ABSTRACT
The GTPase Rab5 and phosphatidylinositol-3 phosphate [PI(3)P] coordinately regulate endosome trafficking. Rab5 recruits Vps34, the class III phosphoinositide 3-kinase (PI3K), to generate PI(3)P and recruit PI(3)P-binding proteins. Loss of Rab5 and loss of Vps34 have opposite effects on endosome size, suggesting that our understanding of how Rab5 and PI(3)P cooperate is incomplete. Here, we report a novel regulatory loop whereby Caenorhabditis elegans VPS-34 inactivates RAB-5 via recruitment of the TBC-2 Rab GTPase-activating protein. We found that loss of VPS-34 caused a phenotype with large late endosomes, as with loss of TBC-2, and that Rab5 activity (mice have two Rab5 isoforms, Rab5a and Rab5b) is increased in Vps34-knockout mouse embryonic fibroblasts (Vps34 is also known as PIK3C3 in mammals). We found that VPS-34 is required for TBC-2 endosome localization and that the pleckstrin homology (PH) domain of TBC-2 bound PI(3)P. Deletion of the PH domain enhanced TBC-2 localization to endosomes in a VPS-34-dependent manner. Thus, PI(3)P binding of the PH domain might be permissive for another PI(3)P-regulated interaction that recruits TBC-2 to endosomes. Therefore, VPS-34 recruits TBC-2 to endosomes to inactivate RAB-5 to ensure the directionality of endosome maturation.
KEY WORDS: Endosome, Rab GTPase, Phosphatidylinositide-3 phosphate, Vps34, TBC-2, Caenorhabditis elegans
Summary: Rab5 recruits the Vps34-containing PI3K to regulate endosome trafficking. We identified a negative feedback loop whereby C. elegans VPS-34 recruits the TBC-2 RAB-5 GAP to endosomes to inactivate RAB-5 during endosome maturation.
INTRODUCTION
The Vps34 (also known as PIK3C3 in mammals) class III phosphoinositide 3-kinase (PI3K) regulates endosome and phagosome trafficking, and the initiation of autophagy (Vieira et al., 2001; Kihara et al., 2001; Peterson et al., 1999; Futter et al., 2001; Schu et al., 1993). Vps34 forms a complex with Vps15 (also known as PIK3R4 in mammals), a serine/threonine kinase (Stack et al., 1993; Volinia et al., 1995), and Beclin1 (yeast Atg6 or Vps30), a key regulator of autophagy (Kihara et al., 2001). Vps34 specifically phosphorylates phosphatidylinositol (PI) to generate phosphatidylinositol 3-phosphate [PI(3)P] (Schu et al., 1993; Volinia et al., 1995), which serves as a substrate for proteins with PI(3)P-binding domains such as FYVE and Phox (PX) domains (Burd and Emr, 1998; Gaullier et al., 1998; Patki et al., 1998; Ellson et al., 2001; Cheever et al., 2001; Kanai et al., 2001; Song et al., 2001). Myotubularins are PI3 phosphatases that counteract the effects of Vps34 by dephosphorylating PI(3)P and PI(3,5)P2 to generate PI and PI(5)P, respectively (Robinson and Dixon, 2006). Thus, Vps34 and myotubularins regulate the lipid and protein composition of endosomes.
The Rab5 and Rab7 GTPases, respectively, localize to early and late endosomes where they drive multiple aspects of endosome trafficking to the lysosome (Huotari and Helenius, 2011; Stenmark, 2009). In their GTP-bound ‘active’ state, they bind and recruit effector proteins to endosomes. Both Rab5 and Rab7 GTPases (note both Rab5 and Rab7 have a and b isoforms in mammals) can interact with Vps15 to recruit and/or activate Vps34 on early and late endosomes, respectively (Christoforidis et al., 1999; Murray et al., 2002; Stein et al., 2003). Many Rab5 and Rab7 effectors also bind PI(3)P, indicating a cooperative function for the Rab GTPases and Vps34 on endosomes (Pankiv et al., 2010; Simonsen et al., 1998; Patki et al., 1997; Nielsen et al., 2000). For example, EEA1 promotes homotypic fusion of early endosomes, and it is recruited to endosomes via interactions with PI(3)P and GTP-bound Rab5 (Mills et al., 1998; Simonsen et al., 1998; Patki et al., 1997). Interestingly, inhibition of Vps34 via wortmannin causes a large late endosome phenotype (Futter et al., 2001; Brown et al., 1995; Reaves et al., 1996) that was reversed by expression of dominant-negative Rab5(S34N) (Fernandez-Borja et al., 1999). Constitutively active Rab5(Q79L) also induces large early and late endosome phenotypes (Wegener et al., 2010; Rink et al., 2005; Stenmark et al., 1994; Lawe et al., 2002), and was able to recruit EEA1 to membranes in the presence of wortmannin (Simonsen et al., 1998). The above data suggests that the enlarged endosomes seen with loss of Vps34/PI(3)P could result from increased Rab5 activity.
The Caenorhabditis elegans intestine consists of a single layer of polarized epithelial cells that provides an in vivo model to study endosome trafficking (Sato et al., 2014). As in mammalian cells, expression of constitutively active RAB-5(Q78L) [analogous to mammalian Rab5(Q79L)] in the C. elegans intestine results in the formation of enlarged late endosomes (Chotard et al., 2010a). Similarly, loss of C. elegans TBC-2, a RAB-5 GTPase-activating protein (GAP), also results in the formation of enlarged late endosomes in the intestine (Chotard et al., 2010a). TBC-2 localizes to late endosomes in a RAB-7-dependent manner, where it may facilitate the replacement of RAB-5 with RAB-7 by inactivating RAB-5. In addition to facilitating early to late endosome maturation, TBC-2 also regulates phagosome maturation during apoptotic cell clearance (Li et al., 2009), endosome recycling (Sun et al., 2012; Liu and Grant, 2015), yolk protein trafficking/storage (Chotard et al., 2010b) and neuropeptide release via dense core vesicle trafficking (Sasidharan et al., 2012). TBC-2 is homologous to human Armus (also known as TBC1D2A) and TBC1D2B proteins, and comprises a unique family of Rab GAPs with an N-terminal pleckstrin homology (PH) domain, a central coiled-coil (CC) domain and a C-terminal Tre2/Bub2/Cdc16 (TBC) catalytic domain (Chotard et al., 2010a). Armus has Rab7 GAP activity and interacts with Rac1 to regulate degradation of E-cadherin and interacts with LC3 proteins (also known as MAP1LC3 proteins) and LRRK1 to regulate autophagy (Carroll et al., 2013; Frasa et al., 2010; Toyofuku et al., 2015). TBC1D2B interacts with Rab22 (also known as RAB22A) in a nucleotide-independent manner, but its cellular function is not known (Kanno et al., 2010).
C. elegans VPS-34 (also known as LET-512) is essential for development. It has been shown to regulate trafficking of the LRP-1 low-density lipoprotein (LDL) receptor, nuclear membrane morphology and phagosome maturation during engulfment of apoptotic cells (Roggo et al., 2002; Kinchen et al., 2008). Loss of vps-34 was reported to result in accumulation of large vesicles, the nature of which has not been described (Roggo et al., 2002). Here, we determine the requirements for VPS-34 PI3K on endosome size in the intestine, and its relationship with RAB-5 and TBC-2. We show that knockdown of the C. elegans class III PI3K components vps-34, vps-15 or bec-1 resulted in the formation of enlarged late endosomes that are similar in nature to those seen upon loss of tbc-2 or expression of constitutively active RAB-5(Q78L). This large late endosome phenotype required the late endosomal RAB-7 GTPase. Overexpression of the MTM-1 myotubularin PI3 phosphatase induced the same phenotype as knockdown of the PI3K components, indicating that the large late endosome phenotype was due to loss of PI(3)P. We show that the PH domain of TBC-2 bound PI(3)P in vitro and that VPS-34 promoted TBC-2 localization to endosomes. Truncation of the PH domain enhanced TBC-2 endosome localization in a VPS-34-dependent manner, and further truncation of the PH and CC domains abrogated TBC-2 endosome localization. We demonstrate that PI(3)P has dual roles in regulating TBC-2 endosome localization. A direct interaction between PI(3)P and the PH domain of TBC-2 may result in a conformational change allowing the CC domain to mediate a protein–protein interaction with an endosomal protein whose localization or activity is regulated by PI(3)P. Consistent with Vps34 antagonizing the activity of Rab5, we found that Rab5 activity is increased in Vps34-knockout (KO) mouse embryonic fibroblasts (MEFs). Therefore, the VPS-34–VPS-15–BEC-1 PI3K complex may negatively regulate RAB-5 via recruitment of TBC-2 to endosomes.
RESULTS
Loss of the VPS-34 PI3K complex phenocopies the effect of constitutively active RAB-5
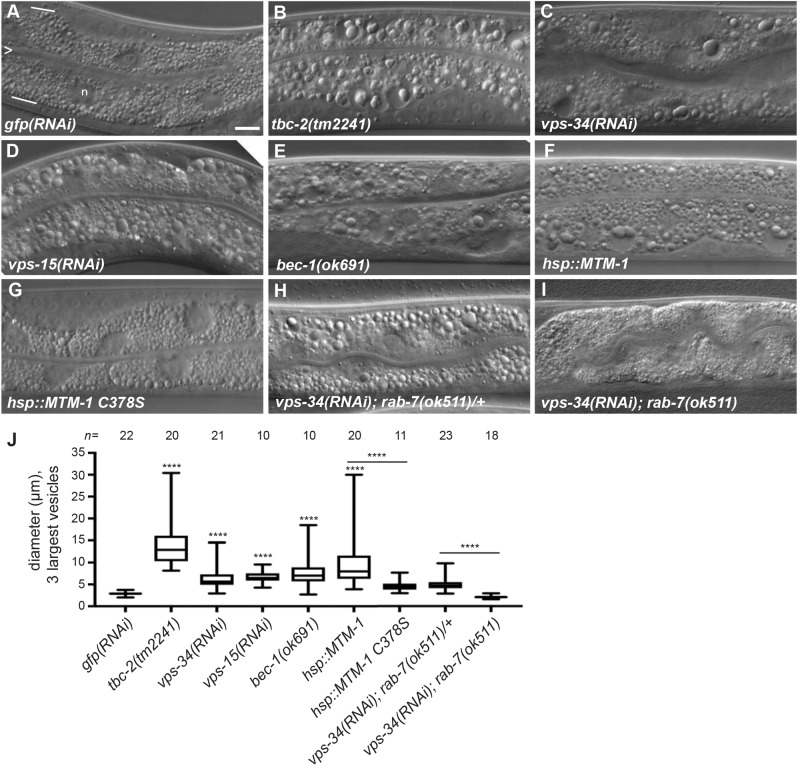
To determine whether C. elegans VPS-34 regulates the size of endosomes in the intestine, we performed an RNAi experiment to knockdown VPS34 [denoted vps-34(RNAi)] in wild-type animals. The intestinal cells of vps-34(RNAi) animals accumulated large vesicles containing globular material, similar to what is seen in tbc-2 mutants, while the negative control, gfp(RNAi), had no effect (Fig. 1A–C,J). In yeast and mammals, Vps34 functions in a complex with Vps15 and Beclin1 (Volinia et al., 1995; Kihara et al., 2001; Stack et al., 1993), therefore we tested whether loss or knockdown of the C. elegans homologs, VPS-15 and BEC-1, had intestinal phenotypes (Manning, 2005; Melendez et al., 2003). We found that vps-15(RNAi) caused a large vesicle phenotype identical to that seen upon loss of vps-34 (Fig. 1D,J). Loss of bec-1, either through RNAi or homozygous bec-1(ok691) mutants derived from heterozygous mothers, also resulted in the accumulation of large vesicles containing globular material (Fig. 1E,J; Fig. S1D). The presence of large late endosomes in the intestine of bec-1– animals has been previously reported, yet the presence of globular material inside the vesicles had not been described (Hars et al., 2007; Ruck et al., 2011). This specific intestinal phenotype has thus far only been described for conditions that increase RAB-5 GTPase activity via loss-of-function mutations in tbc-2, a RAB-5 GAP (Fig. 1B), or expression of the constitutively active RAB-5(Q78L) mutant (Chotard et al., 2010a). Consistent with vps-34 and tbc-2 functioning in a common pathway, vps-34(RNAi) failed to enhance the severity of the large vesicle phenotype of the tbc-2(tm2241) deletion (Fig. S2).
Fig. 1.
Loss of the VPS-34 PI3K complex or overexpression of the MTM-1 PI3 phosphatase causes a RAB-7-dependent large vesicle phenotype in the intestine. (A–I) Representative Nomarski differential interference contrast (DIC) images of the intestine of L4 stage larvae with genotype or RNAi expression: gfp(RNAi) (A), tbc-2(tm2241) (B), vps-34(RNAi) (C), vps-15(RNAi) (D), bec-1(ok691) (E), qxIs156[Phsp::mtm-1] (F), qxIs210[Phsp::mtm-1 C378S] (G), vps-34(RNAi); rab-7(ok511)/mIn1 (H), and vps-34(RNAi); rab-7(ok511) (I). For reference, the intestine is composed primarily of binucleate cells and is one cell in thickness. In A, the intestinal lumen is marked with an ‘>’ and represents the apical membrane, and white lines mark the basal membrane. Although not discernible, approximately four cells are present in each image. ‘n’ denotes a nucleus and is not to be confused with large vesicles. (J) Box-and-whisker plot of the diameters (µm) of the three largest vesicles in the intestines of the indicted genotypes and RNAi conditions. The box represents the 25–75th percentiles, and the median is indicated. The whiskers show the minimum to maximum of all data. ****P<0.0001 (unpaired t-test); all conditions were compared to control gfp(RNAi) unless indicated by a bar. n, number of animals scored. Scale bar: 10 µm.
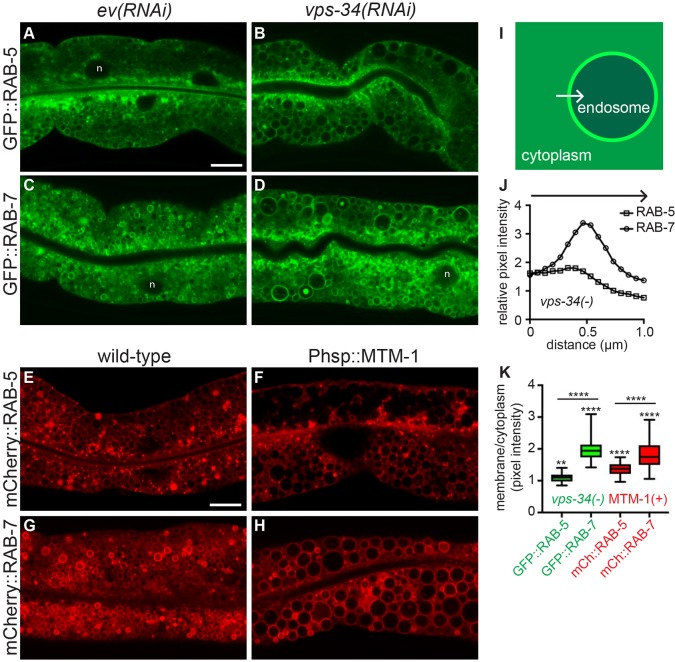
The large intestinal vesicles of tbc-2 and bec-1 mutants, as well as animals expressing RAB-5(Q78L), are primarily RAB-7-positive vesicles (Chotard et al., 2010a; Ruck et al., 2011). To determine the nature of the enlarged vps-34(RNAi) vesicles, we analyzed the localization of RAB-5 and RAB-7. Similar to what is seen upon loss of tbc-2, bec-1 or expression of RAB-5(Q78L), the large vesicles of vps-34(RNAi) animals were positive for GFP::RAB-7, and to a lesser extent GFP::RAB-5 (Fig. 2A–D,I–K). Large RAB-7-positive vesicles were also seen with vps-15(RNAi) and bec-1(RNAi) animals (Fig. S1). Taken together, the loss of the VPS-34–VPS-15–BEC-1 PI3K complex causes a large late endosome phenotype very similar to that of either loss of TBC-2 or expression of constitutively active RAB-5(Q78L) (Chotard et al., 2010a), suggesting that this complex might function closely with TBC-2 to inactivate the RAB-5 GTPase.
Fig. 2.
Loss of VPS-34 or overexpression of MTM-1 induces enlarged endosomes. (A–D) Representative confocal images of GFP::RAB-5 (A,B) and GFP::RAB-7 (C,D) localization in wild-type L4 larvae treated with empty vector (ev) RNAi (A,C) or vps-34(RNAi) (B,D). RNAi was performed a minimum of three times with >20 animals scored per experiment. (E–H) Representative confocal images of mCherry::RAB-5 (E,F) and mCherry::RAB-7 (G,H) in wild-type L4 larvae (E,G) or L4 larvae expressing Phsp::mtm-1 (F,H). n denotes nuclei (A,C,D). (I) Illustration of how cytoplasmic versus membrane fluorescence intensity was measured. A 1 μm line was drawn from the cytoplasm, across the membrane into the endosome lumen using ImageJ, from which the intensity was measured. (J) The average intensity profiles for GFP::RAB-5 and GFP::RAB-7 from a total of 30 vesicles from 5–10 vps-34(RNAi) animals. (K) The average of the three highest intensity points across the membrane divided by the average of the first three cytoplasmic intensity points were used to generate a membrane:cytoplasmic ratio for 30 vesicles per condition to generate a box-and-whisker plot for GFP::RAB-5 and GFP::RAB-7 in vps-34(RNAi) animals (data from J) as well as for mCherry::RAB-5 and mCherry::RAB-7 from 6-10 Phsp::mtm-1 expressing animals. The box represents the 25–75th percentiles, and the median is indicated. The whiskers show the minimum to maximum of all data. **P<0.01, ****P<0.0001 (a paired t-test was used to determine the significance of the difference in membrane versus cytoplasmic intensities for each condition, and an unpaired t-test was used to determine significance of differences in membrane:cytoplasmic ratios between RAB-5 and RAB-7 in each condition). Scale bar: 10 µm.
Overexpression of the MTM-1 myotubularin induces a large late endosome phenotype
Loss of vps-34 results in decreased PI(3)P levels in several tissues (Roggo et al., 2002; Kinchen et al., 2008; Lu et al., 2012; Zou et al., 2009; Neukomm et al., 2011). If the vps-34(RNAi) large late endosome phenotype was caused by a reduction of PI(3)P, then overexpression of a PI3 phosphatase should have the same phenotype. MTM-1 is a myotubularin PI3 phosphatase that has an opposite function to VPS-34 to regulate PI(3)P levels during phagosome maturation and endocytosis in C. elegans (Lu et al., 2012; Neukomm et al., 2011; Xue et al., 2003; Zou et al., 2009). Overexpression of MTM-1 under control of a heat-shock promoter at 23–24°C was sufficient to induce a large vesicle phenotype, similar to that seen upon loss of vps-34 and tbc-2 (Fig. 1F,J). Overexpression of a predicted catalytically inactive MTM-1(C378S) failed to induce a substantial large vesicle phenotype (Fig. 1G,J). As seen with loss of vps-34 and tbc-2, the large vesicles caused by the overexpression of MTM-1 were positive for RAB-5 and RAB-7 (Fig. 2E–H,K). The similar phenotypes caused by loss of vps-34 or overexpression of MTM-1 indicates that reduced PI(3)P or its derivatives [e.g. PI(3,5)P2] are the cause of the large late endosome phenotype.
RAB-7 is required for the vps-34(RNAi)-induced large endosome phenotype
We previously demonstrated that the large late endosome phenotypes induced by loss of tbc-2 or expression of activated RAB-5(Q78L) required RAB-7 (Chotard et al., 2010a). To determine whether RAB-7 is required for the vps-34(RNAi) phenotype, we treated rab-7(ok511)/mIn1 heterozygotes with vps-34(RNAi) and analyzed the F1 progeny. ok511 is a deletion allele of rab-7, and mIn1 is a chromosomal inversion that contains a wild-type copy of the rab-7 gene and functions as a genetic balancer (Edgley and Riddle, 2001; Skorobogata and Rocheleau, 2012). We found that rab-7(ok511)/mIn1 progeny displayed the vps-34(RNAi) large vesicle phenotype, while rab-7(ok511) homozygous siblings did not display the large vesicle phenotype (Fig. 1H–J). Similar results were seen with rab-7 and vps-34 double RNAi or with rab-7(RNAi) in a bec-1(ok691) mutant (Fig. S3). Since rab-7 is not required for the RNAi process (Rocheleau et al., 2008), we conclude that rab-7 is required for the large vesicle phenotype caused by loss of vps-34 or bec-1. This data is consistent with the large late endosome phenotype of vps-34(RNAi) being driven by activated RAB-5 and RAB-7 GTPases as previously proposed for loss of tbc-2.
The VPS-34 PI3K complex is required for TBC-2 membrane localization
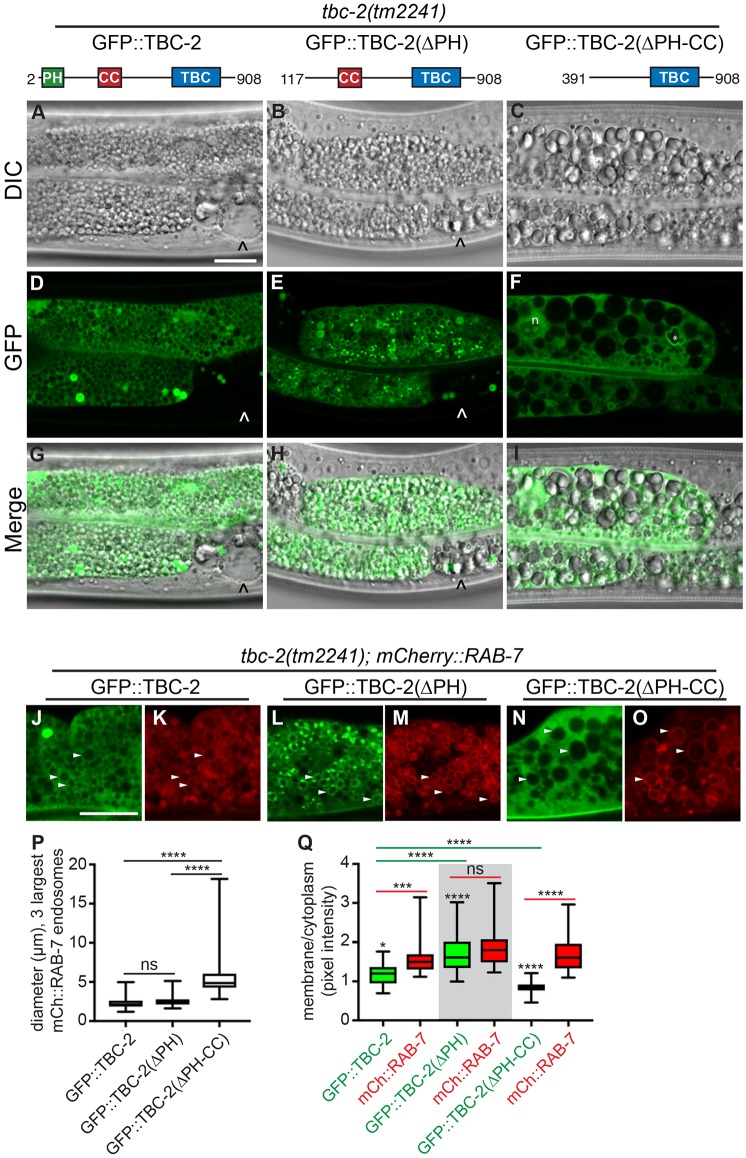
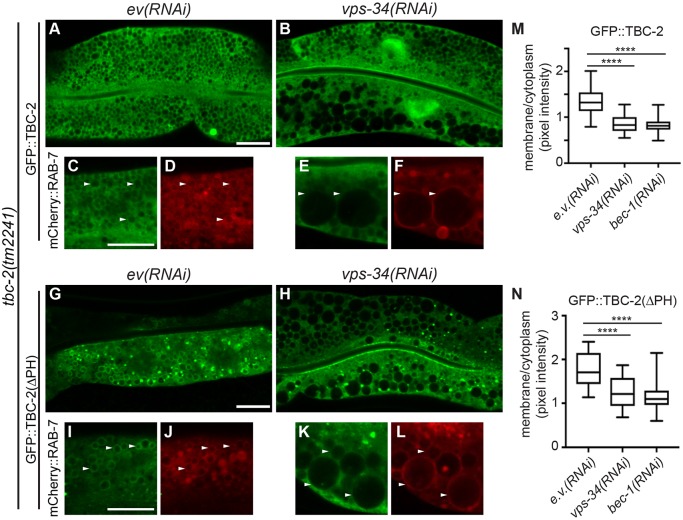
PI(3)P plays an important role in recruiting proteins to the endosome (Corvera et al., 1999), and therefore, might recruit TBC-2 to endosomes. A GFP::TBC-2::Flag fusion expressed from an extrachromosomal array, vhEx12, rescued the tbc-2(tm2241) strong loss-of-function intestinal phenotype, and localized to both the cytoplasm and RAB-7-positive endosomes (Fig. 3A,D,G,J,K,P,Q). We found that in vps-34(RNAi) and bec-1(RNAi) knockdown animals, GFP::TBC-2::Flag was primarily cytoplasmic in cells that exhibited the large vesicle phenotype (Fig. 4A–F,M). These data show that the VPS-34 PI3K complex is required for recruitment of TBC-2 to endosomes.
Fig. 3.
The PH domain of TBC-2 antagonizes endosome localization. Representative DIC (A–C), confocal (D–F) or merged (G–I) images of tbc-2(tm2241) L4 stage animals expressing vhEx12 (GFP::tbc-2::Flag) (A,D,G), vhEx20 (GFP::tbc-2(ΔPH)::Flag) (B,E,H) or vhEx21 (GFP::tbc-2(ΔPH-CC)::Flag) (C,F,I). The GFP::tbc-2::Flag constructs are expressed as extrachromosomal arrays and thus have mosaic expression. ‘^’ marks cells that lack transgene expression and are not rescued for the tbc-2(tm2241) large vesicle phenotype (A–F). n denotes the nucleus; * denotes a GFP-positive irregularly shaped vesicle occasionally seen in this background (F). Representative confocal images (J–O) of tbc-2(tm2241) L4 stage larvae expressing pwIs429 (mCherry::rab-7) and vhEx12 (J,K), vhEx20 (L,M) and vhEx21 (N,O). GFP::TBC-2 is green and mCherry::RAB-7 is red. White arrowheads denote examples of common vesicles in both channels. (P) Box-and-whisker plot of the diameters (µm) of the three largest mCherry::RAB-7-positive endosomes in the intestines of animals (n=14–23) expressing vhEx12 (GFP::tbc-2::Flag), vhEx20 (GFP::tbc-2(ΔPH)::Flag) and vhEx21 (GFP::tbc-2(ΔPH-CC)::Flag). An unpaired t-test was used to determine significance. (Q) The membrane:cytoplasmic ratio of GFP::TBC-2, GFP::TBC-2(ΔPH), GFP::TBC-2(ΔPH-CC) and mCherry::RAB-7 on 30 vesicles in at least ten animals per condition as described in the Fig. 2 legend. A paired t-test was used to determine the significance of the difference in membrane versus cytoplasmic intensities for GFP::TBC-2, GFP::TBC-2(ΔPH), and GFP::TBC-2(ΔPH-CC). An unpaired t-test was used to determine significance of the differences in the membrane:cytoplasmic ratios between GFP::TBC-2 and mCherry::RAB-7 in each strain as well as between full-length GFP::TBC-2 and each truncation mutant. In the box-and-whisker plots (P,Q), the box represents the 25–75th percentiles, and the median is indicated. The whiskers show the minimum to maximum of all data. *P<0.05; ***P<0.001; ****P<0.0001; ns, not significant. Scale bars: 10 µm.
Fig. 4.
VPS-34 PI3K regulates TBC-2 endosome localization. (A,B,G,H) Representative confocal images of GFP::TBC-2::Flag (A,B) and GFP::TBC-2(ΔPH)::Flag (G,H) in the intestine of tbc-2(tm2241) L4 larvae with empty vector RNAi [ev(RNAi)] (A,G) or vps-34(RNAi) (B,H). Note that the vps-34(RNAi) (B) affected the lower cell, based on the large vesicle phenotype, more severely than the upper cell. (C–F,I–L) Representative confocal images of tbc-2(tm2241) L4 stage larvae expressing pwIs429 (mCherry::rab-7) and vhEx12 (C–F) or vhEx20 (I–L) with empty vector(ev)(RNAi) (C,D,I,J) or vps-34(RNAi) (E,F,K,L). GFP::TBC-2 is green and mCherry::RAB-7 is red. White arrowheads denote examples of common vesicles in both channels. RNAi was performed a minimum of three times with >20 animals scored per experiment. (M,N) The membrane:cytoplasmic ratio of GFP::TBC-2 (M) and GFP::TBC-2(ΔPH) (N) on endosomes of animals treated with ev(RNAi), vps-34(RNAi) or bec-1(RNAi). A total of 30 vesicles from ten animals were measured for each condition. The box represents the 25–75th percentiles, and the median is indicated. The whiskers show the minimum to maximum of all data. An unpaired t-test was used to determine the significance of the differences of membrane:cytoplasmic ratios between ev(RNAi) and each vps-34(RNAi) and bec-1(RNAi) conditions. ****P<0.0001. Scale bars: 10 µm.
The PH domain of TBC-2 binds to PI(3)P and PI(4)P in vitro
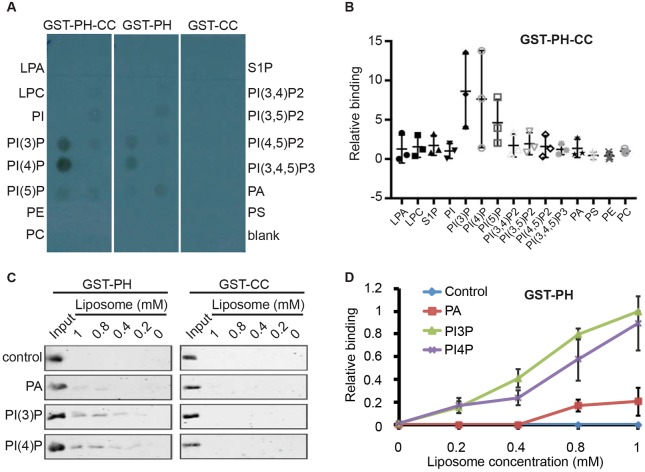
If VPS-34 PI3K is required for TBC-2 endosome localization, then TBC-2 might bind to PI(3)P. Since PH domains can mediate binding to lipids (Lemmon, 2007), we tested whether TBC-2 can directly bind to PI(3)P. We specifically tested whether the N-terminus of TBC-2 containing both the PH and CC domains, as well as either the PH or CC domains individually, could bind to a variety of phosphoinositides in a protein–lipid overlay assay (Fig. 5A,B). We found that the PH–CC domains and the PH domain alone of TBC-2 bound to PI(3)P and PI(4)P. The CC domain of TBC-2 alone failed to bind lipids.
Fig. 5.
The PH domain of TBC-2 binds PI3P and PI4P in vitro. (A) Protein–lipid overlay assay demonstrating that the PH-CC and PH domains, but not the CC domain alone, of TBC-2 bind to PI(3)P and PI(4)P. (B) The relative binding of GST::PH-CC to each lipid as compared to the blank from three independent protein–lipid overlay experiments as assessed with ImageJ. Bars show the mean and range for the three data points. (C) The binding between purified recombinant GST::PH and GST::CC proteins, and liposomes containing different phospholipids were detected by western blotting using an anti-GST antibody. The control liposomes were composed of PC and PE. The PA, PI(3)P and PI(4)P liposomes were composed of PC and PE, with either PA, PI(3)P or PI(4)P, respectively. (D) Quantification of GST::PH lipid-binding results in C. The bindings to different liposomes were normalized to the input. The results are mean±s.d. from three independent experiments. LPA, lysophosphatidic acid; LPC, lysophosphocholine; PE, phosphatidylethanolamine; PC, phosphatidylcholine; S1P, sphingosine-1-phosphate; PA, phosphatidic acid; PS, phosphatidylserine.
To confirm the binding of the PH domain to PI(3)P and PI(4)P, we performed liposome-binding assays. We found that the PH domain, but not the CC domain, bound to liposomes composed of phosphatidylcholine (PC), phosphatidylethanolamine (PE) and PI(3)P, and PC, PE and PI(4)P with similar affinity (Fig. 5C,D), but not to liposomes composed of PC and PE, or PC, PE and phosphatidic acid (PA). We do not know the significance of the PI(4)P binding, but the interaction of the PH domain with PI(3)P suggests that it mediates VPS-34 PI3K-regulated TBC-2 endosome localization.
The PH domain antagonizes TBC-2 localization to endosomes
To test whether the PH domain mediates endosomal localization of TBC-2, we generated a transgenic strain expressing an N-terminal truncation of TBC-2 without the PH domain, GFP::TBC-2(ΔPH)::Flag, and tested whether it could rescue the tbc-2(tm2241) phenotype and localize to endosomes. The GFP::TBC-2(ΔPH)::Flag showed strong rescue of the tbc-2(tm2241) large endosome phenotype (Fig. 3B,H,P). Unexpectedly, GFP::TBC-2(ΔPH)::Flag showed stronger localization to endosomes than the full-length form (Fig. 3E,L,M,Q) and localized to RAB-7-positive endosomes as well as adjacent foci. Therefore, contrary to our hypothesis, the PH domain was not essential for rescue, and had an inhibitory effect on endosome localization of TBC-2.
Further truncation of both the PH and CC domains, GFP::TBC-2(ΔPH-CC)::Flag, failed to efficiently rescue the tbc-2(tm2241) large endosome phenotype (Fig. 3C,I,P) and failed to localize to endosomes (Fig. 3F,N,O,Q). Therefore, at least in the absence of the PH domain, the CC domain and the intervening region between the PH and CC domains are essential for endosome localization and TBC-2 function. Taken together, these data suggest that PI(3)P is important for TBC-2 recruitment to endosomes, but the PH domain, which binds PI(3)P, is not essential and may in fact be inhibitory. Therefore, VPS-34-derived PI(3)P may be permissive for TBC-2 endosome localization, possibly by relieving an inhibitory role of the PH domain and freeing the region containing the CC domain for a protein–protein interaction on the endosome membrane.
VPS-34 PI3K is required for TBC-2(ΔPH) endosome localization
If VPS-34 promotes TBC-2 endosome localization via the PH domain, then we would expect that endosome localization of GFP::TBC-2(ΔPH)::Flag should not require VPS-34. However, this was not the case as vps-34(RNAi) and bec-1(RNAi) still inhibited endosomal localization of GFP::TBC-2(ΔPH)::Flag (Fig. 4G–L,N). Although GFP::TBC-2(ΔPH)::Flag was still present on foci, the amount of this protein was strongly reduced on the enlarged endosomes in vps-34(RNAi) and bec-1(RNAi) animals. This suggests that PI(3)P has an additional role in recruiting TBC-2 to endosomes that could be mediated by a protein–protein interaction via the region containing the CC domain.
Rab5 activity is increased in Vps34 KO MEFs
If VPS-34 negatively regulates RAB-5 activity via recruitment of TBC-2, then we would expect that the levels of RAB-5-GTP will be increased in vps-34 mutants as previously demonstrated in tbc-2 mutants (Liu and Grant, 2015). Unlike tbc-2, vps-34 is required for larval viability (Roggo et al., 2002), thus precluding us from biochemically testing this hypothesis in C. elegans. Therefore, we examined Rab5-GTP levels by using Vps34-null MEFs, which are viable and accumulate large Rab7-positive late endosomes (Jaber et al., 2012). We assessed Rab5 activity in wild-type and Vps34-null MEFs by using a Rab5-effector pulldown assay (Liu et al., 2007). In three independent experiments, we found that Vps34-null MEFs had increased Rab5-GTP in comparison to that in wild-type MEFs (Fig. 6). Thus, Vps34 negatively regulates Rab5 activity in mouse cells, corroborating our genetic data in C. elegans and suggesting a role for the human TBC-2 homologs in regulation of Rab5.
Fig. 6.
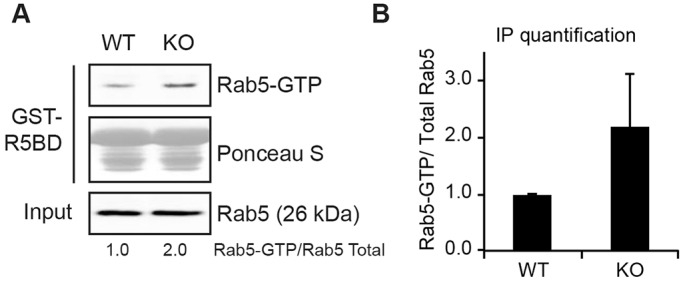
Rab5-GTP level is enhanced in Vps34-null cells. A Rab5 activity assay (antibody recognizes Rab5a) was performed in wild-type (WT) and Vps34−/− MEFs using the Rab5-binding domain (R5BD) of Rabaptin-5 fused to GST, which pulls down GTP-bound Rab5. (A) One of the three independent experiments, all showing increased Rab5-GTP in the Vps34−/− MEFs. (B) The relative intensity of Rab5-GTP versus total Rab5 from all three experiments was quantified by ImageJ.
DISCUSSION
Here, we described the identification of a novel negative-feedback loop in which Rab GTPase-mediated recruitment of VPS-34 leads to RAB-5 inactivation via endosome recruitment of TBC-2, a RAB-5 GAP. We demonstrate that loss of C. elegans vps-34, or other components of the PI3K complex (vps-15 or bec-1), leads to a large late endosome phenotype in the intestinal cells that is very similar to that seen with activated RAB-5(Q78L) or in tbc-2 mutants, and that vps34-null MEFs have increased Rab5 activity. This phenotype is likely due to loss of PI(3)P, as overexpression of the MTM-1 PI3 phosphatase, and not a catalytically inactive MTM-1, results in a similar phenotype. We demonstrate VPS-34 is required for TBC-2 localization to endosomes and that the PH domain of TBC-2 binds PI(3)P in vitro. However, deletion of the PH domain results in enhanced localization of TBC-2 to endosomes and this requires the CC region of TBC-2 and still requires VPS-34. Based on these studies, we propose a working model by which the VPS-34–VPS-15–BEC-1 PI3K complex dually regulates TBC-2 localization to endosomes (Fig. 7).
Fig. 7.
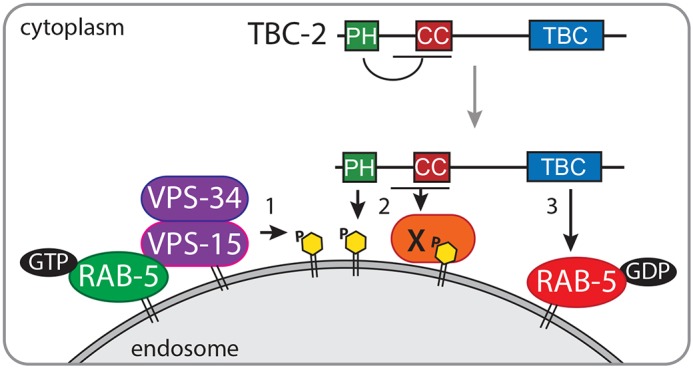
A working model of TBC-2 recruitment to endosomes. (1) Active GTP-bound RAB-5 (green) recruits and activates VPS-34, which phosphorylates PI to generate PI(3)P (yellow) on the endosome membrane. (2) Endosomal PI(3)P creates binding sites for various PI(3)P-binding proteins, such as TBC-2 and others including protein X (orange). Binding of the PH domain of TBC-2 to PI(3)P potentially relieves an intramolecular interaction or interaction with an unknown protein exposing a binding site for TBC-2 with protein X. (3) TBC-2 facilitates the hydrolysis of GTP to GDP by RAB-5, thus inactivating RAB-5 (red).
The PH domain likely plays a permissive role in TBC-2 recruitment to endosomes. Since deletion of the PH domain enhanced endosome localization of TBC-2, and the PH domain bound PI(3)P in vitro, we propose that the PH domain might be involved in either an intramolecular interaction or an interaction with another protein that sequesters TBC-2 in the cytoplasm. When the PH domain binds PI(3)P, a conformational change may occur whereby the region containing the CC domain is free to interact with other proteins and/or lipids on endosomes. During the course of this study, two independent studies on mammalian Armus (TBC1D2A) have corroborated our model that the PH domain is involved in an auto-inhibitory role that is regulated by PI(3)P: Jaber et al. find that the PH of Armus binds PI(3)P and that Vps34 regulates Armus recruitment to endosomes (Jaber et al., 2016), and Toyofuku et al. demonstrate that expression of Armus (ΔPH) in cells results in decreased Rab7 activity as compared to the full-length Armus, suggesting increased activity (Toyofuku et al., 2015). Furthermore, the PH domain binds Armus (ΔPH) in vitro (Toyofuku et al., 2015), suggesting an intramolecular interaction. Together, these data suggest a conserved mechanism of regulation for TBC-2 and Armus.
Since TBC-2(ΔPH) still requires VPS-34 PI3K for endosome localization, it suggests that there is a second requirement for VPS-34 and PI(3)P in addition to the role in PH domain binding. One possibility is that there is a second PI(3)P-binding site. Although the CC domain alone does not bind PI(3)P, it is possible that the region spanning the PH and CC domains could bind PI(3)P. The second possibility is that PI(3)P is not required and that TBC-2(ΔPH) physically interacts with the VPS-34 complex itself. The third possibility is that PI(3)P indirectly recruits TBC-2(ΔPH) to endosomes via a protein intermediate that we refer to as factor ‘X’ (Fig. 7).
While several proteins have been shown to interact with and regulate the localization of TBC-2, none are strong candidates for being factor X. We previously showed that RAB-7 is required for TBC-2 endosome localization (Chotard et al., 2010a). Rab7 interacts with Vps34 in mammalian cells (Stein et al., 2003), and thus, it is possible that RAB-5 and/or RAB-7 regulate TBC-2 localization via recruitment of VPS-34 during early to late endosome maturation. In C. elegans, RAB-10, AMPH-1 (amphiphysin) and CED-10 (Rac1) bind to TBC-2 and regulate its localization in the recycling pathway (Liu and Grant, 2015; Sasidharan et al., 2012; Xin et al., 2013; Sun et al., 2012). However, neither rab-10, amph-1 nor ced-10 mutants have been reported to display phenotypes that would be consistent with regulating early to late endosome trafficking with TBC-2 and VPS-34. We predict factor X will have a PI(3)P-binding domain, and that its loss will result in a large late endosome phenotype similar to loss of TBC-2 or VPS-34. Further analysis will be required to distinguish between these possibilities and if Armus is also dually regulated by Vps34.
We do not know the importance of TBC-2 binding to PI(4)P. However, an endosomal pool of PI(4)P has been shown to regulate recycling tubule formation (Ketel et al., 2016; Jovic et al., 2009). Since TBC-2 is also involved in endosome recycling (Liu and Grant, 2015; Sun et al., 2012), we speculate that PI(4)P binding could be important for TBC-2 regulation in the recycling pathway, while PI(3)P binding regulates TBC-2 during early to late endosome trafficking.
Vps34 negatively regulates Rab5. Although Rab5 recruits Vps34 to generate PI(3)P (Christoforidis et al., 1999; Murray et al., 2002) and PI(3)P recruits proteins to promote Rab5 activity and endosome trafficking (Horiuchi et al., 1997), we show that loss of vps-34 in C. elegans causes phenotypes consistent with increased RAB-5 activity and that Vps34-null MEFs have increased Rab5-GTP levels as compared to wild-type MEFs. Therefore, Vps34 negatively regulates Rab5 activity in C. elegans and in mouse cells. It will be interesting to determine whether Armus and/or TBC1D2B regulate Rab5 activity in mammalian cells.
In conclusion, we identified a novel role for the class III VPS-34 PI3K complex in negatively regulating the RAB-5 GTPase via recruitment of the RAB-5 GAP, TBC-2, to endosomes. The large late endosome phenotype of vps-34(RNAi) animals appears to be the result of increased RAB-5, and possibly, RAB-7 activity. RAB-mediated activation of VPS-34 promotes endosome trafficking via multiple PI(3)P-interacting proteins, of which, TBC-2 represents a negative-feedback loop to limit Rab GTPase activity and promote early to late endosome maturation.
MATERIALS AND METHODS
C. elegans strains and culture
The maintenance and culture of C. elegans was conducted as described in Wormbook (Stiernagle, 2006). The N2 Bristol strain was the wild-type parent of all strains, and the HB101 E. coli strain was used as a food source, both were obtained from the Caenorhabditis Genetics Center (CGC). The following strains were used in this study: QR2 rab-7(ok511)/mIn1 (Skorobogata and Rocheleau, 2012), QR15 tbc-2(tm2241) (Chotard et al., 2010a), QR449 tbc-2(tm2241); vhEx12 [Pvha-6::GFP::tbc-2::Flag+Pttx-3::GFP] (this study), QR457 tbc-2(tm2241); vhEx20 [Pvha-6::GFP::tbc-2(ΔPH)::Flag+Pttx-3::GFP] (this study), QR459 tbc-2(tm2241); vhEx21 [Pvha-6::GFP::tbc-2(ΔPH-CC)::Flag+Pttx-3::GFP] (this study), QR581 pwIs429 [Pvha-6::mCherry::rab-7+Cb-unc-119]; qxIs156 [Phsp::mtm-1+Psur-5::GFP] (this study), QR599 pwIs480 [Pvha-6::mCherry::rab-5+Cb-unc-119]; qxIs156 [Phsp::mtm-1+Psur-5::GFP] (this study), QR640 tbc-2(tm2241); vhEx12 [Pvha-6::GFP::tbc-2::Flag+Pttx-3::GFP]; pwIs429 [Pvha-6::mCherry::rab-7+Cb-unc-119] (this study), QR641 tbc-2(tm2241); vhEx20 [Pvha-6::GFP::tbc-2(ΔPH)::Flag+Pttx-3::GFP]; pwIs429 [Pvha-6::mCherry::rab-7+Cb-unc-119] (this study), QR642 tbc-2(tm2241); vhEx21 [Pvha-6::GFP::tbc-2(ΔPH-CC)::Flag+Pttx-3::GFP]; pwIs429 [Pvha-6::mCherry::rab-7+Cb-unc-119] (this study), RT327 unc-119(ed3) III; pwIs72 [Pvha-6::GFP::rab-5+Cb-unc-119] (Chen et al., 2006), RT476 unc-119(ed3) III; pwIs170 [Pvha-6::GFP::rab-7+Cb-unc-119] (Chen et al., 2006), RT1103 unc-119(ed3) III; pwIs429 [Pvha-6::mCherry::rab-7+Cb-unc-119] (gift from Barth Grant, Department of Molecular Biology and Biochemistry, Rutgers University, NJ), RT1239 unc-119(ed3) III; pwIs480 [Pvha-6::mCherry::rab-5+Cb-unc-119] (gift from Barth Grant), VC517 bec-1(ok691)/nT1[qIs51] (C. elegans Gene Knockout Consortium), XW2335 qxIs156 [Phsp::mtm-1+Psur-5::GFP] (Zou et al., 2009), XW3232 qxIs210 [Phsp::mtm-1 C378S+Psur-5::GFP] (Zou et al., 2009).
RNA interference
RNAi feeding was performed essentially as previously described (Kamath et al., 2001), using the rab-7 (II-8G13), bec-1 (IV-2N20), vps-15 (II-8E15) and vps-34 (I-2F20) RNAi clones from the Ahringer library (Geneservice, Cambridge, UK). The identity of the clones was verified by DNA sequencing. The empty L4440 RNAi feeding vector and vector targeting gfp were used as negative controls. L4 animals were plated on RNAi feeding plates for 24 h, and transferred to fresh plates, from which the progeny were scored when they reached the L4 stage. For the double-RNAi epistasis experiments, two dsRNA-expressing bacterial cultures were mixed before spreading them on the nematode growth plates.
Plasmid construction
Full-length TBC-2 [amino acids (aa) 2–908] and the TBC-2 truncations, ΔPH (aa 117–908) and ΔPH-CC (aa 391–908), fused to GFP at the N-terminus and Flag at the C-terminus were generated by PCR using the vector Pvha-6::GFP::tbc-2 as a template and as a receipt vector (Chotard et al., 2010a) digested with BamHI and EcoRI. The forward primers were: for full-length TBC-2, JHo130 (5′-GTCGACTGGATCCGGTAC-3′), for TBC-2(ΔPH), FLo5 (5′-CACTATGGATCCTGGAAGGCAACGAAATCAAAATC-3′) and for TBC-2(ΔPH-CC), FLo6 (5′-CAGTAAGGATCCCTTCGAATGTGTGAAGAAGAAAATCG-3′). The reverse primer for all constructs was FLo7 (5′-CATATAGAATTCTTACTTATCGTCGTCATCCTTGTAATCCATACAGTGGCCAGTATC-3′) carrying a Flag epitope tag.
GST fusion constructs were generated by PCR with the vector Pvha-6::GFP::tbc-2 (Chotard et al., 2010a) as a template. PH-CC (aa 2–390) was amplified using JHo160F (5′-CATTATGGATCCATTCTGCGGCGCCGGGGACAATC-3′) and JHo154R (5′-GTGTTGGAATTCTTAAATAAGCTCTTCTTTTGTTCGTAATG-3′). PH (aa 2–115) was amplified by using JHo160F and JHo153R (5′-GTGTTGGAATTCTTACTTCTTTTCCGTGTACTTTGAAGAGCTTTCATCC-3′). CC (aa 283–390) was amplified by using JHo161F (5′-CACCATGGATCCGAAGAGAAAGTTATTG-3′) and JHo154R. PH-CC and PH were cloned into pGEX-5X-2, and CC was cloned into pGEX4T-3 by means of the BamHI and EcoRI restriction sites. All clones were verified by Sanger sequencing (Genome Quebec).
Generation of transgenic strains
Extrachromosomal arrays vhEx12, vhEx20 and vhEx21 were generated by microinjection as described previously (Mello et al., 1991). GFP::tbc-2::Flag-containing plasmids were coinjected with the Pttx-3::GFP marker (Hobert et al., 1997) at a concentration of 50 ng/µl each.
Microscopy and phenotype analysis
Intestinal phenotypes of live fourth larval stage (L4) hermaphrodites were imaged at room temperature. Animals were mounted on 2% agarose pads, anesthetized with 100 mM levamisole in M9 buffer and covered with an 18 mm2 coverslip. Differential interference contrast (DIC) microscopy (Fig. 1) was performed using an Axio Imager A1 compound microscope with a 100×1.3 NA Plan-Neofluar oil-immersion objective lens (Zeiss) and images were captured by using an Axio Cam MRm camera and AxioVision software (Zeiss). Confocal microscopy was performed on an Axio Observer Z1 LSM-780 laser scanning confocal microscope with a 63×1.4 NA Plan-Apochromat oil-immersion objective lens (Zeiss) in a single-track mode using an argon multiline laser (488 nm excitation for GFP and a 561 nm excitation for mCherry). Images were captured with a 32 channel GaAsP detector and ZEN2010 image software. Raw data was analyzed using Fiji (ImageJ), and modifications to images for presentation purposes were made by using Fiji (ImageJ) and Adobe Photoshop CS6.
The severity of the large intestinal vesicle phenotype was quantified by measuring the diameter of the three largest vesicles per animal using Fiji (ImageJ) software. Prism 7 (Graphpad) was used to graph the data and determine statistical analysis using an unpaired t-test.
Lipid-binding assays
For the lipid overlay assays, PIP Strips™ (Echelon Biosciences Inc.) were blocked with 3% bovine serum albumin (BSA) in Tris-buffered saline with Tween 20 (1× TBST; 10 mM Tris-HCl, 150 mM NaCl pH 8.0 and 0.1% Tween 20) for 1 h at room temperature, then incubated with 0.5 µg/ml of each GST::TBC-2 fusion protein, or PIP2 Grip™ as positive control and GST as negative control, in blocking solution for 1 h at room temperature. The strips were washed three times with 1× TBST, 10 min per wash and then incubated with a 1:2000 dilution of anti-GST polyclonal antibody (G7781; Sigma-Aldrich) for 1 h at room temperature in the blocking solution. After washing with TBST, the strips were incubated with a 1:10,000 dilution of goat anti-rabbit-IgG horseradish peroxidase conjugate (Sigma-Aldrich) for 1 h at room temperature in blocking solution. After three washes in 1× TBST, the lipid binding was detected with enhanced chemiluminescence substrate (Pierce).
Liposome pulldown assay
Liposomes for the binding assays were prepared as described previously (Zhang et al., 2014; Roach et al., 2012). 1,2-dioleoyl-sn-glycero-3-phosphocholine (PC), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (PE), 1,2-dioleoyl-sn-glycero-3-phosphate (PA), 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-3′-phosphate) (PI(3)P) and 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-4′-phosphate) (PI(4)P) were purchased from Avanti Polar Lipids (Alabaster, AL). Control liposomes were prepared as a mixture of PC and PE at a molar ratio of 67:33. The liposomes containing acidic phospholipids were prepared as a mixture of PC and PE, with either PA, PI(3)P or PI(4)P at a molar ratio of 65:32:3. The lipid mixtures were dried by a rotatory vacuum, re-suspended with the internal buffer (256 mM sucrose, 20 mM Tris-HCl, pH 7.4), followed by 10 cycles of freezing in liquid nitrogen and thawing in a 37°C water bath. To generate small unilamellar liposomes the multilamellar lipids were extruded through polycarbonate membranes (pore size 100 nm, Whatman) ten times and then washed with the binding buffer (150 mM NaCl, 20 mM Tris-HCl, pH 7.4) and centrifuged at 30,000 g for 1 h. The pelleted liposomes were re-suspended with the binding buffer and used within 1 week. 10 pmol purified GST::TBC-2-CC or GST::TBC-2-PH protein was mixed and incubated with the serially diluted liposomes at 4°C for 30 min. After centrifugation at 30,000 g for 30 min, the supernatants were carefully removed. The pellets were resuspended with 1× SDS loading buffer and boiled for 5 min, and then separated by SDS-PAGE. The binding of GST::TBC-2-PH or GST::TBC-2-CC to liposomes was detected by an anti-GST antibody (1:1000, ab111947; Abcam) and followed by an IRDye 680-conjugated secondary antibody (Rockland Immunochemicals, Gilbertsville, PA), and visualized by a LI-COR odyssey imaging system. ImageJ was used to quantify the intensity of western blot results.
Rab5 effector pulldown assays
The GST::R5BD construct and pulldown assays were described previously (Liu et al., 2007), with slight modifications. Briefly, GST::R5BD was expressed in BL21-CodonPlus E. coli and purified with glutathione–agarose beads (Invitrogen). For pulldown of cell lysates, cells in 10-cm plates were lysed in buffer containing 25 mM HEPES pH 7.4, 100 mM NaCl, 1 mM CaCl2, 5 mM MgCl2, 1% NP-40, 10% glycerol, 1 mM DTT, 100 μM PMSF, and EDTA-free protease inhibitor cocktail. After centrifugation, supernatants were incubated with GST::R5BD beads at 4°C, washed with lysis buffer, boiled in 1× SDS sample buffer and subjected to immunoblotting using an anti-Rab5 antibody at a 1:5000 dilution (Cell Signaling #2143).
Acknowledgements
We thank Min Fu in the RI MUHC microscopy facility and Stéphane Laporte for imaging assistance, Richard Roy for reagents, and Xiaochen Wang and Barth Grant for strains. We thank Nadia Jaber for sharing data before publication. We thank İçten Meraş, Olga Skorobogata and Kimberley Gauthier for comments on the manuscript. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: F.L., J.S., C.E.R.; Methodology: F.L., J.S., Z.W., J.L.D., G.D., C.E.R.; Formal analysis: F.L., J.S., Z.W., J.L.D., Y.B., A.B., W.Z., G.D., C.E.R.; Investigation: F.L., J.S., Z.W., J.L.D., Y.B., A.B., W.Z., G.D., C.E.R.; Writing - original draft: F.L., J.S., Z.W., J.L.D., W.Z., G.D., C.E.R.; Writing - review & editing: F.L., J.S., Z.W., W.Z., G.D., C.E.R.; Visualization: F.L., J.S., Z.W., J.L.D., C.E.R.; Supervision: W.Z., G.D., C.E.R.; Funding acquisition: W.Z., G.D., C.E.R.
Funding
This work was funded by National Institutes of Health R01 grants (HL119478) to G.D. and (GM97355) to W.-X.Z., and a Canadian Institutes of Health Research operating grant (MOP-114935) to C.E.R. The Research Institute of the McGill University Health Centre is funded in part by the Fonds de Recherche du Québec - Santé and the imaging facility is supported by the Canada Foundation for Innovation. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.194746.supplemental
References
- Brown W. J., Dewald D. B., Emr S. D., Plutner H. and Balch W. E. (1995). Role for phosphatidylinositol 3-kinase in the sorting and transport of newly synthesized lysosomal enzymes in mammalian cells. J. Cell Biol. 130, 781-796. 10.1083/jcb.130.4.781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C. G. and Emr S. D. (1998). Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol. Cell 2, 157-162. 10.1016/S1097-2765(00)80125-2 [DOI] [PubMed] [Google Scholar]
- Carroll B., Mohd-Naim N., Maximiano F., Frasa M. A., Mccormack J., Finelli M., Thoresen S. B., Perdios L., Daigaku R., Francis R. E. et al. (2013). The TBC/RabGAP Armus coordinates Rac1 and Rab7 functions during autophagy. Dev. Cell 25, 15-28. 10.1016/j.devcel.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever M. L., Sato T. K., de Beer T., Kutateladze T. G., Emr S. D. and Overduin M. (2001). Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat. Cell Biol. 3, 613-618. 10.1038/35083000 [DOI] [PubMed] [Google Scholar]
- Chen C. C.-H., Schweinsberg P. J., Vashist S., Mareiniss D. P., Lambie E. J. and Grant B. D. (2006). RAB-10 is required for endocytic recycling in the Caenorhabditis elegans intestine. Mol. Biol. Cell 17, 1286-1297. 10.1091/mbc.E05-08-0787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotard L., Mishra A. K., Sylvain M.-A., Tuck S., Lambright D. G. and Rocheleau C. E. (2010a). TBC-2 regulates RAB-5/RAB-7-mediated endosomal trafficking in Caenorhabditis elegans. Mol. Biol. Cell 21, 2285-2296. 10.1091/mbc.E09-11-0947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotard L., Skorobogata O., Sylvain M.-A., Shrivastava S. and Rocheleau C. E. (2010b). TBC-2 is required for embryonic yolk protein storage and larval survival during L1 diapause in Caenorhabditis elegans. PLoS ONE 5, e15662 10.1371/journal.pone.0015662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis S., Miaczynska M., Ashman K., Wilm M., Zhao L., Yip S.-C., Waterfield M. D., Backer J. M. and Zerial M. (1999). Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat. Cell Biol. 1, 249-252. 10.1038/12075 [DOI] [PubMed] [Google Scholar]
- Corvera S., D'arrigo A. and Stenmark H. (1999). Phosphoinositides in membrane traffic. Curr. Opin. Cell Biol. 11, 460-465. 10.1016/S0955-0674(99)80066-0 [DOI] [PubMed] [Google Scholar]
- Edgley M. and Riddle D. (2001). LG II balancer chromosomes in Caenorhabditis elegans: mT1(II;III) and the mIn1 set of dominantly and recessively marked inversions. Mol. Genet. Genomics 266, 385-395. 10.1007/s004380100523 [DOI] [PubMed] [Google Scholar]
- Ellson C. D., Gobert-Gosse S., Anderson K. E., Davidson K., Erdjument-Bromage H., Tempst P., Thuring J. W., Cooper M. A., Lim Z.-Y., Holmes A. B. et al. (2001). PtdIns(3)P regulates the neutrophil oxidase complex by binding to the PX domain of p40(phox). Nat. Cell Biol. 3, 679-682. 10.1038/35083076 [DOI] [PubMed] [Google Scholar]
- Fernandez-Borja M., Wubbolts R., Calafat J., Janssen H., Divecha N., Dusseljee S. and Neefjes J. (1999). Multivesicular body morphogenesis requires phosphatidyl-inositol 3-kinase activity. Curr. Biol. 9, 55-58. 10.1016/S0960-9822(99)80048-7 [DOI] [PubMed] [Google Scholar]
- Frasa M. A. M., Maximiano F. C., Smolarczyk K., Francis R. E., Betson M. E., Lozano E., Goldenring J., Seabra M. C., Rak A., Ahmadian M. R. et al. (2010). Armus is a Rac1 effector that inactivates Rab7 and regulates E-cadherin degradation. Curr. Biol. 20, 198-208. 10.1016/j.cub.2009.12.053 [DOI] [PubMed] [Google Scholar]
- Futter C. E., Collinson L. M., Backer J. M. and Hopkins C. R. (2001). Human VPS34 is required for internal vesicle formation within multivesicular endosomes. J. Cell Biol. 155, 1251-1264. 10.1083/jcb.200108152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaullier J.-M., Simonsen A., D'arrigo A., Bremnes B., Stenmark H. and Aasland R. (1998). FYVE fingers bind PtdIns(3)P. Nature 394, 432-433. 10.1038/28767 [DOI] [PubMed] [Google Scholar]
- Hars E. S., Qi H., Ryazanov A. G., Jin S., Cai L., Hu C. and Liu L. F. (2007). Autophagy regulates ageing in C. elegans. Autophagy 3, 93-95. 10.4161/auto.3636 [DOI] [PubMed] [Google Scholar]
- Hobert O., Mori I., Yamashita Y., Honda H., Ohshima Y., Liu Y. and Ruvkun G. (1997). Regulation of interneuron function in the C. elegans thermoregulatory pathway by the ttx-3 LIM homeobox gene. Neuron 19, 345-357. 10.1016/S0896-6273(00)80944-7 [DOI] [PubMed] [Google Scholar]
- Horiuchi H., Lippé R., Mcbride H. M., Rubino M., Woodman P., Stenmark H., Rybin V., Wilm M., Ashman K., Mann M. et al. (1997). A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell 90, 1149-1159. 10.1016/S0092-8674(00)80380-3 [DOI] [PubMed] [Google Scholar]
- Huotari J. and Helenius A. (2011). Endosome maturation. EMBO J. 30, 3481-3500. 10.1038/emboj.2011.286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaber N., Dou Z., Chen J.-S., Catanzaro J., Jiang Y.-P., Ballou L. M., Selinger E., Ouyang X., Lin R. Z., Zhang J. et al. (2012). Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc. Natl. Acad. Sci. USA 109, 2003-2008. 10.1073/pnas.1112848109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaber N., Mohd-Naim N., Wang Z., Deleon J. L., Kim S., Zhong H., Sheshadri N., Dou Z., Edinger A. L., Du G. et al. (2016). Vps34 regulates Rab7 and late endocytic trafficking through recruitment of the GTPase-activating protein Armus. J. Cell Sci. 129, 4424-4435. 10.1242/jcs.192260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovic M., Kieken F., Naslavsky N., Sorgen P. L. and Caplan S. (2009). Eps15 homology domain 1-associated tubules contain phosphatidylinositol-4-phosphate and phosphatidylinositol-(4,5)-bisphosphate and are required for efficient recycling. Mol. Biol. Cell 20, 2731-2743. 10.1091/mbc.E08-11-1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Martinez-Campos M., Zipperlen P., Fraser A. G. and Ahringer J. (2001). Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2, RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai F., Liu H., Field S. J., Akbary H., Matsuo T., Brown G. E., Cantley L. C. and Yaffe M. B. (2001). The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat. Cell Biol. 3, 675-678. 10.1038/35083070 [DOI] [PubMed] [Google Scholar]
- Kanno E., Ishibashi K., Kobayashi H., Matsui T., Ohbayashi N. and Fukuda M. (2010). Comprehensive screening for novel rab-binding proteins by GST pull-down assay using 60 different mammalian Rabs. Traffic 11, 491-507. 10.1111/j.1600-0854.2010.01038.x [DOI] [PubMed] [Google Scholar]
- Ketel K., Krauss M., Nicot A.-S., Puchkov D., Wieffer M., Müller R., Subramanian D., Schultz C., Laporte J. and Haucke V. (2016). A phosphoinositide conversion mechanism for exit from endosomes. Nature 529, 408-412. 10.1038/nature16516 [DOI] [PubMed] [Google Scholar]
- Kihara A., Noda T., Ishihara N. and Ohsumi Y. (2001). Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152, 519-530. 10.1083/jcb.152.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen J. M., Doukoumetzidis K., Almendinger J., Stergiou L., Tosello-Trampont A., Sifri C. D., Hengartner M. O. and Ravichandran K. S. (2008). A pathway for phagosome maturation during engulfment of apoptotic cells. Nat. Cell Biol. 10, 556-566. 10.1038/ncb1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawe D. C., Chawla A., Merithew E., Dumas J., Carrington W., Fogarty K., Lifshitz L., Tuft R., Lambright D. and Corvera S. (2002). Sequential roles for phosphatidylinositol 3-phosphate and Rab5 in tethering and fusion of early endosomes via their interaction with EEA1. J. Biol. Chem. 277, 8611-8617. 10.1074/jbc.M109239200 [DOI] [PubMed] [Google Scholar]
- Lemmon M. A. (2007). Pleckstrin homology (PH) domains and phosphoinositides. Biochem. Soc. Symp. 74, 81-93. 10.1042/BSS2007c08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zou W., Zhao D., Yan J., Zhu Z., Lu J. and Wang X. (2009). C. elegans Rab GTPase activating protein TBC-2 promotes cell corpse degradation by regulating the small GTPase RAB-5. Development 136, 2445-2455. 10.1242/dev.035949 [DOI] [PubMed] [Google Scholar]
- Liu O. and Grant B. D. (2015). Basolateral endocytic recycling requires RAB-10 and AMPH-1 mediated recruitment of RAB-5 GAP TBC-2 to endosomes. PLoS Genet. 11, e1005514 10.1371/journal.pgen.1005514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Lamb D., Chou M. M., Liu Y.-J. and Li G. (2007). Nerve growth factor-mediated neurite outgrowth via regulation of Rab5. Mol. Biol. Cell 18, 1375-1384. 10.1091/mbc.E06-08-0725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu N., Shen Q., Mahoney T. R., Neukomm L. J., Wang Y. and Zhou Z. (2012). Two PI 3-kinases and one PI 3-phosphatase together establish the cyclic waves of phagosomal PtdIns(3)P critical for the degradation of apoptotic cells. PLoS Biol. 10, e1001245 10.1371/journal.pbio.1001245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G. (2005). Genomic overview of protein kinases. WormBook, 1-19. doi:10.1895/wormbook.1.60.1 10.1895/wormbook.1.60.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez A., Talloczy Z., Seaman M., Eskelinen E. L., Hall D. H. and Levine B. (2003). Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301, 1387-1391. 10.1126/science.1087782 [DOI] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D. and Ambros V. (1991). Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills I. G., Jones A. T. and Clague M. J. (1998). Involvement of the endosomal autoantigen EEA1 in homotypic fusion of early endosomes. Curr. Biol. 8, 881-884. 10.1016/S0960-9822(07)00351-X [DOI] [PubMed] [Google Scholar]
- Murray J. T., Panaretou C., Stenmark H., Miaczynska M. and Backer J. M. (2002). Role of Rab5 in the recruitment of hVps34/p150 to the early endosome. Traffic 3, 416-427. 10.1034/j.1600-0854.2002.30605.x [DOI] [PubMed] [Google Scholar]
- Neukomm L. J., Nicot A.-S., Kinchen J. M., Almendinger J., Pinto S. M., Zeng S., Doukoumetzidis K., Tronchere H., Payrastre B., Laporte J. F. et al. (2011). The phosphoinositide phosphatase MTM-1 regulates apoptotic cell corpse clearance through CED-5-CED-12 in C. elegans. Development 138, 2003-2014. 10.1242/dev.060012 [DOI] [PubMed] [Google Scholar]
- Nielsen E., Christoforidis S., Uttenweiler-Joseph S., Miaczynska M., Dewitte F., Wilm M., Hoflack B. and Zerial M. (2000). Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J. Cell Biol. 151, 601-612. 10.1083/jcb.151.3.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S., Alemu E. A., Brech A., Bruun J.-A., Lamark T., Øvervatn A., Bjørkøy G. and Johansen T. (2010). FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 188, 253-269. 10.1083/jcb.200907015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patki V., Virbasius J., Lane W. S., Toh B.-H., Shpetner H. S. and Corvera S. (1997). Identification of an early endosomal protein regulated by phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. USA 94, 7326-7330. 10.1073/pnas.94.14.7326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patki V., Lawe D. C., Corvera S., Virbasius J. V. and Chawla A. (1998). A functional PtdIns(3)P-binding motif. Nature 394, 433-434. 10.1038/28771 [DOI] [PubMed] [Google Scholar]
- Peterson M. R., Burd C. G. and Emr S. D. (1999). Vac1p coordinates Rab and phosphatidylinositol 3-kinase signaling in Vps45p-dependent vesicle docking/fusion at the endosome. Curr. Biol. 9, 159-162. 10.1016/S0960-9822(99)80071-2 [DOI] [PubMed] [Google Scholar]
- Reaves B. J., Bright N. A., Mullock B. M. and Luzio J. P. (1996). The effect of wortmannin on the localisation of lysosomal type I integral membrane glycoproteins suggests a role for phosphoinositide 3-kinase activity in regulating membrane traffic late in the endocytic pathway. J. Cell Sci. 109, 749-762. [DOI] [PubMed] [Google Scholar]
- Rink J., Ghigo E., Kalaidzidis Y. and Zerial M. (2005). Rab conversion as a mechanism of progression from early to late endosomes. Cell 122, 735-749. 10.1016/j.cell.2005.06.043 [DOI] [PubMed] [Google Scholar]
- Roach A. N., Wang Z., Wu P., Zhang F., Chan R. B., Yonekubo Y., Di Paolo G., Gorfe A. A. and Du G. (2012). Phosphatidic acid regulation of PIPKI is critical for actin cytoskeletal reorganization. J. Lipid Res. 53, 2598-2609. 10.1194/jlr.M028597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson F. L. and Dixon J. E. (2006). Myotubularin phosphatases: policing 3-phosphoinositides. Trends Cell Biol. 16, 403-412. 10.1016/j.tcb.2006.06.001 [DOI] [PubMed] [Google Scholar]
- Rocheleau C. E., Cullison K., Huang K., Bernstein Y., Spilker A. C. and Sundaram M. V. (2008). The Caenorhabditis elegans ekl (enhancer of ksr-1 lethality) genes include putative components of a germline small RNA pathway. Genetics 178, 1431-1443. 10.1534/genetics.107.084608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roggo L., Bernard V., Kovacs A. L., Rose A. M., Savoy F., Zetka M., Wymann M. P. and Muller F. (2002). Membrane transport in Caenorhabditis elegans: an essential role for VPS34 at the nuclear membrane. EMBO J. 21, 1673-1683. 10.1093/emboj/21.7.1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruck A., Attonito J., Garces K. T., Nuñez L., Palmisano N. J., Rubel Z., Bai Z., Nguyen K. C. Q., Sun L., Grant B. D. et al. (2011). The Atg6/Vps30/Beclin 1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy 7, 386-400. 10.4161/auto.7.4.14391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan N., Sumakovic M., Hannemann M., Hegermann J., Liewald J. F., Olendrowitz C., Koenig S., Grant B. D., Rizzoli S. O., Gottschalk A. et al. (2012). RAB-5 and RAB-10 cooperate to regulate neuropeptide release in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 109, 18944-18949. 10.1073/pnas.1203306109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Norris A., Sato M. and Grant B. D. (2014). C. elegans as a model for membrane traffic. WormBook, 1-47. doi:10.1895/wormbook.1.77.2 10.1895/wormbook.1.77.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schu P. V., Takegawa K., Fry M. J., Stack J. H., Waterfield M. D. and Emr S. D. (1993). Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 260, 88-91. 10.1126/science.8385367 [DOI] [PubMed] [Google Scholar]
- Simonsen A., Lippé R., Christoforidis S., Gaullier J.-M., Brech A., Callaghan J., Toh B.-H., Murphy C., Zerial M. and Stenmark H. (1998). EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 394, 494-498. 10.1038/28879 [DOI] [PubMed] [Google Scholar]
- Skorobogata O. and Rocheleau C. E. (2012). RAB-7 antagonizes LET-23 EGFR signaling during vulva development in Caenorhabditis elegans. PLoS ONE 7, e36489 10.1371/journal.pone.0036489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Xu W., Zhang A., Huang G., Liang X., Virbasius J. V., Czech M. P. and Zhou G. W. (2001). Phox homology domains specifically bind phosphatidylinositol phosphates. Biochemistry 40, 8940-8944. 10.1021/bi0155100 [DOI] [PubMed] [Google Scholar]
- Stack J. H., Herman P. K., Schu P. V. and Emr S. D. (1993). A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 12, 2195-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M.-P., Feng Y., Cooper K. L., Welford A. M. and Wandinger-Ness A. (2003). Human VPS34 and p150 are Rab7 interacting partners. Traffic 4, 754-771. 10.1034/j.1600-0854.2003.00133.x [DOI] [PubMed] [Google Scholar]
- Stenmark H. (2009). Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513-525. 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- Stenmark H., Parton R. G., Steele-Mortimer O., Lutcke A., Gruenberg J. and Zerial M. (1994). Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 13, 1287-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T. (2006). Maintenance of C. elegans. WormBook, 1-11. doi:10.1895/wormbook.1.101.1 10.1895/wormbook.1.101.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Liu O., Desai J., Karbassi F., Sylvain M.-A., Shi A., Zhou Z., Rocheleau C. E. and Grant B. D. (2012). CED-10/Rac1 regulates endocytic recycling through the RAB-5 GAP TBC-2. PLoS Genet. 8, e1002785 10.1371/journal.pgen.1002785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku T., Morimoto K., Sasawatari S. and Kumanogoh A. (2015). Leucine-rich repeat kinase 1 regulates autophagy through turning on TBC1D2-dependent Rab7 inactivation. Mol. Cell. Biol. 35, 3044-3058. 10.1128/MCB.00085-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira O. V., Botelho R. J., Rameh L., Brachmann S. M., Matsuo T., Davidson H. W., Schreiber A., Backer J. M., Cantley L. C. and Grinstein S. (2001). Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J. Cell Biol. 155, 19-26. 10.1083/jcb.200107069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S., Dhand R., Vanhaesebroeck B., Macdougall L. K., Stein R., Zvelebil M. J., Domin J., Panaretou C. and Waterfield M. D. (1995). A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 14, 3339-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener C. S., Malerød L., Pedersen N. M., Progida C., Bakke O., Stenmark H. and Brech A. (2010). Ultrastructural characterization of giant endosomes induced by GTPase-deficient Rab5. Histochem. Cell Biol. 133, 41-55. 10.1007/s00418-009-0643-8 [DOI] [PubMed] [Google Scholar]
- Xin X., Gfeller D., Cheng J., Tonikian R., Sun L., Guo A., Lopez L., Pavlenco A., Akintobi A., Zhang Y. et al. (2013). SH3 interactome conserves general function over specific form. Mol. Syst. Biol. 9, 652 10.1038/msb.2013.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Fares H., Grant B., Li Z., Rose A. M., Clark S. G. and Skolnik E. Y. (2003). Genetic analysis of the myotubularin family of phosphatases in Caenorhabditis elegans. J. Biol. Chem. 278, 34380-34386. 10.1074/jbc.M303259200 [DOI] [PubMed] [Google Scholar]
- Zhang F., Wang Z., Lu M., Yonekubo Y., Liang X., Zhang Y., Wu P., Zhou Y., Grinstein S., Hancock J. F. et al. (2014). Temporal production of the signaling lipid phosphatidic acid by phospholipase D2 determines the output of extracellular signal-regulated kinase signaling in cancer cells. Mol. Cell. Biol. 34, 84-95. 10.1128/MCB.00987-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W., Lu Q., Zhao D., Li W., Mapes J., Xie Y. and Wang X. (2009). Caenorhabditis elegans myotubularin MTM-1 negatively regulates the engulfment of apoptotic cells. PLoS Genet. 5, e1000679 10.1371/journal.pgen.1000679 [DOI] [PMC free article] [PubMed] [Google Scholar]