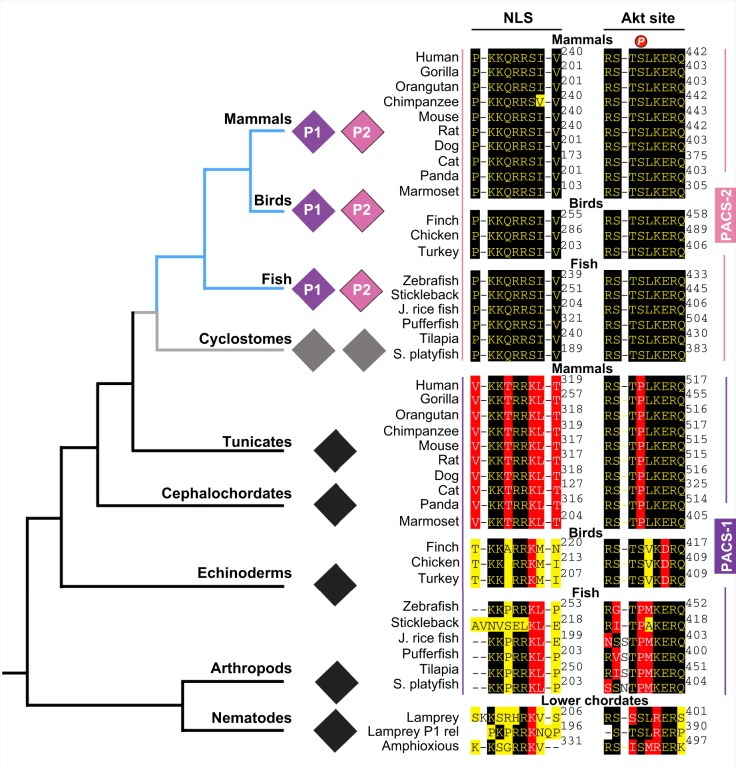
The PACS genes are a recent addition to the eukaryotic genome, appearing first in lower metazoans (Fig. 2). Invertebrates, including nematodes, arthropods and echinoderms, possess a single PACS locus that is apparently dedicated to membrane trafficking. Notably, Caernorhabditis elegans PACS (cePACS, T18H9.7a) localizes to early endosomes at the presynaptic terminus of the neuromuscular junction where it mediates synaptic transmission (Sieburth et al., 2005). Lower chordates, including Amphioxus and tunicates also possess only a single gene. The PACS gene was duplicated with the appearance of the vertebrates, resulting in the PACS-1 and PACS-2 genes.
Fig. 2.
Phylogenetic analysis of the PACS genes. Non-redundant protein sequences of the PACS family members were obtained from the UniProt and NCBI databases. The protein sequences were aligned using Muscle and were manually examined/modified for their accuracy within the non-conserved domains that flank conserved domains. The program Gblocks was used to curate and eliminate poorly aligned positions and divergent regions with the protein alignment prior to the phylogenetic analysis (Castresana, 2000). The program PhyML was used to estimate the maximum likelihood phylogenies from alignments of amino acid sequences (Guindon et al., 2005). The tree (A) and multiple alignments of the NLS and Akt sites (B) were visualized using Mega7. Black diamonds indicate invertebrate PACS proteins expressed from a single gene. Gray diamonds indicate cyclostome PACS proteins, which may be precursors to PACS-1 and PACS-2 and expressed from duplicated genes. Purple and magenta diamonds represent subfunctionalized PACS-1 and PACS-2 paralogs, respectively, that are expressed from duplicated genes in jawed vertebrates. A black background to the amino acid residue indicates identical residues, a red background to the amino acid residue indicates similar residues, and a yellow background divergent residues.
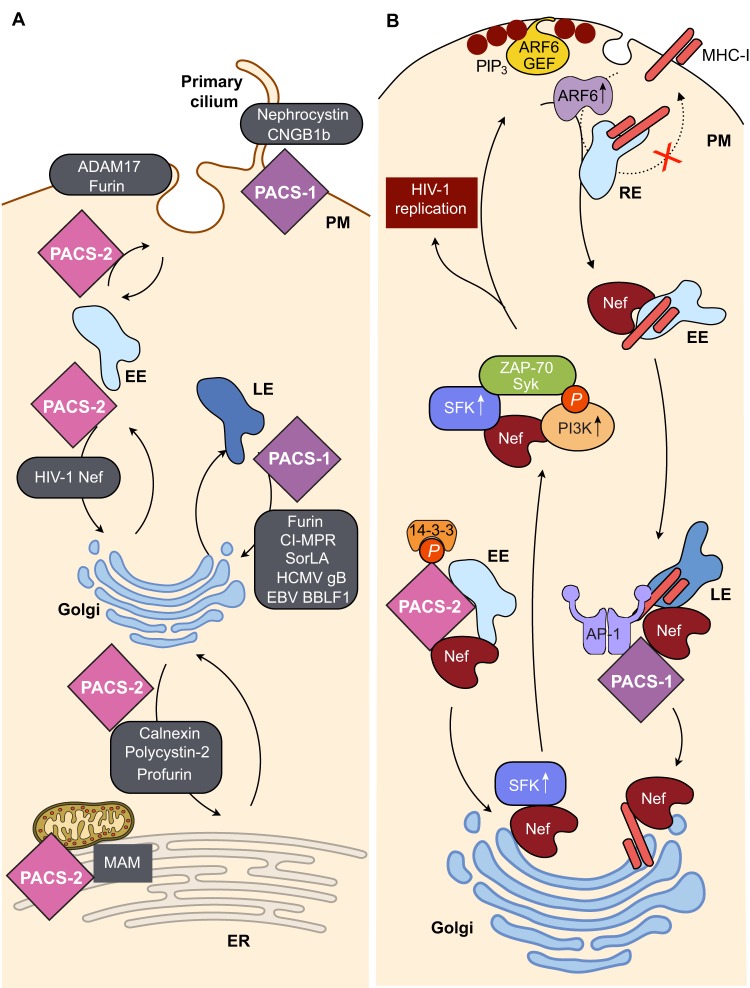
The regulation of membrane traffic remains a role that is conserved among the vertebrate PACS proteins. The PACS-1 FBR binds to acidic clusters that can be phosphorylated by casein kinase 2 (CK2), as well as α-helices on a large number of client proteins (Wan et al., 1998; Youker et al., 2009; Dikeakos et al., 2012). Thus, PACS-1, which interacts with the clathrin adaptors AP-1 and AP-3, as well as the monomeric adaptor GGA3, mediates localization of furin and other client proteins to the trans-Golgi network (TGN) and also targets a subset of client proteins to the primary cilium, including the adaptor protein nephrocystin (also known as NPHP1) and the olfactory cyclic-nucleotide-gated ion channel (CNG), the latter by binding to the β1 subunit (CNGB1) (Fig. 3A) (Wan et al., 1998; Schermer et al., 2005; Jenkins et al., 2009; Youker et al., 2009). PACS-1 acquired a CK2-phosphorylated acidic cluster of its own, which is located in the disordered MR (see Fig. 1). CK2 phosphorylation of Ser278 in the PACS-1 autoregulatory domain controls intramolecular binding to the FBR, which regulates the interaction with client proteins (Scott et al., 2003). PACS-2, which interacts with the coatomer COPI, mediates the localization of cargo to the endoplasmic reticulum (ER) and, similar to cePACS, directs trafficking from early endosomes (Youker et al., 2009) (Fig. 3A, see also Fig. 4A). PACS-2 stabilizes a pool of the metalloproteinase ADAM17 (also known as TACE) on early endosomes, from where the enzyme is trafficked to the cell surface to shed ErbB ligands, including those that ligate the epidermal growth factor receptor (EGFR) (Dombernowsky et al., 2015). In the absence of PACS-2, ADAM17 is diverted to lysosomes, which reduces ErbB shedding. Correspondingly, EGFR signaling is reduced in the intestinal epithelium of PACS2−/− mice (Dombernowsky et al., 2015). The role of the candidate autoregulatory domain in the PACS-2 MR has not been established.
Fig. 3.
Protein traffic steps mediated by PACS-1 and PACS-2. (A) PACS-1 mediates the sorting of client proteins from late endosomes (LE) to the TGN, from early endosomes (EE) to the plasma membrane (PM), as well as delivery to the primary cilium. PACS-2 mediates the localization of cargo proteins to the ER, from early endosomes to the TGN or plasma membrane, and also promotes MAM integrity. (B) HIV-1 Nef usurps the sorting steps mediated by PACS-2 and PACS-1 to downregulate the levels of cell surface MHC-I in CD4+ T-cells. Nef binds to Akt-phosphorylated PACS-2 on early endosomes (Atkins et al., 2008; Dikeakos et al., 2012, and L.T. and G.T., unpublished results). This allows Nef to traffic to the TGN region where it binds and activates a Src family kinase (SFK; Hck, Src or Lyn). The Nef–SFK complex then recruits ZAP-70 (in T-cells) or Syk (in monocytes and other cell types) and a class I PI3K, which increases the level of PIP3 (maroon circles) at the plasma membrane. This recruits an ARF6 GEF that accelerates MHC-I internalization through an ARF6-regulated endocytic pathway. Nef diverts the internalized MHC-I molecules from their local recycling compartment (dashed line) and combines with AP-1 and PACS-1 to transport MHC-I through early and late endosomes and sequester it in the TGN. The identity of the precise compartment containing Nef, MHC-I, AP-1 and PACS-1 is under investigation. This MHC-I downregulation pathway protects HIV-1 from CD8+ T-cell killing thereby allowing the virus to evade immune surveillance. Thus, small-molecule inhibition of the multi-kinase complex re-exposes MHC-I on the cells surface and sensitizes HIV-1-infected cells to CD8+ T-cell killing. The steps shown here depict the ‘signaling mode’ of HIV-1 Nef-induced immune evasion, which HIV-1 implements during the first 48 h post infection. A detailed discussion of Nef-induced immune evasion is presented elsewhere (Dikeakos et al., 2010; Pawlak and Dikeakos 2015).
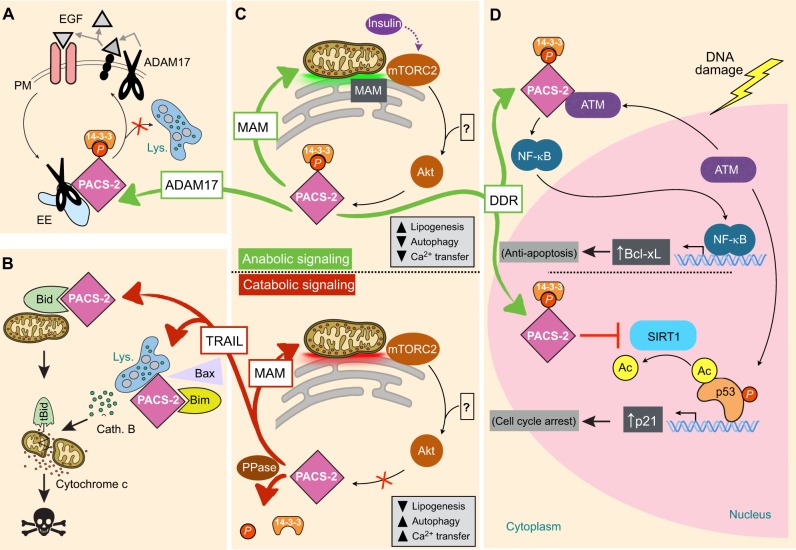
Fig. 4.
The PACS-2 Akt site and NLS together modulate membrane traffic, TRAIL-induced apoptosis, MAM integrity and the response to DNA damage. (A) Protein trafficking. Akt-phosphorylated pSer437-PACS-2 (pPACS-2) interacts with ADAM17 on early endosomes (EE) and mediates delivery of the protease to the cell surface where it sheds EGF ligands to stimulate EGFR signaling. In the absence of PACS-2, ADAM17 is degraded in lysosomes (Lys.). (B) TRAIL-induced MOMP. TRAIL triggers dephosphorylation of PACS-2 Ser437, which mediates two trafficking steps required for MOMP. In one trafficking step, PACS-2 binds full-length Bid and translocates Bid to mitochondria. In the other trafficking step, PACS-2 forms a complex with Bim and Bax on lysosomes called the PIXosome, which is required for lysosome membrane permeabilization to release cathepsin B (cath. B). (C) MAMS. Top panel: insulin or growth factors trigger activation of mTORC2 on mitochondria-associated membranes (MAMs; green shading at the ER–mitochondria contact site), which activates Akt to phosphorylate PACS-2. In turn, pPACS-2 increases MAM contacts, which may modulate ER–mitochondria exchange and support increased lipogenesis. The ? denotes signaling pathways that may lead to Akt-dependent phosphorylation of PACS-2 independent of MAM-localized TORC2. Bottom panel: in starved cells or cells treated with TRAIL, Akt is inhibited and PACS-2 Ser437 is dephosphorylated by a protein phosphatase (PPase). Dephosphorylated PACS-2 in turn remodels MAMs (red shading at the ER–mitochondria contact site), which may reduce lipogenesis but increase ER–mitochondrial Ca2+ exchange as well as induction of autophagy. (D) DNA damage response. Top panel: to support induction of the NF-κB and Bcl-xL anti-apoptotic pathway, cytoplasmic PACS-2 interacts with a pool of ATM released from the nucleus and maintains the DNA damage kinase in the cytoplasm. The cytoplasmic ATM then triggers activation of the canonical IκBα–NF-κB pathway that leads to induction of anti-apoptotic Bcl-xL. Bottom panel: to support induction of the p53–p21 cell cycle arrest pathway, pPACS-2 traffics to the nucleus where it binds and inhibits SIRT1 to protect acetylation of p53 bound to the p21 promoter, promoting p21 induction and cell cycle arrest. Green arrows, pro-survival-anabolic pathways mediated by pPACS-2. Red arrows, apoptotic or catabolic pathways mediated by dephosphorylated PACS-2. Ac, acetylation; DDR, DNA-damage response.
Several pathogenic viruses acquired furin-like acidic clusters to exploit the endosomal sorting steps mediated by PACS-1 and PACS-2. This viral mimicry (see Davey et al., 2011) enables the viruses to assemble progeny, escape immune surveillance and prevent apoptosis. For example, PACS-1 binds to acidic clusters in the human cytomegalovirus (HCMV) envelope glycoprotein gB and the Epstein–Barr virus (EBV) tegument protein BBLF1 and localizes them to the TGN to support virus assembly (Crump et al., 2003; Chiu et al., 2012) (see Fig. 3A). Furthermore, PACS-2 interacts with a pair of small acidic clusters in the ubiquitin ligase K5 of Kaposi sarcoma herpesvirus (KSHV) at the ER. This interaction enables KSHV to downregulate the cell adhesion molecule CD31 (also known as PECAM1), which may contribute to KSHV-induced cancer (Mansouri et al., 2006). The HIV-1 accessory protein Nef uses a bipartite motif composed of a short acidic cluster and the αB helix to interact with both PACS-1 and PACS-2 (Piguet et al., 2000; Atkins et al., 2008; Dikeakos et al., 2012). This bipartite binding enables HIV-1 Nef to downregulate major histocompatibility complex class I (MHC-I) from the cell surface, which allows the virus to escape immune detection (Pawlak and Dikeakos, 2015) (Fig. 3B). To this end, Nef interacts with PACS-2 on early endosomes, which enables the HIV-1 protein to assemble a multi-kinase complex consisting of a Src family kinase (SFK), ζ-chain-associated protein kinase 70 (ZAP-70) and a class I phosphoinositide-3 kinase (PI3K) (Blagoveshchenskaya et al., 2002; Hung et al., 2007; Atkins et al., 2008). The activated multi-kinase complex increases the amount of phosphatidylinositol (3,4,5)-trisphosphate (PIP3) underneath the plasma membrane, which recruits an ARF6 GEF to activate ARF6 and accelerate endocytosis of cell-surface MHC-I (Blagoveshchenskaya et al., 2002). Nef then connects the internalized MHC-I molecules to PACS-1 and AP-1 on an endosomal compartment to prevent their recycling to the cell surface and instead sequester them in the TGN, thereby protecting the virus from immune surveillance (Blagoveshchenskaya et al., 2002; Chaudhry et al., 2008; Noviello et al., 2008; Dikeakos et al., 2010; Dirk et al., 2016). Unlike KSHV and many other viruses, HIV-1 Nef does not induce degradation of downregulated MHC-I (Blagoveshchenskaya et al., 2002; Dikeakos et al., 2010; Dirk et al., 2016). This chink in the armor of HIV-1 provides an alternative approach to combat the virus by reversing the Nef-mediated immune evasion pathway. In support of this possibility, treatment of HIV-1-infected primary CD4+ T-cells with small-molecule inhibitors of the multi-kinase complex restores cell surface expression of MHC-I and sensitizes them to killing by CD8+ T-cells (Hung et al., 2007; Dikeakos et al., 2010; and M. Ostrowski, personal communication).
Links between mammalian PACS proteins and TRAIL-induced apoptosis
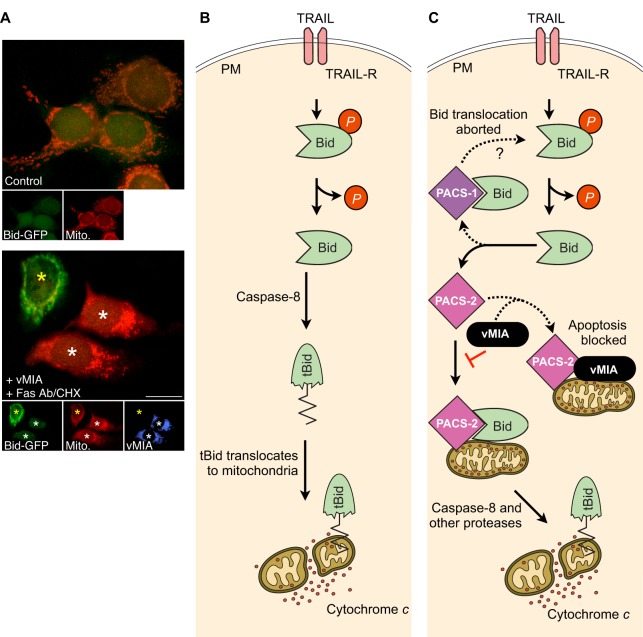
Surprisingly, while PACS-2 mediates trafficking of many pro-survival signaling molecules, it is also tasked with mediating death-ligand-induced apoptosis induced by TNF-related apoptosis-inducing ligand (TRAIL, also known as TNFSF10). This clinically important death ligand is an in vivo metastasis inhibitor that selectively kills diseased cells, including cancer cells and virally infected cells (Johnstone et al., 2008). TRAIL-induced cell death requires the coordinated permeabilization of multiple organelles, including mitochondria and lysosomes (Aslan and Thomas, 2009). TRAIL triggers mitochondrial outer membrane permeabilization (MOMP) by inducing dephosphorylation and proteolytic cleavage of the pro-apoptotic Bcl-2 protein Bid to form truncated Bid (tBid), which drives Bax-dependent MOMP and the consequent activation of executioner caspases (Aslan et al., 2009).
PACS-2 has essential roles in TRAIL-induced MOMP (Fig. 4B). In response to TRAIL, PACS-2 binds full-length dephosphorylated Bid and traffics it to mitochondria where it can be converted into tBid (Simmen et al., 2005). In parallel, TRAIL triggers assembly of a complex that contains PACS-2, Bim (also known as BCL2L11) and Bax, called the PIXosome, on lysosomal membranes (Werneburg et al., 2012). The PIXosome drives lysosomal membrane permeabilization (LMP), which releases cathepsin B and other hydrolases that are required for MOMP into the cytosol (Boya and Kroemer, 2008). Several mechanisms interfere with these proapoptotic functions of PACS-2. In hepatocellular carcinoma cells, inhibitors of apoptosis (IAPs) target PACS-2 for proteasomal degradation, thereby protecting the cancer cells from being killed by TRAIL (Guicciardi et al., 2014). In cardiomyocytes, miR499 prevents Bid-dependent apoptosis by downregulating PACS-2 (Wang et al., 2014). Not surprisingly, herpesviruses also exploit the ability of PACS-2 to traffic proteins to mitochondria. The HCMV protein viral inhibitor of apoptosis (vMIA) prevents cell death by trapping pro-apoptotic Bax on mitochondria (Arnoult et al., 2004). vMIA interacts with PACS-2 but not PACS-1 (our unpublished data), and the vMIA–PACS-2 interaction is required for efficient translocation of vMIA to mitochondria (Salka et al., 2017). Similar to the fate of Bax, vMIA traps PACS-2 on mitochondria. Notably, this sequestration of PACS-2 is coupled with a blunted translocation of Bid to mitochondria in response to death ligands, suggesting vMIA may trap both inactivated Bax and PACS-2 on mitochondria to inhibit apoptosis in HCMV-infected cells (Fig. 5A; L.T., T.S., J.A. and G.T., unpublished results). The PACS-2-dependent recruitment of Bid to mitochondria prior to formation of tBid suggests this translocation step may be a checkpoint for apoptotic regulation. In support of this possibility, PACS-1 also binds dephosphorylated Bid, but prevents translocation of Bid to mitochondria (T.S., L.T., J.A. and G.T., unpublished results).
Fig. 5.
Possible models for the regulation of Bid translocation to mitochondria by PACS proteins and vMIA. (A) Example of an experiment showing that vMIA prevents translocation of Bid–GFP to mitochondria (L.T., T.S., J.A. and G.T., unpublished results). MCF-7 cells expressing Bid–GFP were left untreated (control, top) or transfected with a vector expressing vMIA (blue) followed by treatment with anti-Fas antibody (1 µg/ml) plus cycloheximide (CHX, 20 µg/ml) for 3 h (bottom) to induce Bid translocation. Mitochondria were then labeled with Mitotracker Red. Image analysis showed that anti-Fas antibody concentrated Bid–GFP staining on mitochondria in untransfected cells (yellow asterisk) but not in cells expressing vMIA (white asterisks). Scale bar: 20 µm. (B) Conventional model of Bid regulation. Dephosphorylation of full-length Bid exposes a cleavage site for caspase-8. Caspase-8-mediated cleavage generates tBid, which is then myristoylated and traffics to mitochondria to promote MOMP. (C) Alternative model of Bid regulation. Dephosphorylation of full-length Bid exposes a binding site for PACS-2 or PACS-1. Binding to PACS-2 promotes Bid translocation to mitochondria (solid lines), whereas binding of Bid to PACS-1 interrupts its translocation to mitochondria (upper dashed lines). In HCMV-infected cells, vMIA sequesters PACS-2 to mitochondria (lower dashed lines), thereby preventing Bid recruitment and, ultimately, MOMP. TRAIL-R, TRAIL receptor.
PACS-2 regulates MAM-localized Ca2+ signaling, lipid metabolism and autophagy
Mitochondria-associated membranes (MAMs) are ER–mitochondria contacts, which were discovered as lipogeneic platforms in liver that are responsive to feeding and starvation (Bernhard et al., 1952; Vance, 1990). Subsequent studies further revealed that MAMs are a dynamic communication center that regulates Ca2+ signaling, lipid metabolism and autophagy (Raturi and Simmen, 2013; Sood et al., 2014; Theurey et al., 2016). Mammalian MAMs are formed by protein tethers, similar to the tetrameric tethering complexes in fungi known as ER–mitochondria encounter structures (ERMES) (Kornmann et al., 2009). However, of the more than 30 proteins involved in vertebrate MAM function, only two of them, GRAMD1A, which corresponds to yeast Lam6p, and MIRO (also known as RHOT1), which corresponds to yeast Gem1p, are conserved in ERMES (Kornmann et al., 2011; Elbaz-Alon et al., 2015). This increased complexity of animal MAMs suggests they acquired new roles beyond those controlled by ERMES (Herrera-Cruz and Simmen, 2017). This increased function correlates with the emerging realization that disturbances in MAM integrity are associated with diseases ranging from obesity to neurodegenerative disorders (Arruda and Hotamisligil, 2015; Paillusson et al., 2016).
Knockdown or knockout of PACS-2 detaches the ER from mitochondria and interferes with a number of key MAM functions (Simmen et al., 2005) (see Fig. 4C). For example, PACS-2 is required for starvation-induced autophagy because it promotes the recruitment of the early autophagy marker Atg14 to MAMs (Hamasaki et al., 2013). PACS-2 knockdown also blocks Ca2+-mediated apoptosis progression, which suggests that PACS-2 is required for efficient Ca2+ transfer between the ER and mitochondria (Simmen et al., 2005). This PACS-2 function may be coupled to its reported dynamic roles in the trafficking of calnexin and perhaps other ER Ca2+ regulatory proteins that modulate ER–mitochondria Ca2+ flux (Myhill et al., 2008). Mechanistically, PACS-2 may modulate MAM formation by preventing cleavage of the ER–mitochondria tethering protein BAP31 (also known as BCAP31) (Iwasawa et al., 2011). Disturbances in these PACS-2 functions could contribute to disease. For example, in obese mice challenged by a high-fat diet, PACS-2 is responsible for a chronic increase in MAM formation (Arruda et al., 2014), leading to toxic mitochondrial Ca2+ overload that consequently impairs mitochondrial oxidative capacity, exacerbates insulin resistance and disrupts glucose homeostasis (Arruda et al., 2014). Conversely, PACS-2 knockdown protects liver from overnutrition-induced steatosis and optimizes mitochondrial respiration and insulin sensitivity (Arruda et al., 2014). Aberrantly elevated PACS-2 and MAM formation are also found in neurons from Alzheimer's patients and Alzheimer's mouse models, suggesting that the deleterious consequences from abnormally strong MAM formation in this disease may also result from dysregulated PACS-2 expression (Area-Gomez et al., 2012; Hedskog et al., 2013).
Vertebrate PACS-2 acquired an Akt phosphorylation site to switch between its anabolic and catabolic roles
The seemingly incongruous roles for PACS-2 in mediating pro-survival protein traffic and MAM function, as well as TRAIL-induced cell death, arose with the acquisition of an Akt phosphorylation site that switches the function of PACS-2 between these divergent roles. Akt phosphorylates PACS-2 on Ser437, which promotes high-affinity binding to 14-3-3 proteins (Aslan et al., 2009). In response to insulin, MAM-localized mTORC2 activates Akt, which phosphorylates PACS-2 to modulate MAM integrity (Betz et al., 2013) (Fig. 4C, top). Akt-mediated phosphorylation of Ser437 is also required for PACS-2-dependent membrane trafficking steps (Fig. 4A) (Aslan et al., 2009). By contrast, dephosphorylation of PACS-2 Ser437, which prevents 14-3-3 binding, blocks pro-survival membrane traffic and disrupts MAMs, but is required for apoptotic trafficking to mitochondria and lysosomes and may support autophagy induction (Fig. 4B,C, bottom) (Aslan et al., 2009; Werneburg et al., 2012; Betz et al., 2013; Hamasaki et al., 2013; and L.T. and G.T., unpublished data).
Akt and 14-3-3 proteins not only repress the apoptotic roles of PACS-2 but also regulate other proapoptotic proteins, including the Bcl-2 protein Bad and FOXO transcription factors (Datta et al., 1997, 2002; Brunet et al., 1999; Singh et al., 2010; Feehan and Shantz, 2016). The regulation of Bad and FOXO proteins by Akt and 14-3-3 is akin to a simple molecular ‘on or off’ switch (Van Roey et al., 2012). However, the regulation of the anabolic (pro-survival) roles of PACS-2 versus the catabolic (apoptotic) roles exerted by Akt and 14-3-3 appears to be more complex. We suggest that the Akt site in PACS-2 resembles a ‘bifurcation’ switch such that both the phosphorylated and dephosphorylated states of PACS-2 are active, but in opposing directions (anabolic versus catabolic pathways, respectively). This model is supported by the ability of 14-3-3 proteins to induce conformational changes in their partners that may enable PACS-2 to selectively bind to client proteins that are involved in anabolic (including pro-survival trafficking) pathways, which would be mediated by binding of 14-3-3 to phosphorylated (p)Ser437, versus catabolic (including autophagic and apoptotic trafficking) pathways, which would be mediated by PACS-2 that is dephosphorylated on Ser437 (Yaffe, 2002).
PACS-2 acquired nuclear trafficking signals to modulate gene expression
The extrinsic and intrinsic apoptotic pathways induced by TRAIL or DNA damage, respectively, use different molecular steps to trigger MOMP. Nonetheless, these pathways are intimately coupled, as p53 (also known as TP53) induces expression of the TRAIL receptor DR5 (also known as TNFRSF10B), and chemotherapeutics and TRAIL synergize to kill cancer cells (Sheikh et al., 1998; Ifeadi and Garnett-Benson, 2012). Surprisingly, PACS-2 has markedly different roles in its response to TRAIL compared with DNA damage; PACS2−/− mice are impaired in TRAIL-induced apoptosis, but are sensitized to DNA-damage-induced apoptosis (Aslan et al., 2009; Barroso-Gonzalez et al., 2016). These opposing roles for PACS-2 are partly explained by the phosphorylation state of Ser437, which is reduced by TRAIL, but increased upon DNA damage (Barroso-Gonzalez et al., 2016). Whereas TRAIL uses dephosphorylated PACS-2 to induce MOMP and LMP, the DNA damage response uses phosphorylated PACS-2 to support cytostasis by coordinating NF-κB and Bcl-xL-dependent anti-apoptosis with p53- and p21 (CDKN1A)-dependent cell cycle arrest (Aslan et al., 2009; Barroso-Gonzalez et al., 2016).
Clues to understanding the roles of PACS-2 in mediating the p53–p21 and NF-κB–Bcl-xL pathways were provided by the DNA repair kinase ATM (Shiloh and Ziv, 2013). In response to DNA damage, nuclear ATM phosphorylates p53, which stabilizes the multi-functional transcription factor to induce its target genes, thus favoring cell cycle arrest and senescence over apoptosis (Xu and Baltimore, 1996; Sperka et al., 2012). Concurrently, a small pool of activated ATM translocates from the nucleus into the cytoplasm where it initiates a novel ‘nucleus-to-cytoplasm’ signaling pathway that promotes the NF-κB-dependent induction of anti-apoptotic Bcl-xL through an as-yet-unresolved mechanism (Miyamoto, 2011).
PACS-2 intersects both ATM pathways to coordinate induction of the NF-κB–Bcl-xL and p53–p21 pathways. Following DNA damage, cytoplasmic PACS-2 interacts with ATM, thereby sequestering the kinase in the cytoplasm where it induces TGF-β-activating kinase 1 (TAK1; also known as MAP3K7) to trigger activation of IκB kinase (IκK) (Wu et al., 2010; Barroso-Gonzalez et al., 2016) (Fig. 4D, top). ATM-activated IκK then stimulates the canonical IκBα–NF-κB induction of Bcl-xL. In the absence of PACS-2, ATM fails to accumulate in the cytoplasm, which reduces the induction of Bcl-xL. Consequently, induction of the apoptotic p53–Puma (Puma is also known as BBC3) pathway remains, thereby increasing MOMP and cell death (Barroso-Gonzalez et al., 2016).
To promote p53–p21-dependent cell cycle arrest over p53–Puma-dependent cell death, PACS-2 traffics to the nucleus where it increases the transactivation of p53 bound to the p21 promoter (Atkins et al., 2014) (Fig. 4D, bottom). Notably, in addition to the ATM-mediated phosphorylation of p53 within its N-terminal region, acetylation of critical lysine residues near the C-terminus of p53 is also required for it to promote maximal transcriptional activity of its target genes (Gu and Roeder, 1997). The class III histone deacetylase (HDAC) SIRT1, in turn, represses p53-mediated transcriptional activation by deacetylating p53 following DNA damage (Kruse and Gu, 2009). In response to DNA damage, Akt-phosphorylated PACS-2 enters the nucleus where it binds to and inhibits SIRT1, which protects acetylated p53 bound to the p21 promoter and, consequently, increases transcriptional output of p21 (Atkins et al., 2014; and L.T. and G.T., unpublished results). In the absence of PACS-2, the excessive SIRT1 activity reduces the p53-dependent induction of p21, which impedes cell cycle arrest and sensitizes cells to p53–Puma-dependent apoptosis. To access the nucleus, vertebrate PACS-2 acquired a polybasic nuclear localization signal (NLS) and a Crm1-dependent nuclear export signal (Atkins et al., 2014). Similar to the Ser437 Akt site of PACS-2, its NLS is located within a predicted IDR and is present only in vertebrate PACS proteins (Figs 1 and 2).
PACS proteins as models of evolutionary protein adaptation
The acquisition of the Ser437 Akt site enabled cytoplasmic PACS-2 to switch between its homeostatic (pSer437) and apoptotic (dephosphorylated Ser437) roles (see Fig. 4). PACS-2 proteins in jawed vertebrates share an identical Akt motif nestled in a disordered region that apparently became fixed by positive selection (Fig. 2). Phylogenetic studies suggest this SLiM evolved rapidly, albeit along a circuitous path (Figs 1 and 2). An Akt consensus motif first appeared in lamprey PACS-1-related which coincided with an apparent gene duplication (Fig. 2). This consensus phosphorylation site was disrupted in fish PACS-1 by the acquisition of a proline residue at the +2 position. Surprisingly, in birds, the Akt motif reappeared in both PACS-1 and PACS-2. However, in mammals, the PACS-1 site again acquired a proline residue, this time in place of the phosphorylatable serine residue (Fig. 2). These findings suggest that PACS-2 acquired the Akt site to act as a vital ‘bifurcation’ switch between its anabolic versus catabolic pathways and that evolutionary pressure assigned this switch to mammalian PACS-2 by negatively selecting against the phosphorylation site in mammalian PACS-1 (Figs 2 and 4) (Davey et al., 2015).
Like the Akt site, the PACS-2 NLS, which is also located within a disordered region, is highly conserved across all jawed vertebrates (Figs 1 and 2). Phylogenetic studies suggest that a polybasic site with limited similarity to the vertebrate PACS NLS first appeared in amphioxus and highly homologous NLS sequences were first acquired in fish (Fig. 2). Acquisition of the NLS by jawed vertebrate PACS-2 parallels the evolutionary expansion of roles of p53 in directing apoptosis versus cell cycle arrest. In worms and flies, p53 is dedicated to driving apoptosis (Schumacher et al., 2001). However, vertebrate p53 additionally induces p21-dependent cell cycle arrest (Lu et al., 2009). Consistent with this finding, vertebrate PACS-2, but not C. elegans PACS, can localize in the nucleus (Atkins et al., 2014). Thus, the ability of PACS-2 to traffic to the nucleus, where it modulates SIRT1 to promote p53-dependent induction of p21, appears to have resulted from an evolutionary adaptation required to support the more complex demands of vertebrate p53 in responding to mild DNA damage by supporting p21-dependent cell cycle arrest. The predicted PACS-1 NLS is not fixed across all vertebrates. Mammals, birds and fish each express a characteristic NLS in their PACS-1 sequences. The significance of this variation, as well as validation of the PACS-1 polybasic site as a bona fide NLS, remains to be determined.
Conclusions and perspectives
The PACS proteins first appeared in lower metazoans as membrane trafficking regulators and then adapted to evolutionary pressure by acquiring motifs to support the increasingly complex interorganellar communication pathways required for vertebrate cellular homeostasis (Figs 1 and 2). The association of mutations in the PACS1 and PACS2 genes with diseases ranging from obesity to epilepsy and cancer underscores the broad and important roles this gene family plays in vertebrate biology. As membrane trafficking regulators, the PACS proteins interact with sorting adaptors as well as a variety of trafficking motifs on client proteins, ranging from CK2-phosphorylatable acidic clusters to α-helices, in order to modulate endomembrane protein traffic (Figs 3 and 4). The PACS-1 and PACS-2 sorting pathways are also hijacked by pathogenic viruses to support production of progeny, immune evasion and protection from apoptosis.
The acquisition of the Ser437 Akt site enabled cytoplasmic PACS-2 to act as a bifurcation switch that separates its roles between endomembrane trafficking (phosphorylated PACS-2) and apoptotic signaling (unphosphorylated PACS-2) (see Fig. 4). The essential role for PACS-2 in mediating TRAIL-induced translocation of Bid to mitochondria was surprising as it expanded the prevailing model of Bid action. This conventional model posits that death receptors trigger dephosphorylation of Bid, which permits caspase-8-dependent proteolytic cleavage and myristoylation of human Bid at Asp↓-Gly61 to form tBid (Li et al., 1998; Desagher et al., 2001; Kaufmann et al., 2012). However, and congruent with live-cell imaging analyses demonstrating that full-length Bid can translocate to mitochondria prior to the generation of tBid, we found that PACS-2 binds to and traffics full-length dephosphorylated Bid to mitochondria (Simmen et al., 2005; Ward et al., 2006). These findings suggest that myristoylation of tBid at Gly61 may not be absolutely required for Bid action. Indeed, granzyme B, which cleaves human Bid at Asp↓-Ser76 to generate a non-myristoylated Bid species, requires PACS-2 to recruit Bid to mitochondria (Li et al., 1998; Brasacchio et al., 2014). These findings suggest that following dephosphorylation, full-length Bid follows one of two distinct pathways leading to MOMP. One pathway is irreversible and relies on cleavage by caspase-8 to generate tBid, which obligates cleaved Bid to trigger MOMP (Fig. 5B). The other pathway is reversible and relies on the PACS-2-dependent translocation of full-length dephosphorylated Bid to mitochondria (Fig. 5C). Following translocation, Bid can be cleaved by multiple proteases, including caspase-8 and granzyme B, to trigger MOMP. Conversely, PACS-1 can bind de-phosphorylated Bid and block translocation to mitochondria, suggesting a mechanism to impede the apoptotic program. In HCMV-infected cells, vMIA traps PACS-2 and Bax on mitochondria, suppressing Bid recruitment and MOMP (Arnoult et al., 2004; Salka et al., 2017; Fig. 5A,C).
The PACS-2 Ser437 Akt site may also act as a bifurcation switch between anabolic and catabolic roles of the MAM. In response to insulin, MAM-localized mTORC2 activates Akt proteins to phosphorylate PACS-2 Ser437, which increases ER–mitochondria contacts (Betz et al., 2013). Accordingly, by increasing MAM integrity, Akt-phosphorylated PACS-2 may increase lipogenesis, modulate ER–mitochondria Ca2+ transfer and repress autophagy induction (Bernhard et al., 1952; Simmen et al., 2005; Betz et al., 2013). By contrast, starvation triggers PACS-2 Ser437 dephosphorylation, which would remodel MAMs to reduce lipogenesis but may increase ER–mitochondria Ca2+ transfer and autophagy (Fig. 4). Future work will investigate to what the extent the phosphorylation state of PACS-2 modulates MAM ‘thickness’ or changes in contacts between mitochondria with smooth or rough endoplasmic reticulum, which may in turn regulate the switch between anabolic versus catabolic functions (Giacomello and Pellegrini, 2016). It will also be interesting to determine whether PACS-2 (as well PACS-1) has roles at additional intermembrane contact sites in other contexts (Schrader et al., 2015). Finally, it will be important to determine to what extent PACS-2 Ser437 phosphorylation is regulated by mTORC2 and Akt localized to MAMs versus other compartments, and whether other signaling pathways might converge on Akt to phosphorylate PACS-2 (Hers et al., 2011; Betz and Hall, 2013). It will also be important to identify the PACS-2 Ser437 phosphatase.
Since autophagy is generally viewed as a survival mechanism, it may at first appear surprising that dephosphorylation of PACS-2 Ser437 is also required for TRAIL to induce Bid translocation to mitochondria and cell death (Simmen et al., 2005; Levine and Kroemer, 2008; Aslan et al., 2009; Rubinstein and Kimchi, 2012). Recent studies show, however, that autophagy is intimately coupled with TRAIL-induced cell death and can influence whether TRAIL kills cells by apoptosis (a death pathway in which cells are removed without membrane disruption) or necroptosis (an inflammatory necrotic-like death pathway that involves cell breakage) (Goodall et al., 2016). In response to TRAIL, induction of early stage autophagic machinery can promote assembly of the necrosome, which diverts the TRAIL-induced mode of killing from apoptosis to necroptosis. It will therefore be interesting to determine to what extent dephosphorylated PACS-2 modulates common or separate steps in the decision between autophagy and autophagy-modulated cell death.
The evolutionally recent acquisition of nuclear trafficking signals by PACS-1 and PACS-2 expanded their roles to also modulate nuclear gene expression (Atkins et al., 2014). The role of the PACS proteins in the nucleus is just beginning to be understood. Notably, PACS-2 is the most recent addition to a small collection of proteins that regulate SIRT1, which include deleted in breast cancer 1 (DBC1; also known as CCAR2), active regulator of SIRT1 (AROS; also known as RPS19BP1) and the moonlighting protein GAPDH (Kim et al., 2007, 2008; Atkins et al., 2014; Chang et al., 2015). It will be important to determine whether PACS-2 acts in the same or different pathways to the other SIRT1 regulators and whether PACS-2 or PACS-1 regulates HDACs in addition to SIRT1. Interestingly, the description of PACS-2 as an in vivo modulator of SIRT1 in response to DNA damage suggests that it may be involved in additional pathways controlled by SIRT1 (Brooks and Gu, 2009). For example, in response to fasting, liver SIRT1 increases the expression of genes that encode proteins involved in fatty acid oxidation and ketogenesis – pathways that protect from diet-induced obesity (Purushotham et al., 2009). Thus, the recent report that PACS-2 knockdown in liver protects mice from diet-induced obesity (Arruda et al., 2014), raises the possibility that the acquired NLS and Akt sites enable PACS-2 to coordinate SIRT1-dependent nuclear gene expression with MAM-dependent changes in mitochondrial respiration to modulate liver metabolism.
In summary, studies of the PACS proteins illustrate the vital role of protein adaptation in coordinating the seemingly autonomous actions of the nucleus, mitochondria and endomembrane systems in response to the complex challenges faced by vertebrate organ systems. Moreover, the findings that mutation or altered expression of PACS proteins are associated with pathologies ranging from cancer, obesity and viral pathogenesis to epilepsy and neurodegeneration points to a rich opportunity for insight into the molecular basis of disease through analysis of this adaptable gene family.
Acknowledgements
The authors thank R. Chaillet and J. Dacks for critically reading the manuscript, J. D. Dikeakos for helpful suggestions and C. H. Hung and C. Crump for contributions to studies described here. We also thank M. Ostrowski, C. Thauvin and H. Olson for communication of unpublished results.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This work in our laboratories was supported by the National Institutes of Health (NIH) (grants CA151564 and DK112844 to G.T.), Natural Sciences and Engineering Research Council of Canada (NSERC) (grant RGPIN-2015-04105 to T.S.) and the American Heart Association (AHA) (grant 17SDG33350075 to J.E.A.). Deposited in PMC for release after 12 months.
References
- Anderson G. R., Brenner B. M., Swede H., Chen N., Henry W. M., Conroy J. M., Karpenko M. J., Issa J. P., Bartos J. D., Brunelle J.K. et al. (2001). Intrachromosomal genomic instability in human sporadic colorectal cancer measured by genome-wide allelotyping and inter-(simple sequence repeat) PCR. Cancer Res. 61, 8274-8283. [PubMed] [Google Scholar]
- Area-Gomez E., del Carmen Lara Castillo M., Tambini M. D., Guardia-Laguarta C., de Groof A. J. C., Madra M., Ikenouchi J., Umeda M., Bird T. D., Sturley S. L. et al. (2012). Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. EMBO J. 31, 4106-4123. 10.1038/emboj.2012.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult D., Bartle L. M., Skaletskaya A., Poncet D., Zamzami N., Park P. U., Sharpe J., Youle R. J. and Goldmacher V. S. (2004). Cytomegalovirus cell death suppressor vMIA blocks Bax- but not Bak-mediated apoptosis by binding and sequestering Bax at mitochondria. Proc. Natl. Acad. Sci. USA 101, 7988-7993. 10.1073/pnas.0401897101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arruda A. P. and Hotamisligil G. S. (2015). Calcium homeostasis and organelle function in the pathogenesis of obesity and diabetes. Cell Metab. 22, 381-397. 10.1016/j.cmet.2015.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arruda A. P., Pers B. M., Parlakgül G., Güney E., Inouye K. and Hotamisligil G. (2014). Chronic enrichment of hepatic endoplasmic reticulum-mitochondria contact leads to mitochondrial dysfunction in obesity. Nat. Med. 20, 1427-1435. 10.1038/nm.3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslan J. E. and Thomas G. (2009). Death by committee: organellar trafficking and communication in apoptosis. Traffic 10, 1390-1404. 10.1111/j.1600-0854.2009.00951.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslan J. E., You H., Williamson D. M., Endig J., Youker R. T., Thomas L., Shu H., Du Y., Milewski R. L., Brush M. H. et al. (2009). Akt and 14-3-3 control a PACS-2 homeostatic switch that integrates membrane traffic with TRAIL-induced apoptosis. Mol. Cell 34, 497-509. 10.1016/j.molcel.2009.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins K. M., Thomas L., Youker R. T., Harriff M. J., Pissani F., You H. and Thomas G. (2008). HIV-1 Nef binds PACS-2 to assemble a multikinase cascade that triggers major histocompatibility complex class I (MHC-I) down-regulation: analysis using short interfering RNA and knock-out mice. J. Biol. Chem. 283, 11772-11784. 10.1074/jbc.M707572200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins K. M., Thomas L. L., Barroso-González J., Thomas L., Auclair S., Yin J., Kang H., Chung J. H., Dikeakos J. D. and Thomas G. (2014). The multifunctional sorting protein PACS-2 regulates SIRT1-mediated deacetylation of p53 to modulate p21-dependent cell-cycle arrest. Cell Rep 8, 1545-1557. 10.1016/j.celrep.2014.07.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso-Gonzalez J., Auclair S., Luan S., Thomas L., Atkins K. M., Aslan J. E., Thomas L. L., Zhao J., Zhao Y. and Thomas G. (2016). PACS-2 mediates the ATM and NF-kappaB-dependent induction of anti-apoptotic Bcl-xL in response to DNA damage. Cell Death Differ. 23, 1448-1457. 10.1038/cdd.2016.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle G. W. and Tatum E. L. (1941). Genetic control of biochemical reactions in neurospora. Proc. Natl. Acad. Sci. USA 27, 499-506. 10.1073/pnas.27.11.499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard W., Haguenau F., Gautier A. and Oberling C. (1952). [Submicroscopical structure of cytoplasmic basophils in the liver, pancreas and salivary gland; study of ultrafine slices by electron microscope]. Z Zellforsch Mikrosk Anat 37, 281-300. 10.1007/BF00343816 [DOI] [PubMed] [Google Scholar]
- Betz C. and Hall M. N. (2013). Where is mTOR and what is it doing there? J. Cell Biol. 203, 563-574. 10.1083/jcb.201306041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz C., Stracka D., Prescianotto-Baschong C., Frieden M., Demaurex N. and Hall M. N. (2013). Feature Article: mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc. Natl. Acad. Sci. USA 110, 12526-12534. 10.1073/pnas.1302455110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoveshchenskaya A. D., Thomas L., Feliciangeli S. F., Hung C.-H. and Thomas G. (2002). HIV-1 Nef downregulates MHC-I by a PACS-1- and PI3K-regulated ARF6 endocytic pathway. Cell 111, 853-866. 10.1016/S0092-8674(02)01162-5 [DOI] [PubMed] [Google Scholar]
- Boya P. and Kroemer G. (2008). Lysosomal membrane permeabilization in cell death. Oncogene 27, 6434-6451. 10.1038/onc.2008.310 [DOI] [PubMed] [Google Scholar]
- Brasacchio D., Noori T., House C., Brennan A. J., Simpson K. J., Susanto O., Bird P. I., Johnstone R. W. and Trapani J. A. (2014). A functional genomics screen identifies PCAF and ADA3 as regulators of human granzyme B-mediated apoptosis and Bid cleavage. Cell Death Differ. 21, 748-760. 10.1038/cdd.2013.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks C. L. and Gu W. (2009). How does SIRT1 affect metabolism, senescence and cancer? Nat. Rev. Cancer 9, 123-128. 10.1038/nrc2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J. and Greenberg M. E. (1999). Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96, 857-868. 10.1016/S0092-8674(00)80595-4 [DOI] [PubMed] [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540-552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Chang C., Su H., Zhang D., Wang Y., Shen Q., Liu B., Huang R., Zhou T., Peng C., Wong C. C. L. et al. (2015). AMPK-dependent phosphorylation of GAPDH triggers Sirt1 activation and is necessary for autophagy upon glucose starvation. Mol. Cell 60, 930-940. 10.1016/j.molcel.2015.10.037 [DOI] [PubMed] [Google Scholar]
- Chaudhry A., Das S. R., Jameel S., George A., Bal V., Mayor S. and Rath S. (2008). HIV-1 Nef induces a Rab11-dependent routing of endocytosed immune costimulatory proteins CD80 and CD86 to the Golgi. Traffic 9, 1925-1935. 10.1111/j.1600-0854.2008.00802.x [DOI] [PubMed] [Google Scholar]
- Chiu Y.-F., Sugden B., Chang P.-J., Chen L.-W., Lin Y.-J., Lan Y.-C., Lai C.-H., Liou J.-Y., Liu S.-T. and Hung C.-H. (2012). Characterization and intracellular trafficking of Epstein-Barr virus BBLF1, a protein involved in virion maturation. J. Virol. 86, 9647-9655. 10.1128/JVI.01126-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump C. M., Hung C.-H., Thomas L., Wan L. and Thomas G. (2003). Role of PACS-1 in trafficking of human cytomegalovirus glycoprotein B and virus production. J. Virol. 77, 11105-11113. 10.1128/JVI.77.20.11105-11113.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S. R., Dudek H., Tao X., Masters S., Fu H., Gotoh Y. and Greenberg M. E. (1997). Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 91, 231-241. 10.1016/S0092-8674(00)80405-5 [DOI] [PubMed] [Google Scholar]
- Datta S. R., Ranger A. M., Lin M. Z., Sturgill J. F., Ma Y.-C., Cowan C. W., Dikkes P., Korsmeyer S. J. and Greenberg M. E. (2002). Survival factor-mediated BAD phosphorylation raises the mitochondrial threshold for apoptosis. Dev. Cell 3, 631-643. 10.1016/S1534-5807(02)00326-X [DOI] [PubMed] [Google Scholar]
- Davey N. E., Travé G. and Gibson T. J. (2011). How viruses hijack cell regulation. Trends Biochem. Sci. 36, 159-169. 10.1016/j.tibs.2010.10.002 [DOI] [PubMed] [Google Scholar]
- Davey N. E., Van Roey K., Weatheritt R. J., Toedt G., Uyar B., Altenberg B., Budd A., Diella F., Dinkel H. and Gibson T. J. (2012). Attributes of short linear motifs. Mol. Biosyst. 8, 268-281. 10.1039/C1MB05231D [DOI] [PubMed] [Google Scholar]
- Davey N. E., Cyert M. S. and Moses A. M. (2015). Short linear motifs - ex nihilo evolution of protein regulation. Cell Commun Signal 13, 43 10.1186/s12964-015-0120-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deciphering Developmental Disorders Study (2017). Prevalence and architecture of de novo mutations in developmental disorders. Nature. 542, 433-438. 10.1038/nature21062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desagher S., Osen-Sand A., Montessuit S., Magnenat E., Vilbois F., Hochmann A., Journot L., Antonsson B. and Martinou J.-C. (2001). Phosphorylation of bid by casein kinases I and II regulates its cleavage by caspase 8. Mol. Cell 8, 601-611. 10.1016/S1097-2765(01)00335-5 [DOI] [PubMed] [Google Scholar]
- Dikeakos J. D., Atkins K. M., Thomas L., Emert-Sedlak L., Byeon I.-J. L., Jung J., Ahn J., Wortman M. D., Kukull B., Saito M. et al. (2010). Small molecule inhibition of HIV-1-induced MHC-I down-regulation identifies a temporally regulated switch in Nef action. Mol. Biol. Cell 21, 3279-3292. 10.1091/mbc.E10-05-0470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikeakos J. D., Thomas L., Kwon G., Elferich J., Shinde U. and Thomas G. (2012). An interdomain binding site on HIV-1 Nef interacts with PACS-1 and PACS-2 on endosomes to down-regulate MHC-I. Mol. Biol. Cell 23, 2184-2197. 10.1091/mbc.E11-11-0928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirk B. S. Pawlak E. N., Johnson A. L., Van Nynatten L. R., Jacob R. A., Heit B. and Dikeakos J. D. (2016). HIV-1 Nef sequesters MHC-I intracellularly by targeting early stages of endocytosis and recycling. Sci. Rep. 6, 37021 10.1038/srep37021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombernowsky S. L., Samsøe-Petersen J., Petersen C. H., Instrell R., Hedegaard A.-M. B., Thomas L., Atkins K. M., Auclair S., Albrechtsen R., Mygind K. J. et al. (2015). The sorting protein PACS-2 promotes ErbB signalling by regulating recycling of the metalloproteinase ADAM17. Nat. Commun. 6, 7518 10.1038/ncomms8518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker A. K., Cortese M. S., Romero P., Iakoucheva L. M. and Uversky V. N. (2005). Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 272, 5129-5148. 10.1111/j.1742-4658.2005.04948.x [DOI] [PubMed] [Google Scholar]
- Elbaz-Alon Y., Eisenberg-Bord M., Shinder V., Stiller S. B., Shimoni E., Wiedemann N., Geiger T. and Schuldiner M. (2015). Lam6 regulates the extent of contacts between organelles. Cell Rep 12, 7-14. 10.1016/j.celrep.2015.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feehan R. P. and Shantz L. M. (2016). Negative regulation of the FOXO3a transcription factor by mTORC2 induces a pro-survival response following exposure to ultraviolet-B irradiation. Cell. Signal. 28, 798-809. 10.1016/j.cellsig.2016.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomello M. and Pellegrini L. (2016). The coming of age of the mitochondria-ER contact: a matter of thickness. Cell Death Differ. 23, 1417-1427. 10.1038/cdd.2016.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall M. L., Fitzwalter B. E., Zahedi S., Wu M., Rodriguez D., Mulcahy-Levy J. M., Green D. R., Morgan M., Cramer S. D. and Thorburn A. (2016). The autophagy machinery controls cell death switching between apoptosis and necroptosis. Dev. Cell 37, 337-349. 10.1016/j.devcel.2016.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W. and Roeder R. G. (1997). Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595-606. 10.1016/S0092-8674(00)80521-8 [DOI] [PubMed] [Google Scholar]
- Guicciardi M. E., Werneburg N. W., Bronk S. F., Franke A., Yagita H., Thomas G. and Gores G. J. (2014). Cellular inhibitor of apoptosis (cIAP)-mediated ubiquitination of phosphofurin acidic cluster sorting protein 2 (PACS-2) negatively regulates tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) cytotoxicity. PLoS One 9, e92124 10.1371/journal.pone.0092124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Lethiec F., Duroux P. and Gascuel O. (2005). PHYML Online--a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33, W557-W559. 10.1093/nar/gki352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y. et al. (2013). Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389-393. 10.1038/nature11910 [DOI] [PubMed] [Google Scholar]
- Hedskog L., Pinho C. M., Filadi R., Ronnback A., Hertwig L., Wiehager B., Larssen P., Gellhaar S., Sandebring A., Westerlund M. et al. (2013). Modulation of the endoplasmic reticulum-mitochondria interface in Alzheimer's disease and related models. Proc. Natl. Acad. Sci. USA 110, 7916-7921. 10.1073/pnas.1300677110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson B. and Martin A. C. (2014). Protein moonlighting: a new factor in biology and medicine. Biochem. Soc. Trans. 42, 1671-1678. 10.1042/BST20140273 [DOI] [PubMed] [Google Scholar]
- Herrera-Cruz M. S. and Simmen T. (2017). Of yeast, mice and men: MAMs come in two flavors. Biol. Direct 12, 3 10.1186/s13062-017-0174-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hers I., Vincent E. E. and Tavaré J. M. (2011). Akt signalling in health and disease. Cell. Signal. 23, 1515-1527. 10.1016/j.cellsig.2011.05.004 [DOI] [PubMed] [Google Scholar]
- Hornbeck P. V., Zhang B., Murray B., Kornhauser J. M., Latham V. and Skrzypek E. (2015). PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512-D520. 10.1093/nar/gku1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C. H., Thomas L., Ruby C. E., Atkins K. M., Morris N. P., Knight Z. A., Scholz I., Barklis E., Weinberg A. D., Shokat K. M. et al. (2007). HIV-1 Nef assembles a Src family kinase-ZAP-70/Syk-PI3K cascade to downregulate cell-surface MHC-I. Cell Host Microbe 1, 121-133. 10.1016/j.chom.2007.03.004 [DOI] [PubMed] [Google Scholar]
- Ifeadi V. and Garnett-Benson C. (2012). Sub-lethal irradiation of human colorectal tumor cells imparts enhanced and sustained susceptibility to multiple death receptor signaling pathways. PLoS One 7, e31762 10.1371/journal.pone.0031762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T. and Kinoshita K. (2007). PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 35, W460-W464. 10.1093/nar/gkm363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasawa R., Mahul-Mellier A.-L., Datler C., Pazarentzos E. and Grimm S. (2011). Fis1 and Bap31 bridge the mitochondria-ER interface to establish a platform for apoptosis induction. EMBO J. 30, 556-568. 10.1038/emboj.2010.346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery C. J. (1999). Moonlighting proteins. Trends Biochem. Sci. 24, 8-11. 10.1016/S0968-0004(98)01335-8 [DOI] [PubMed] [Google Scholar]
- Jenkins P. M., Zhang L., Thomas G. and Martens J. R. (2009). PACS-1 mediates phosphorylation-dependent ciliary trafficking of the cyclic-nucleotide-gated channel in olfactory sensory neurons. J. Neurosci. 29, 10541-10551. 10.1523/JNEUROSCI.1590-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone R. W., Frew A. J. and Smyth M. J. (2008). The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat. Rev. Cancer 8, 782-798. 10.1038/nrc2465 [DOI] [PubMed] [Google Scholar]
- Jonas S. and Izaurralde E. (2013). The role of disordered protein regions in the assembly of decapping complexes and RNP granules. Genes Dev. 27, 2628-2641. 10.1101/gad.227843.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann T., Strasser A. and Jost P. J. (2012). Fas death receptor signalling: roles of Bid and XIAP. Cell Death Differ. 19, 42-50. 10.1038/cdd.2011.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelves D. and Hood L. (eds) (1992). The Code of Codes: Scientific and Social Issues in the Human Genome Project Harvard University Press, Cambridge, MA [Google Scholar]
- Kim E.-J., Kho J.-H., Kang M.-R. and Um S.-J. (2007). Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol. Cell 28, 277-290. 10.1016/j.molcel.2007.08.030 [DOI] [PubMed] [Google Scholar]
- Kim J. E., Chen J. and Lou Z. (2008). DBC1 is a negative regulator of SIRT1. Nature 451, 583-586. 10.1038/nature06500 [DOI] [PubMed] [Google Scholar]
- Kornmann B., Currie E., Collins S. R., Schuldiner M., Nunnari J., Weissman J. S. and Walter P. (2009). An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477-481. 10.1126/science.1175088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B., Osman C. and Walter P. (2011). The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc. Natl. Acad. Sci. USA 108, 14151-14156. 10.1073/pnas.1111314108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse J.-P. and Gu W. (2009). Modes of p53 regulation. Cell 137, 609-622. 10.1016/j.cell.2009.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B. and Kroemer G. (2008). Autophagy in the pathogenesis of disease. Cell 132, 27-42. 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Zhu H., Xu C.-j. and Yuan J. (1998). Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94, 491-501. 10.1016/S0092-8674(00)81590-1 [DOI] [PubMed] [Google Scholar]
- Lu W. J., Amatruda J. F. and Abrams J. M. (2009). p53 ancestry: gazing through an evolutionary lens. Nat. Rev. Cancer 9, 758-762. 10.1038/nrc2732 [DOI] [PubMed] [Google Scholar]
- Mansouri M., Douglas J., Rose P. P., Gouveia K., Thomas G., Means R. E., Moses A. V. and Früh K. (2006). Kaposi sarcoma herpesvirus K5 removes CD31/PECAM from endothelial cells. Blood 108, 1932-1940. 10.1182/blood-2005-11-4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S. (2011). Nuclear initiated NF-kappaB signaling: NEMO and ATM take center stage. Cell Res. 21, 116-130. 10.1038/cr.2010.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myhill N., Lynes E. M., Nanji J. A., Blagoveshchenskaya A. D., Fei H., Carmine Simmen K., Cooper T. J., Thomas G. and Simmen T. (2008). The subcellular distribution of calnexin is mediated by PACS-2. Mol. Biol. Cell 19, 2777-2788. 10.1091/mbc.E07-10-0995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noviello C. M., Benichou S. and Guatelli J. C. (2008). Cooperative binding of the class I major histocompatibility complex cytoplasmic domain and human immunodeficiency virus type 1 Nef to the endosomal AP-1 complex via its mu subunit. J. Virol. 82, 1249-1258. 10.1128/JVI.00660-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillusson S., Stoica R., Gomez-Suaga P., Lau D. H. W., Mueller S., Miller T. and Miller C. C. J. (2016). There's something wrong with my MAM; the ER-mitochondria axis and neurodegenerative diseases. Trends Neurosci. 39, 146-157. 10.1016/j.tins.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancsa R. and Tompa P. (2016). Coding regions of intrinsic disorder accommodate parallel functions. Trends Biochem. Sci. 41, 898-906. 10.1016/j.tibs.2016.08.009 [DOI] [PubMed] [Google Scholar]
- Pawlak E. N. and Dikeakos J. D. (2015). HIV-1 Nef: a master manipulator of the membrane trafficking machinery mediating immune evasion. Biochim. Biophys. Acta 1850, 733-741. 10.1016/j.bbagen.2015.01.003 [DOI] [PubMed] [Google Scholar]
- Piguet V., Trono D., Piguet V., Wan L., Borel C., Mangasarian A. and Demaurex N. (2000). HIV-1 Nef protein binds to the cellular protein PACS-1 to downregulate class I major histocompatibility complexes. Nat. Cell Biol. 2, 163-167. 10.1038/35004038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushotham A., Schug T. T., Xu Q., Surapureddi S., Guo X. and Li X. (2009). Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 9, 327-338. 10.1016/j.cmet.2009.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raturi A. and Simmen T. (2013). Where the endoplasmic reticulum and the mitochondrion tie the knot: the mitochondria-associated membrane (MAM). Biochim. Biophys. Acta 1833, 213-224. 10.1016/j.bbamcr.2012.04.013 [DOI] [PubMed] [Google Scholar]
- Rubinstein A. D. and Kimchi A. (2012). Life in the balance - a mechanistic view of the crosstalk between autophagy and apoptosis. J. Cell Sci. 125, 5259-5268. 10.1242/jcs.115865 [DOI] [PubMed] [Google Scholar]
- Salka K., Bhuvanendran S., Wilson K., Bozidis P., Mehta M., Rainey K., Sesaki H., Patterson G. H., Jaiswal J. K. and Colberg-Poley A. M. (2017). Superresolution imaging identifies that conventional trafficking pathways are not essential for endoplasmic reticulum to outer mitochondrial membrane protein transport. Sci. Rep. 7, 16 10.1038/s41598-017-00039-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermer B., Höpker K., Omran H., Ghenoiu C., Fliegauf M., Fekete A., Horvath J., Köttgen M., Hackl M., Zschiedrich S. et al. (2005). Phosphorylation by casein kinase 2 induces PACS-1 binding of nephrocystin and targeting to cilia. EMBO J. 24, 4415-4424. 10.1038/sj.emboj.7600885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger A., Schaefer C., Vicedo E., Schmidberger M., Punta M. and Rost B. (2011). Protein disorder--a breakthrough invention of evolution? Curr. Opin. Struct. Biol. 21, 412-418. 10.1016/j.sbi.2011.03.014 [DOI] [PubMed] [Google Scholar]
- Schrader M., Godinho L. F., Costello J. L. and Islinger M. (2015). The different facets of organelle interplay-an overview of organelle interactions. Front Cell Dev Biol 3, 56 10.3389/fcell.2015.00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B., Hofmann K., Boulton S. and Gartner A. (2001). The C. elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis. Curr. Biol. 11, 1722-1727. 10.1016/S0960-9822(01)00534-6 [DOI] [PubMed] [Google Scholar]
- Schuurs-Hoeijmakers J. H., Oh E. C., Vissers L. E. L. M., Swinkels M. E. M., Gilissen C., Willemsen M. A., Holvoet M., Steehouwer M., Veltman J. A., de Vries B. B. A. et al. (2012). Recurrent de novo mutations in PACS1 cause defective cranial-neural-crest migration and define a recognizable intellectual-disability syndrome. Am. J. Hum. Genet. 91, 1122-1127. 10.1016/j.ajhg.2012.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuurs-Hoeijmakers J. H., Landsverk M. L., Foulds N., Kukolich M. K., Gavrilova R. H., Greville-Heygate S., Hanson-Kahn A., Bernstein J. A., Glass J., Chitayat D. et al. (2016). Clinical delineation of the PACS1-related syndrome--Report on 19 patients. Am. J. Med. Genet. A 170, 670-675. 10.1002/ajmg.a.37476 [DOI] [PubMed] [Google Scholar]
- Scott G. K., Gu F., Crump C. M., Thomas L., Wan L., Xiang Y. and Thomas G. (2003). The phosphorylation state of an autoregulatory domain controls PACS-1-directed protein traffic. EMBO J. 22, 6234-6244. 10.1093/emboj/cdg596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh M. S., Burns T. F., Huang Y., Wu G. S., Amundson S., Brooks K. S., Fornace A. J. Jr and el-Deiry W. S. (1998). p53-dependent and -independent regulation of the death receptor KILLER/DR5 gene expression in response to genotoxic stress and tumor necrosis factor alpha. Cancer Res. 58, 1593-1598. [PubMed] [Google Scholar]
- Shiloh Y. and Ziv Y. (2013). The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 14, 197-210. 10.1038/nrm3546 [DOI] [PubMed] [Google Scholar]
- Sieburth D., Ch'ng Q. L., Dybbs M., Tavazoie M., Kennedy S., Wang D., Dupuy D., Rual J.-F., Hill D. E., Vidal M. et al. (2005). Systematic analysis of genes required for synapse structure and function. Nature 436, 510-517. 10.1038/nature03809 [DOI] [PubMed] [Google Scholar]
- Simmen T., Aslan J. E., Blagoveshchenskaya A. D., Thomas L., Wan L., Xiang Y., Feliciangeli S. F., Hung C.-H., Crump C. M. and Thomas G. (2005). PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J. 24, 717-729. 10.1038/sj.emboj.7600559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Ye M., Bucur O., Zhu S., Tanya Santos M., Rabinovitz I., Wei W., Gao D., Hahn W. C. and Khosravi-Far R. (2010). Protein phosphatase 2A reactivates FOXO3a through a dynamic interplay with 14-3-3 and AKT. Mol. Biol. Cell 21, 1140-1152. 10.1091/mbc.E09-09-0795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirover M. A. (2011). On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochim. Biophys. Acta 1810, 741-751. 10.1016/j.bbagen.2011.05.010 [DOI] [PubMed] [Google Scholar]
- Sood A., Jeyaraju D. V., Prudent J., Caron A., Lemieux P., Mcbride H. M., Laplante M., Tóth K. and Pellegrini L. (2014). A Mitofusin-2-dependent inactivating cleavage of Opa1 links changes in mitochondria cristae and ER contacts in the postprandial liver. Proc. Natl. Acad. Sci. USA 111, 16017-16022. 10.1073/pnas.1408061111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperka T., Wang J. and Rudolph K. L. (2012). DNA damage checkpoints in stem cells, ageing and cancer. Nat. Rev. Mol. Cell Biol. 13, 579-590. 10.1038/nrm3420 [DOI] [PubMed] [Google Scholar]
- Stern D., Cho M. T., Chikarmane R., Willaert R., Retterer K., Kendall F., Deardorff M., Hopkins S. and Bedoukian E. and Slavotinek A. et al. (2017). Association of the missense variant p.Arg203Trp in PACS1 as a cause of intellectual disability and seizures. Clin. Genet. doi:10.1111/cge.12956 [Epub] 10.1111/cge.12956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurey P., Tubbs E., Vial G., Jacquemetton J., Bendridi N., Chauvin M. A., Alam M. R., Le Romancer M., Vidal H. and Rieusset J. (2016). Mitochondria-associated endoplasmic reticulum membranes allow adaptation of mitochondrial metabolism to glucose availability in the liver. J .Mol. Cell Biol. 8, 129-143. 10.1093/jmcb/mjw004 [DOI] [PubMed] [Google Scholar]
- Thomas G. (2002). Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat. Rev. Mol. Cell Biol. 3, 753-766. 10.1038/nrm934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P., Davey N. E., Gibson T. J. and Babu M. M. (2014). A million peptide motifs for the molecular biologist. Mol. Cell 55, 161-169. 10.1016/j.molcel.2014.05.032 [DOI] [PubMed] [Google Scholar]
- Van Roey K., Gibson T. J. and Davey N. E. (2012). Motif switches: decision-making in cell regulation. Curr. Opin. Struct. Biol. 22, 378-385. 10.1016/j.sbi.2012.03.004 [DOI] [PubMed] [Google Scholar]
- Vance J. E. (1990). Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 265, 7248-7256. [PubMed] [Google Scholar]
- Wan L., Molloy S. S., Thomas L., Liu G., Xiang Y., Rybak S. L. and Thomas G. (1998). PACS-1 defines a novel gene family of cytosolic sorting proteins required for trans-Golgi network localization. Cell 94, 205-216. 10.1016/S0092-8674(00)81420-8 [DOI] [PubMed] [Google Scholar]
- Wang J., Jia Z., Zhang C., Sun M., Wang W., Chen P., Ma K., Zhang Y., Li X. and Zhou C. (2014). miR-499 protects cardiomyocytes from H 2O 2-induced apoptosis via its effects on Pdcd4 and Pacs2. RNA Biol. 11, 339-350. 10.4161/rna.28300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward M. W., Rehm M., Duessmann H., Kacmar S., Concannon C. G. and Prehn J. H. (2006). Real time single cell analysis of Bid cleavage and Bid translocation during caspase-dependent and neuronal caspase-independent apoptosis. J. Biol. Chem. 281, 5837-5844. 10.1074/jbc.M511562200 [DOI] [PubMed] [Google Scholar]
- Werneburg N. W., Bronk S. F., Guicciardi M. E., Thomas L., Dikeakos J. D., Thomas G. and Gores G. J. (2012). Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) protein-induced lysosomal translocation of proapoptotic effectors is mediated by phosphofurin acidic cluster sorting protein-2 (PACS-2). J. Biol. Chem. 287, 24427-24437. 10.1074/jbc.M112.342238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler E., Huang N., Bochukova E. G., Keogh J. M., Lindsay S., Garg S., Henning E., Blackburn H., Loos R. J. F., Wareham N. J. et al. (2013). Genome-wide SNP and CNV analysis identifies common and low-frequency variants associated with severe early-onset obesity. Nat. Genet. 45, 513-517. 10.1038/ng.2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z.-H., Wong E. T., Shi Y., Niu J., Chen Z., Miyamoto S. and Tergaonkar V. (2010). ATM- and NEMO-dependent ELKS ubiquitination coordinates TAK1-mediated IKK activation in response to genotoxic stress. Mol. Cell 40, 75-86. 10.1016/j.molcel.2010.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wygrecka M., Marsh L. M., Morty R. E., Henneke I., Guenther A., Lohmeyer J., Markart P. and Preissner K. T. (2009). Enolase-1 promotes plasminogen-mediated recruitment of monocytes to the acutely inflamed lung. Blood 113, 5588-5598. 10.1182/blood-2008-08-170837 [DOI] [PubMed] [Google Scholar]
- Xu Y. and Baltimore D. (1996). Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes Dev. 10, 2401-2410. 10.1101/gad.10.19.2401 [DOI] [PubMed] [Google Scholar]
- Yaffe M. B. (2002). How do 14-3-3 proteins work?-- Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett. 513, 53-57. 10.1016/S0014-5793(01)03288-4 [DOI] [PubMed] [Google Scholar]
- Yang X., Coulombe-Huntington J., Kang S., Sheynkman G. M., Hao T., Richardson A., Sun S., Yang F., Shen Y. A., Murray R. R. et al. (2016). Widespread expansion of protein interaction capabilities by alternative splicing. Cell 164, 805-817. 10.1016/j.cell.2016.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youker R. T., Shinde U., Day R. and Thomas G. (2009). At the crossroads of homoeostasis and disease: roles of the PACS proteins in membrane traffic and apoptosis. Biochem. J. 421, 1-15. 10.1042/BJ20081016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. L., Zou Y., Yu R. T., Gage F. H. and Evans R. M. (2006). Nuclear receptor TLX prevents retinal dystrophy and recruits the corepressor atrophin1. Genes Dev. 20, 1308-1320. 10.1101/gad.1413606 [DOI] [PMC free article] [PubMed] [Google Scholar]