Abstract
Genetic factors are known to strongly influence susceptibility to allergic inflammation. The Th2 cytokine IL-13 is a central mediator of allergy and asthma, and common single-nucleotide polymorphisms in IL13 are associated with allergic phenotypes in several ethnically diverse populations. In particular, IL13+2044G→A is expected to result in the nonconservative replacement of arginine 130 (R130) with glutamine (Q). We examined the impact of IL13+2044G→A on the functional properties of IL-13 by directly comparing the activity of WT IL-13 and IL-13 R130Q on primary human cells involved in the effector mechanisms of allergic inflammation. Our results show that IL-13 R130Q was significantly more active than WT IL-13 in inducing STAT6 phosphorylation and CD23 expression in monocytes and hydrocortisone-dependent IgE switching in B cells. Notably, IL-13 R130Q was neutralized less effectively than WT IL-13 by an IL-13Rα2 decoy. Decreased neutralization of the minor variant could contribute to its enhanced in vivo activity. Neither IL-13 variant was able to engage T cells, which suggests that increased allergic inflammation in carriers of IL13+2044A depends on enhanced IL-13–mediated Th2 effector functions rather than increased Th2 differentiation. Collectively, our data indicate that natural variation in the coding region of IL13 may be an important genetic determinant of susceptibility to allergy.
Introduction
IL-13, a cytokine typically produced by Th2 cells, is a central mediator of allergic inflammation and is sufficient to induce most if not all of the key features of experimental asthma (reviewed in ref. 1). IL-13 promotes allergen-induced airway hyperreactivity (2), epithelial cell damage, goblet cell hyperplasia with mucus hyperproduction, and eosinophilia. All of these effects are IL-4Rα/STAT6 mediated but IL-4 independent (3–5). Furthermore, IL-13 stimulates airway fibrosis, largely through the ability of matrix metalloproteinases 9 and 12 to activate latent TGF-β1, which favors the accumulation of eosinophils and macrophages in the lung (6, 7). Inflammation is amplified by local responses of the epithelium, smooth muscle, fibroblasts, and macrophages through the production of chemokines, cytokines (8), and other effector molecules such as acidic mammalian chitinase (9). The pivotal role of IL-13 in allergic inflammation has been further emphasized by the finding that IL-13 secreted by non–T cells, particularly eosinophils, is essential in inducing airway hyperresponsiveness and augmenting inflammation. Eosinophils, recruited to the lung through the concerted action of Th2-derived IL-5 and eotaxins released by epithelial cells, become an important cellular source of IL-13 (10–13).
Human allergic inflammation and experimental asthma share several IL-13–related signatures (reviewed in ref. 1). However, IL-13 induces class switch recombination to IgE in human but not murine B cells (14) and upregulates the expression of CD23, an IgE receptor, on human monocyte/macrophages (15). Since events mediated by the binding of IgE to its receptors are integral components of allergic reactions in tissues (reviewed in ref. 16), the induction of IgE synthesis and IgE receptors is likely to be critical for the proinflammatory role played by human IL-13 in allergy and asthma. Of note, IL-13 is readily detectable in the human placenta (17) and is vigorously secreted by neonatal T cells (18). The early timing of IL-13 expression is in line with the established importance of early life immunoregulatory events in controlling the inception of asthma (19).
Genetic factors are major determinants of susceptibility to allergic inflammation in humans (reviewed in refs. 20, 21). The IL13 locus on chromosome 5q31 contains a block of common single-nucleotide polymorphisms (SNPs) in virtually complete linkage disequilibrium (LD), which spans the third intron (+1923C→T), the fourth exon (+2044G→A), and the 3′ untranslated region of the gene (+2525G→A, +2580C→A, and +2749C→T) (22). Two SNPs in the promoter (–1512A→C and –1112C→T) are also in strong, albeit not complete, LD with the distal polymorphisms. Consistent with the prominent role of IL-13 in experimental asthma, robust associations have been detected between IL13 polymorphisms and allergic phenotypes. IL13+2044G→A is strongly associated with increased total serum IgE (22–25), asthma (26), atopy (27), atopic dermatitis (23, 27, 28), and a grouped phenotype that includes eosinophilia, IgE, and positive skin tests (29). IL13–1112C→T is associated with asthma, bronchial hyperreactivity and skin test responsiveness (30, 31), total IgE (32), and sensitization to food and outdoor allergens (32, 33). Interestingly, a significant gene/gene interaction has been detected between an IL-4Rα coding variant (S478P) and IL13–1112C→T. Individuals with the risk genotype for both genes had increased risk of developing allergic inflammation compared with that of individuals with both nonrisk genotypes (32, 34).
Although the overall association between allergic inflammation and SNPs in IL13 is extremely robust and has been replicated in several ethnically diverse populations (35), the interpretation of the mechanisms underlying this association is made problematic by the extended LD blocks found in chromosome 5q31 (36). Functional studies are required to determine which polymorphisms within a complex haplotype affect gene expression and/or function and to decipher the molecular mechanisms by which genetic variation in IL13 increases susceptibility to allergic inflammation. IL13+2044G→A is of particular interest because this SNP is found in approximately 25% of the general population (22) and is expected to result in the nonconservative replacement of a positively charged arginine (R) with a neutral glutamine (Q) at position 130 (this numbering includes the signal peptide; 130 is referred to as position 110 when numbering does not include the signal peptide; refs. 25, 26, 37, 38). The R130Q substitution occurs in α-helix D, the region of IL-13 that is thought to interact with IL-4Rα/IL-13Rα1 heterodimers (39). Therefore, IL13+2044G→A has the potential to affect IL-13–dependent signaling events.
We examined the impact of IL13+2044G→A on the functional properties of IL-13 by directly comparing the activity of the 2 variants on primary effector cells of human allergic inflammation. IL-13 R130Q was significantly more active than WT IL-13 in multiple effector assays and was neutralized less effectively by an IL-13Rα2 decoy. Our results strongly suggest that natural variation in the coding region of IL13 may be an important genetic determinant of susceptibility to allergy.
Results
Functional comparison between prokaryotic and eukaryotic WT IL-13.
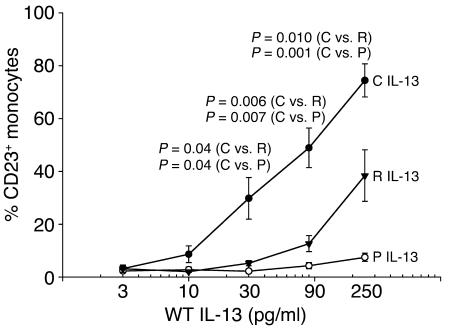
The native eukaryotic IL-13 molecule undergoes glycosylation (40), a posttranslational modification commonly missing in prokaryotic expression systems. Furthermore, C-terminal truncation immediately downstream of R130 has been reported to occur in E. coli–expressed IL-13 (F.D. Vladich et al., unpublished observations). Therefore, preliminary experiments were performed to compare the activity of 2 commercial WT IL-13 preparations from E. coli with that of COS-7 cell–derived WT IL-13 and to determine which was best suited for subsequent functional studies. Figure 1 shows that a direct comparison of the ability of these IL-13 preparations to upregulate CD23 expression in normal human monocytes revealed a marked difference in potency. Eukaryotic IL-13 was significantly more active than either prokaryotic IL-13 preparation over the 30–250 pg/ml concentration range, with a difference in EC50 of approximately 5-fold. In view of the IL-13 levels measured in supernatants of PBMCs stimulated with phytohemagglutinin for 48 hours (less than 300 pg/ml; M. Halonen et al., unpublished observations) and serum (less than 15 pg/ml; ref. 37), the activity of COS-7 cell–derived WT IL-13 was optimal within the physiologic range. Because of these results, all subsequent functional studies were performed using recombinant IL-13 variants expressed in eukaryotic cells. Since the R130Q substitution appeared to affect the recognition of IL-13 epitopes by our ELISA antibodies, which resulted in underestimation of the minor variant, concentrations of COS-7 cell–derived IL-13 R130Q were adjusted using a correction factor developed as discussed in Methods. Supernatants from mock-transfected COS-7 cells were used as negative controls throughout the study and consistently failed to elicit a response (data not shown).
Figure 1.
Functional comparison between prokaryotic and eukaryotic WT IL-13. Normal human PBMCs were stimulated with increasing concentrations of prokaryotic IL-13 (from Peprotech [P], open circles, or R&D [R], triangles) or eukaryotic IL-13 (COS-7 cell–derived [C], closed circles) for 48 hours. Surface CD23 expression was then assessed by direct immunofluorescence using PE-conjugated anti-CD23 mAb or isotype control, gating on the monocyte population. Two-tailed paired Student’s t test was used to compare responses to different IL-13 preparations. P values are noted for each IL-13 concentration at which a statistically significant difference in the response was detected. The figure shows the mean ± SE of the results obtained from 4 experiments.
IL-13 R130Q is more active than WT IL-13 in inducing STAT6 phosphorylation in primary monocytes.
Next we compared recombinant WT IL-13 and IL-13 R130Q for their impact on key events along the IL-13–dependent signaling pathway. To maximize the relevance of our experimental model to IL-13–mediated events occurring in vivo, we chose to test the activity of the IL-13 variants on primary immune cells (lymphocytes, monocytes) involved in the pathogenesis of allergic inflammation.
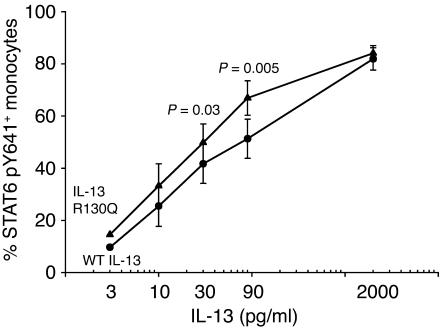
The most upstream event examined was STAT6 phosphorylation, an essential step in the IL-13–dependent regulation of gene expression (41). PBMCs from normal individuals (n = 7) were stimulated with increasing concentrations of WT IL-13 or IL-13 R130Q for 1 hour. STAT6 phosphorylation was then quantitatively assessed by intracellular immunofluorescence, which measured the percentage of monocytes that became positive for Y641-phosphorylated STAT6. Figure 2 shows that STAT6 phosphorylation increased dose-dependently and was consistently more robust in IL-13 R130Q–treated cells than in cells stimulated with WT IL-13. The difference in potency reached high statistical significance for cultures stimulated with physiologically relevant concentrations of IL-13 variants (P = 0.03 at 30 pg/ml and P = 0.005 at 90 pg/ml, 2-tailed paired Student’s t test). Analysis of the results using an additional statistical approach based on random effects models (42) confirmed the increased biological potency of IL-13 R130Q relative to WT IL-13 (Supplemental Section 1; supplemental material available online with this article; doi:10.1172/JCI200522818DS1). Furthermore, the EC50 for IL-13 R130Q was 5.6-fold lower than the one for WT IL-13 (P = 0.03; see Supplemental Table 1). In contrast, the 2 dose-response curves converged at supraphysiologic concentrations (2 ng/ml). Baseline STAT6 phosphorylation in monocytes was negligible (data not shown). Of note, the introduction of a correction factor for IL-13 R130Q did not artificially amplify the difference in potency between the 2 variants because the dose-response curve for IL-13 R130Q would shift further to the left if calculated using uncorrected IL-13 concentrations (see Supplemental Figure 1, top panel) and the EC50 for the minor variant would become 7.6-fold lower than the one for WT IL-13 (P = 0.02; see Supplemental Table 1). Collectively, these results strongly suggest that IL-13 R130Q is more active than WT IL-13 in initiating IL-13–dependent signaling in primary human inflammatory cells.
Figure 2.
IL-13 R130Q is more active than WT IL-13 in inducing STAT6 phosphorylation in primary monocytes. PBMCs from normal donors were stimulated with increasing concentrations of WT IL-13 (circles) or IL-13 R130Q (triangles) for 1 hour, then stained intracellularly with an Alexa Fluor 488–conjugated anti-STAT6(pY641) antibody. The percentage of STAT6+ cells was assessed by flow cytometry, gating on the monocyte population. Responses to the IL-13 variants were compared using a 2-tailed paired Student’s t test. P values are noted for each IL-13 concentration at which a statistically significant difference in the response was detected. The figure shows the mean ± SE of the results obtained from 7 experiments.
IL-13 R130Q is more active than WT IL-13 in upregulating CD23 expression in primary monocytes.
We have previously shown that IL-4 and IL-13 are key regulators of CD23 gene expression in human monocytes (15, 43). STAT6-dependent upregulation of CD23 expression in these cells (44) complements the unique ability of Th2 cytokines to induce IgE class switch recombination in B cells and is likely to be critical for the pathogenesis of Th2 inflammation in tissues (16).
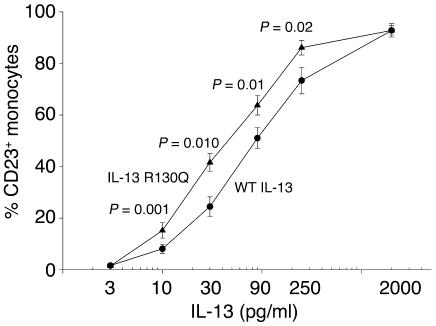
These considerations prompted us to assess whether regulation of CD23 expression in monocytes is differentially affected by IL-13 R130Q and WT IL-13. Human PBMCs from normal donors (n = 11) were incubated with increasing concentrations of recombinant WT IL-13 or IL-13 R130Q for 48 hours. CD23 expression on monocytes was then analyzed by direct immunofluorescence. Figure 3 shows that induction of CD23 by IL-13 R130Q was significantly more intense over the physiologic concentration range (10–250 pg/ml). The EC50 for the minor variant was 2.3-fold lower than the one for WT IL-13 (P = 0.0002), and this pattern was highly reproducible across experiments (see Supplemental Table 2). As already discussed for the induction of STAT6 phosphorylation, the introduction of a correction factor did not amplify the difference in potency between the 2 variants, which would have been even greater for uncorrected IL-13 R130Q concentrations (see Supplemental Figure 1, bottom panel, and Supplemental Table 2). Random effects model analysis again confirmed the enhanced potency of IL-13 R130Q (Supplemental Section 2). These results suggest that increased IL-13–dependent expression of CD23 in carriers of IL13+2044A may contribute to increased IgE-dependent inflammation.
Figure 3.
IL-13 R130Q is more active than WT IL-13 in upregulating CD23 expression in primary monocytes. Normal human PBMCs were incubated with increasing concentrations of WT IL-13 (circles) or IL-13 R130Q (triangles) for 48 hours. Surface CD23 expression was then assessed by direct immunofluorescence using PE-conjugated anti-CD23 mAb or isotype control, gating on the monocyte population. Responses to the IL-13 variants were compared using a 2-tailed paired Student’s t test. P values are noted for each IL-13 concentration at which a statistically significant difference in the percentage of CD23+ cells was detected. The figure shows the mean ± SE of the results obtained from 11 experiments.
IL-13 R130Q is more active than WT IL-13 in inducing hydrocortisone-dependent, but not CD40-dependent, IgE synthesis.
Together with IL-4, IL-13 is the only cytokine which has been shown to induce IgE class switching. This effect requires synergistic interactions with a second signal typically provided by CD40 engagement (reviewed in ref. 45). Hydrocortisone (HC) has also been shown to potently synergize with IL-4 in inducing IgE synthesis, largely through its ability to upregulate CD40 ligand expression in T and possibly B cells (46). The interactions between HC and IL-13 have not been investigated.
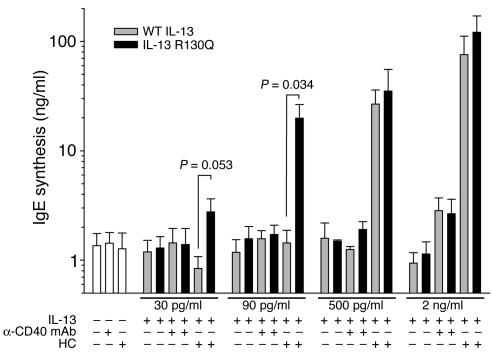
Increased serum IgE levels have been found in carriers of IL13+2044A in several populations (22–25), raising the possibility that expression of IL-13 R130Q and increased IgE class switching might be mechanistically linked. In order to explore this possibility, PBMCs from nonallergic donors were incubated with increasing concentrations of WT IL-13 or IL-13 R130Q in the presence or absence of anti-CD40 mAb or HC, and IgE secretion was measured after 12–14 days of culture. Figure 4 shows that, consistent with the modest IgE-inducing effects previously attributed to IL-13 (14), negligible levels of IgE synthesis were detected in cultures stimulated with WT IL-13 or IL-13 R130Q and anti-CD40 mAb, regardless of the concentration of IL-13 variant added to the cells. Low IgE induction did not result from impaired activity of the anti-CD40 antibody because mAb 626.1 elicited vigorous IgE synthesis (20.52 ± 7.26 ng/ml) in parallel cultures when added with IL-4. Surprisingly, a strong synergism occurred between HC and IL-13, particularly in IL-13 R130Q–stimulated cultures. Substantial IgE synthesis, with a highly significant increase in the response to IL-13 R130Q relative to WT IL-13 (P = 0.034), was detected in cultures stimulated with 90 pg/ml of variant. The difference in the response to a lower IL-13 concentration (30 pg/ml) approached statistical significance. Confirming the trend observed in the other functional assays, IgE induction by supraphysiologic IL-13 concentrations (0.5 and 2 ng/ml) was brisk but not significantly different between the 2 variants. Collectively, these data highlight a novel, selective synergism between IL-13 and HC and furthermore suggest that expression of the minor IL-13 variant may play a role in the dysregulation of IgE synthesis found in carriers of IL13+2044A.
Figure 4.
IL-13 R130Q is more active than WT IL-13 in inducing HC-dependent, but not CD40-dependent, IgE synthesis. Normal human PBMCs were stimulated with increasing concentrations of WT IL-13 (gray bars) or IL-13 R130Q (black bars) in the presence or absence of anti-CD40 mAb 626.1 (1 μg/ml) or HC (1 μM) for 12–14 days. Supernatants were then harvested and assessed for IgE concentrations by ELISA. Net IgE synthesis was calculated by subtracting the IgE concentrations detected in cycloheximide-treated cultures from the values detected in untreated cultures. Responses to the IL-13 variants were compared using a 2-tailed paired Student’s t test. P values are noted for each IL-13 concentration at which a statistically significant difference in IgE response was detected or approached. The figure shows the mean ± SE of results obtained from 4–7 experiments.
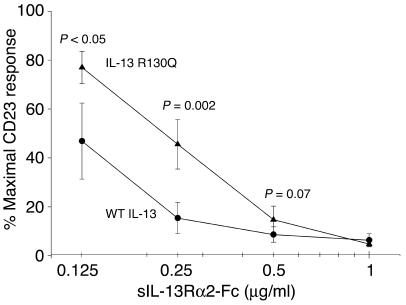
Soluble IL-13R α2 neutralizes WT IL-13 more effectively than IL-13 R130Q.
Several mechanisms may account for the increased potency of IL-13 R130Q relative to WT IL-13. The minor variant may signal more effectively through the IL-13Rα1/IL-4Rα heterodimer and/or may be neutralized less effectively by IL-13Rα2, a protein which binds IL-13 with higher affinity than IL-13Rα1 but does not signal and is thought to act as a natural decoy (reviewed in ref. 47). The C-terminal portion of IL-13 has been shown to be responsible for the high-affinity interaction with IL-13Rα2 (39), raising the possibility that the R130Q replacement may affect IL-13Rα2 binding. In an effort to elucidate the mechanisms underlying the enhanced activity of IL-13 R130Q, we compared the 2 IL-13 variants for their ability to be neutralized by IL-13Rα2. PBMCs from normal donors (n = 5) were incubated with WT IL-13 or IL-13 R130Q (90 pg/ml) for 48 hours in the presence or absence of increasing concentrations of a recombinant soluble IL-13Rα2-Fc chimera. Surface CD23 expression on monocytes was then assessed by immunofluorescence. Figure 5 shows that soluble IL-13Rα2-Fc was sufficient to reduce the WT IL-13–dependent CD23 response by more than 50% when added at a concentration of 125 ng/ml and that it virtually abrogated the response at 250 ng/ml. In contrast, neutralization of IL-13 R130Q was significantly less effective with an overall difference in potency of more than 2-fold. The addition of soluble human IgG (1 μg/ml) had no effect on CD23 expression (data not shown). Our results show that R130Q replacement strongly impaired the ability of the minor IL-13 variant to interact with IL-13Rα2. Decreased neutralization by the endogenous decoy could contribute to enhanced activity of the minor variant in vivo.
Figure 5.
Soluble IL-13Rα2 neutralizes WT IL-13 more effectively than IL-13 R130Q. Normal human PBMCs were incubated with WT IL-13 (circles) or IL-13 R130Q (triangles) (90 pg/ml) for 48 hours in the presence or absence of increasing concentrations of a recombinant soluble IL-13Rα2-Fc (sIL-13Rα2-Fc) chimera. Surface CD23 expression on monocytes was then assessed as described for Figure 3. Neutralization of the 2 IL-13 variants was expressed as a percentage of maximal CD23 response and compared using a 2-tailed paired Student’s t test. P values are noted for each sIL-13Rα2-Fc concentration at which a statistically significant difference in the response was detected or approached. The figure shows the mean ± SE of the results obtained from 5 experiments.
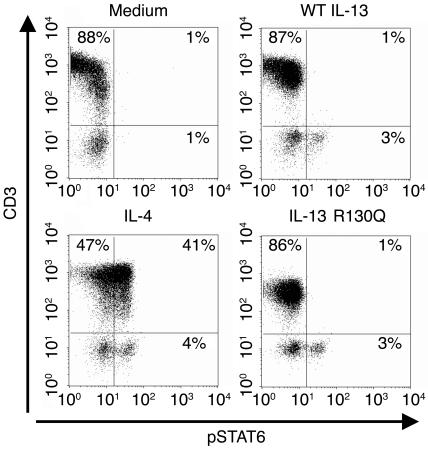
IL-13 R130Q fails to engage T cells.
Although IL-4 and WT IL-13 share several functional properties, only IL-4 controls Th2 differentiation. T cells fail to respond to WT IL-13 because they lack expression of functional IL-4Rα/IL-13Rα1 heterodimers (reviewed in ref. 47) and because IL-13 is unable to bind the IL-4Rα/γc complexes expressed on T cells (48). Thus IL-4 is involved in the initiation of Th2 responses whereas the impact of WT IL-13 remains confined to Th2 effector functions. However, structure/function analyses raise the possibility that the R130Q replacement may confer on the minor IL-13 variant the ability to bind novel cellular targets through a γc-containing receptor. IL-4 and IL-2 interact with γc through a cluster of uncharged residues in the D helix. The presence of charged amino acids in the corresponding region of IL-13 is thought to prevent γc engagement (49). Replacement of the positively charged R130 with a neutral glutamine could alter the electrostatic potential of the surface enough to allow γc binding. Acquisition of the ability to directly engage the IL-4Rα/γc complex on T cells would in turn endow IL-13 R130Q with the potential to affect Th2 differentiation, thus contributing to the enhanced allergic inflammation associated with IL13+2044A.
In view of the potential mechanistic importance of T cell/IL-13 R130Q interactions, we sought to directly explore the ability of the minor IL-13 variant to activate T cells. To this purpose, we took advantage of the ability of immunofluorescence to reveal cytokine-induced STAT6 Y641 phosphorylation in individual CD3+ T cells within a PBMC population. Figure 6 shows that virtually no intracellular STAT6 Y641 phosphorylation was detectable in unstimulated PBMCs isolated from normal donors (top left panel). As expected, incubation with IL-4 (2 ng/ml) resulted in substantial STAT6 Y641 phosphorylation in both CD3+ and CD3– cells (bottom left panel) whereas stimulation with WT IL-13 (2 ng/ml) failed to activate STAT6 in CD3+ cells, even though a vigorous response was detected in non–T cells (top right panel). Most importantly, IL-13 R130Q (2 ng/ml) elicited a response in CD3– but not CD3+ cells (bottom right panel). These data clearly rule out IL-13 R130Q–mediated signaling in T cells and suggest that increased allergic inflammation in carriers of the IL13+2044A allele results from enhanced IL-13–mediated Th2 effector functions rather than increased IL-13 variant-dependent modulation of Th2 differentiation.
Figure 6.
IL-13 R130Q does not induce STAT6 phosphorylation in human T cells. PBMCs from normal donors were incubated with complete medium, IL-4 (2 ng/ml), WT IL-13 (2 ng/nl), or IL-13 R130Q (2 ng/ml) for 1 hour. Cells were then surface stained with PE-conjugated anti-CD3, stained intracellularly with Alexa Fluor 488–conjugated anti-STAT6(pY641) antibody, and analyzed by flow cytometry gating on live cells. The figure shows the results of 1 representative experiment out of 3.
Discussion
The importance of genetic factors in influencing the risk of developing allergic inflammation is well established (20, 21). Polymorphisms that may dysregulate the function and/or expression of IL13 occur frequently in the population and have been consistently associated with allergic phenotypes (35). However, these polymorphisms are linked with one another in extended haplotype blocks that can mask the effects of individual SNPs. Therefore, the mechanisms underlying the association between allergic disease and variation in IL13 will remain unclear unless multiple functional facets of the genotype/phenotype association are explored and dissected in depth.
SNPs in coding regions constitute the majority of disease alleles in mendelian disorders (50), and a recent analysis suggests that common disease variants are likely to show a similar trend (51). Our results show that IL-13 R130Q, a common variant encoded by IL13+2044A and associated with elevated serum IgE levels and other allergy-related phenotypes in individuals of multiple ethnic backgrounds (35), is significantly more active than WT IL-13 in enhancing essential effector pathways of allergic inflammation in primary human cells.
Structure/function analyses provide mechanistic insights into the increased activity of IL-13 R130Q. The replacement of R130 with a glutamine occurs in α helix D, a region of the molecule critical for its interactions with IL-13 receptors. Alanine scanning mutagenesis recently revealed R130 to be important for IL-13 binding to IL-13Rα2 (39), the decoy receptor expressed both on cells and as a soluble protein, which binds IL-13 with high affinity but does not signal (52). IL-13Rα2 is a key negative regulator of IL-13 responses in vivo (53), and its expression is strongly enhanced by IL-13 itself (54), which points to the existence of complex feedback loops designed to tightly control IL-13–dependent events. Consistent with this scenario, IL-13 R130Q was neutralized by a soluble IL-13Rα2-Fc chimera much less effectively than WT IL-13, which suggests the minor IL-13 variant might to some extent escape the dampening mechanisms that normally restrain the activity of WT IL-13 in vivo.
The presence of a positively charged arginine at position 130 also alters the distribution of charges in the region of the D helix that is primarily responsible for binding the IL-4Rα1/IL-13Rα1 heterodimer (39). Substitution of R130 with the negatively charged aspartic acid created an artificial agonist which bound the IL-13 receptor with 5- to 10-fold improved affinity (55). While the effects of the R130Q substitution are more subtle, overall it is clear that IL13+2044G→A targets an amino acid critical for IL-13/IL-13 receptor interactions and IL-13–mediated signaling.
In comparison with the often drastic effects obtained by genetic manipulation in animal models, the functional differences between the common and the minor IL-13 variant may appear too modest to influence disease susceptibility, but several considerations argue against this conclusion. Our results are quantitatively in line with those recently obtained in several other functional studies of human polymorphic genes such as CD14 (56), IL3 (57), SLC22A4 (58), TGF-β (59), LGALS2 (60), and LTA (61), all of which show subtle effects of individual common risk alleles. Furthermore, functional differences between the IL-13 variants became manifest within a physiologically relevant concentration range and were shown to be highly significant by 2 complementary statistical approaches. Finally, IL13+2044G→A is in strong LD with a promoter SNP, IL13–1112C→T, which results in increased IL13 transcription in CD4+ Th2 cells (L. Cameron et al., manuscript submitted for publication). The transcriptional enhancement conferred by IL13–1112T is relatively modest as well, but the increase in IL-13 activity caused by the R→Q replacement, combined with the concomitant increase in IL13 transcription associated with the –1112T allele may effectively synergize to amplify IL-13–dependent events. The functional impact of SNP/SNP interactions within the same gene could be further amplified by gene/gene interactions along the same pathway (62), e.g., when IL-13 R130Q is expressed in carriers of the gain-of-function IL4R V50R551 variant (38).
Excessive production of the Th2 cytokines IL-4 and IL-5 leads to allergic inflammation in transgenic models (63, 64). WT IL-13, unlike IL-4, is unable to initiate Th2 differentiation because both human and murine T cells lack functional IL-13 receptors (47). IL-13 R130Q, albeit more active than WT IL-13 on monocytes and B cells, also failed to engage T cells. Therefore, in the absence of a genetic predisposition to enhanced Th2 responses determined through a different pathway, expression of IL-13 R130Q would not be expected to affect Th2 differentiation. However, IL-13 is a potent effector of Th2-mediated responses (reviewed in ref. 1), and these could be augmented by IL13+2044A, especially when combined with other disease risk alleles. IL13+2044A may contribute to determining an allergic phenotype both in the context of otherwise balanced Th2 responses and in cases in which Th2 differentiation and IL13 expression are independently enhanced by other genetic mechanisms.
IL-4–dependent IgE responses to CD40 crosslinking and HC typically follow similar profiles (46, 65), which is consistent with the notion that HC induces IgE synthesis by upregulating CD40 ligand expression on T cells, thus promoting crosslinking of CD40 on the B cell membrane (46). Our experiments unveiled novel aspects of the interaction between IL-13 and the B cell–activating signals required for IgE switching. IL-13 (particularly IL-13 R130Q) strongly synergized with HC under conditions in which antibody-mediated CD40 engagement had no effect. The positive crosstalk between IL-13 and HC, but not between IL-13 and CD40, may reflect previously unsuspected functional differences in the B cell–signaling pathways that lead to IgE synthesis. Since neither IL-13 variant was able to engage T cells, the synergism between IL-13 and HC was likely to have occurred at the B cell level. We explored 2 events critical for IgE class switch recombination in B cells and found that increased IgE synthesis in cells treated with IL-13 and HC was paralleled by increased ε germ-line (data not shown) and activation-induced deaminase (AID) transcription (see Supplemental Figure 2). However, the enhancement was modest compared with the vigorous upregulation of IgE synthesis detected in the same cultures. This suggests that the combination of IL-13 and HC may target additional, more distal molecular events in IgE switching, the nature of which remains to be determined. Inasmuch as the HC pathway recapitulates mechanisms relevant to IgE regulation in vivo, the enhanced ability of physiologic IL-13 R130Q concentrations to engage this pathway may contribute to enhanced IgE-dependent allergic reactions.
Arima et al. (37) recently compared the activities of WT IL-13 and IL-13 R130Q and found them to be indistinguishable in terms of STAT6 activation and induction of ε germ-line transcription. IgE synthesis was not tested. The discrepancies between these results and ours are likely to reflect significant differences in the experimental design. Arima et al. used recombinant IL-13 expressed in a prokaryotic system and a target B cell line overexpressing IL-13Rα1 (37). Utilization of target cells overexpressing a receptor is likely to mask subtle differences in the affinity of its ligand because the overall strength of ligand/receptor interactions will be dictated by the artificially increased number of receptors rather than the affinity of individual ligand-binding events. Furthermore, unlike eukaryotic proteins, bacterially expressed IL-13 molecules are unstable and undergo rapid degradation at the C-terminus (F.D. Vladich et al., unpublished observations), the same region which contains the R130Q substitution under investigation. We would argue that our approach, which relies on eukaryotic IL-13 proteins and primary human cells, is more sensitive to subtle differences in the properties of the natural IL-13 variants.
The lesson which emerges from these studies is somewhat sobering. Experimental strategies which are successful in classical immunology may not be readily applicable to functional genomics work, the targets of which are inherently elusive. When studying the effects of human genetic variation, we actually explore complex interactions between polymorphic genes (and their products) and the cellular milieu. The latter needs to be modeled as faithfully as the former because the effects of genetic variation are likely to be context-dependent. Thus, functional studies need to recreate the specific biological conditions under which natural genetic variation exerts its subtle effects — and these conditions may be different for different polymorphisms. It is possible, but by no means certain, that in vivo modeling of polymorphisms in mice might circumvent some of these hurdles and provide definitive answers. An effort along these lines is currently ongoing in our laboratory. Regardless, even at this early stage of functional genomics studies, it is clear that unraveling the molecular mechanisms whereby natural genetic variation shapes the pathogenesis of complex diseases will require more adequate conceptual frameworks as well as novel experimental and analytical tools.
Methods
Expression of recombinant IL-13.
To construct eukaryotic expression vectors for WT IL-13 and IL-13 R130Q, PBMCs were isolated from subjects homozygous for IL13+2044G or IL13+2044A and stimulated with ConA (10 μg/ml) and anti-CD28 mAb (1 μg/ml) for 48 hours. Total RNA was then extracted and reverse transcribed. Full-length (473 bp) IL13 cDNA was PCR amplified using primers 5′-CACTGGATCCATGGCGCTTTTGTTGACC and 5′-CGACTTAAGGCAACTTCAATAGTCAGGTC (position 48,483–48,500 and 46,395–46,414 in Genbank accession number AC004039). The primers contain a BamHI and an EcoRI site (underlined) at their 5′ and 3′ ends. Amplification products of the expected size were digested with BamHI and EcoRI and cloned into pcDNA3 (Invitrogen Corp.) to generate pIL13WT and pIL13R130Q. Plasmids were verified by sequencing and proven to contain a G or an A at position +2044.
COS-7 cells were transfected with empty pcDNA3, pIL13WT, or pIL13R130Q using Lipofectamine 2000 (Invitrogen Corp.) as recommended by the manufacturer. Supernatants were harvested 48–72 hours later and assessed for IL-13 concentrations by ELISA (R&D Systems). A correction factor was developed to adjust for potential differences in the ability of the ELISA to detect WT IL-13 and IL-13 R130Q (see below). Metabolic labeling with 35S-methionine/cysteine followed by immunoprecipitation with anti–IL-13 mAb (R&D Systems), SDS-PAGE, and autoradiography revealed that both IL-13 variants were expressed as 2 major species of 19 kDa and 22 kDa (data not shown). No IL-13 protein was detected by ELISA or Western blotting in supernatants from COS-7 cells transfected with empty pcDNA3. E. coli–expressed WT IL-13 was obtained from commercial sources (R&D Systems and PeproTech Inc.). Recombinant human IL-4 was purchased from R&D Systems.
A correction factor for IL-13 R130Q quantification.
Replacement of R130 with glutamine in an exposed region of the molecule (66) may affect recognition of IL-13 R130Q by antibodies generated against WT IL-13. Since functional analysis and comparison of the 2 forms of IL-13 require the utilization of identical cytokine concentrations, we assessed whether a correction factor was needed to adjust IL-13 R130Q concentrations as measured by an existing ELISA. To this end, the 2 IL-13 variants were translated in vitro using rabbit reticulocyte lysates (TNT T7 Coupled Reticulocyte Lysate System; Promega Corp.) and metabolically labeled with S35-methionine, an amino acid which is equally represented in both molecules. Each preparation translated in vitro was divided into 2 aliquots of equal volume. One was assayed for IL-13 concentrations with a commercially available ELISA (R&D Systems) developed to measure WT IL-13. The other was analyzed by SDS-PAGE; this analysis was followed by densitometry of the bands corresponding to WT IL-13 or IL-13 R130Q. We reasoned that if the IL-13 ELISA measured the concentrations of WT IL-13 and IL-13 R130Q with comparable accuracy, then WT IL-13/IL-13 R130Q (ELISA) would be equal to WT IL-13/IL-13 R130Q (densitometry). Otherwise, the difference between the 2 ratios would provide a correction factor to adjust ELISA-based measurements of IL-13 R130Q. The results of 3 consecutive experiments showed that the ELISA results underestimated the concentration of eukaryotic IL-13 R130Q by 25.7% ± 2.9%. This correction factor was subsequently used to adjust the concentrations of COS-7 cell–derived IL-13 R130Q tested in functional studies.
Analysis of STAT6 phosphorylation and CD23 expression.
To assess IL-13–dependent STAT6 phosphorylation, PBMCs were isolated from normal nonallergic donors by density gradient centrifugation and resuspended in RPMI 1640 — 10% heat inactivated FCS (HyClone), 2 mM L-glutamine, and antibiotics (complete medium) at 2 × 106 cells/ml. After stimulation with increasing concentrations of WT IL-13 or IL-13 R130Q for 1 hour, cells were fixed in 2% paraformaldehyde/PBS at 37°C, permeabilized by adding cold 90% methanol, incubated on ice for 30 minutes, and stained intracellularly with an Alexa Fluor 488–conjugated anti-STAT6(pY641) antibody specific for the tyrosine 641 phosphorylated form of STAT6 (BD Biosciences). Immunofluorescence was analyzed on a FACSCalibur flow cytometer (BD Biosciences), gating on the monocyte population on forward and side scatter.
To detect IL-4– or IL-13–dependent STAT6 phosphorylation in T cells, PBMCs (2 × 106 cells/ml) were incubated in the presence of the relevant cytokine (2 ng/ml) for 1 hour. Cells were surface stained with PE-conjugated anti-CD3 (BD Biosciences), permeabilized with cold 90% methanol, and stained intracellularly with Alexa Fluor 488–conjugated anti-STAT6(pY641) antibody. Percentages of positive cells were determined by gating on live cells.
To evaluate IL-13–dependent induction of CD23 expression, PBMCs (2 × 106 cells/ml) were incubated with increasing concentrations of WT IL-13 or IL-13 R130Q for 48 hours, then assayed by direct immunofluorescence with PE-conjugated anti-CD23 mAb or isotype control (BD Biosciences), gating on the monocyte population on forward and side scatter. In additional experiments, expression of CD23 on monocytes was assessed after a 48-hour incubation with the IL-13 variants in the presence or absence of recombinant soluble IL-13Rα2-Fc chimera (R&D Systems) or normal human IgG (Baxter).
Semiquantitative RT-PCR analysis of ε germ-line and AID transcription.
Normal human PBMCs (3 × 106 cells/ml) were incubated with WT IL-13 or IL-13 R130Q (30–90 pg/ml) in the presence or absence of HC (1 μM; Sigma-Aldrich). After 72 hours, total RNA was extracted using RNAeasy Protect (QIAGEN), and cDNA was reverse transcribed using Omniscript (QIAGEN). Amplification of ε germ-line and control GAPDH transcripts was performed as previously described (67). The complete coding region of AID was amplified as detailed in ref. 68. Amplicons were resolved on 1–1.2% agarose gels and semiquantitated by scanning densitometry. Expression of ε germ-line and AID transcripts was normalized relative to the intensity of GAPDH gene expression.
In vitro IgE production.
PBMCs (3 × 106 cells/ml) from normal nonallergic donors were resuspended in complete medium and stimulated with increasing concentrations of WT IL-13 or IL-13 R130Q in the presence or absence of anti-CD40 mAb 626.1 (1 μg/ml; a kind gift of S.M. Fu, University of Virginia, Charlottesville, Virginia, USA) or HC (1 μM). In selected experiments, control cultures were set up in the presence of recombinant IL-4. Culture supernatants were harvested after 12–14 days and assessed for IgE concentrations by ELISA. To this purpose, 96-well plates (MaxiSorp; Nunc) were coated with purified anti-IgE mAb (2 μg/ml; BD Biosciences — Pharmingen) in 0.1 M carbonate buffer, pH 9.0, overnight at 4°C. Wells were then washed 3 times with PBS-0.05% Tween and blocked with 3% milk-PBS-0.01% azide for 4 hours at room temperature. After extensive washing with PBS-0.05% Tween, dilutions of an IgE standard curve (Serotec) or samples were added to the wells overnight at 4°C. Following extensive washing with PBS-0.05% Tween, a horseradish peroxidase–conjugated goat anti-human IgE polyclonal antibody (1:1000 in 0.1 M carbonate buffer, pH 9.0; BioSource International Inc.) was added to the wells for 4 hours at room temperature. After washing extensively with PBS-0.05% Tween, the reaction was developed with OPD Fast Tabs (Sigma-Aldrich) for 30 minutes at room temperature in the dark. After stopping the reaction with 10% sulphuric acid, OD was read at 492 nm. The lower limit of sensitivity of this assay was 0.5 ng/ml. Control cultures for the evaluation of preformed IgE were set up in the presence of cycloheximide (100 μg/ml; Sigma-Aldrich) as previously described (69). Net IgE synthesis was calculated by subtracting the IgE concentrations detected in cycloheximide-treated cultures from the values detected in untreated cultures.
Statistical analysis.
Two-tailed paired Student’s t test was used to compare responses to WT IL-13 and IL-13 R130Q. The effects of the 2 variants on STAT6 phosphorylation and CD23 induction were also analyzed using a random effects model (42). To calculate EC50 values for induction of STAT6 phosphorylation and CD23 expression by WT IL-13, IL-13 R130Q (corrected values), and IL-13 R130Q (uncorrected values), IL-13 concentration and percentage of maximal response for each experiment were log-transformed, a linear regression equation was generated, and log EC50 values were calculated and compared between groups using a paired Student’s t test. Geometric means (95% confidence intervals) EC50 are reported for each IL-13 group.
Human subjects.
Approval for human studies protocols was obtained from the Institutional Review Board of the University of Arizona. All participants gave written, informed consent.
Supplementary Material
Acknowledgments
We are grateful to the other members of the Functional Genomics Laboratory, the investigators of the Arizona Respiratory Center, Michael Cusanovich, and Michael Kabesch for their insightful comments and advice. We thank John Fitch and Marty Cisneros for technical assistance. This work was supported by NIH grants R01 HL66391 and P50 HL67672, Project 3 (to D. Vercelli).
Footnotes
Raffaella Ghittoni’s present address is: Department of Evolutionary Biology, University of Siena, Siena, Italy.
Frank D. Vladich and Susan M. Brazille contributed equally to this work.
Nonstandard abbreviations used: AID, activation-induced deaminase; HC, hydrocortisone; LD, linkage disequilibrium; SNP, single-nucleotide polymorphism.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Cohn L, Elias JA, Chupp GL. Asthma: mechanisms of disease persistence and progression. Annu. Rev. Immunol. 2004;22:789–815. doi: 10.1146/annurev.immunol.22.012703.104716. [DOI] [PubMed] [Google Scholar]
- 2.Walter DM, et al. Critical role for IL-13 in the development of allergen-induced airway hyperreactivity. J. Immunol. 2001;167:4668–4675. doi: 10.4049/jimmunol.167.8.4668. [DOI] [PubMed] [Google Scholar]
- 3.Wills-Karp M, et al. Interleukin-13: Central mediator of allergic asthma. Science. 1998;282:2258–2261. doi: 10.1126/science.282.5397.2258. [DOI] [PubMed] [Google Scholar]
- 4.Grünig G, et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998;282:2261–2263. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu Z, et al. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J. Clin. Invest. 1999;103:779–788. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee C, et al. Interleukin-13 induces tissue fibrosis by selectively stimulating and activating transforming growth factor β-1. J. Exp. Med. 2001;194:809–821. doi: 10.1084/jem.194.6.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanone S, et al. Overlapping and enzyme-specific contributions of matrix metalloproteinases-9 and -12 in IL-13–induced inflammation and remodeling. J. Clin. Invest. 2002;110:463–474. doi:10.1172/JCI200214136. doi: 10.1172/JCI14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JH, et al. Interleukin-13 induces dramatically different transcriptional programs in three human airway cell types. Am. J. Respir. Cell. Mol. Biol. 2001;25:474–485. doi: 10.1165/ajrcmb.25.4.4522. [DOI] [PubMed] [Google Scholar]
- 9.Zhu Z, et al. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science. 2004;304:1678–1682. doi: 10.1126/science.1095336. [DOI] [PubMed] [Google Scholar]
- 10.Mattes J, et al. Intrinsic defect in T cell production of IL-13 in the absence of both IL-5 and eotaxin precludes the development of eosinophilia and airways hyperreactivity in experimental asthma. J. Exp. Med. 2002;195:1433–1444. doi: 10.1084/jem.20020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shinkai K, Mohrs M, Locksley RM. Helper T cells regulate type-2 innate immunity in vivo. Nature. 2002;420:825–829. doi: 10.1038/nature01202. [DOI] [PubMed] [Google Scholar]
- 12.Yang M, et al. Eotaxin-2 and IL-5 cooperate in the lung to regulate IL-13 production and airway eosinophilia and hyperreactivity. J. Allergy Clin. Immunol. 2003;112:935–943. doi: 10.1016/j.jaci.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Voehringer D, Shinkai K, Locksley RM. Type 2 immunity reflects orchestrated recruitment of cells committed to IL-4 production. Immunity. 2004;20:267–277. doi: 10.1016/s1074-7613(04)00026-3. [DOI] [PubMed] [Google Scholar]
- 14.Punnonen J, et al. Interleukin 13 induces interleukin 4-independent IgG4 and IgE synthesis and CD23 expression by human B cells. Proc. Natl. Acad. Sci. U. S. A. 1993;90:3730–3734. doi: 10.1073/pnas.90.8.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cosentino G, et al. IL-13 downregulates CD14 expression and TNF-α secretion in human monocytes. J. Immunol. 1995;155:3145–3151. [PubMed] [Google Scholar]
- 16.Gould HJ, et al. The biology of IgE and the basis of allergic disease. Annu. Rev. Immunol. 2003;21:579–628. doi: 10.1146/annurev.immunol.21.120601.141103. [DOI] [PubMed] [Google Scholar]
- 17.Williams TJ, Jones CA, Miles EA, Warner JO, Warner JA. Fetal and neonatal IL-13 production during pregnancy and at birth and subsequent development of atopic symptoms. J. Allergy Clin. Immunol. 2000;105:951–959. doi: 10.1067/mai.2000.106211. [DOI] [PubMed] [Google Scholar]
- 18.Ribeiro-do-Couto LM, et al. High IL-13 production by human neonatal T cells: neonate immune system regulator? Eur. J. Immunol. 2001;31:3394–3402. doi: 10.1002/1521-4141(200111)31:11<3394::aid-immu3394>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 19.Martinez FD. Toward asthma prevention--does all that really matters happen before we learn to read? N. Engl. J. Med. 2003;349:1473–1475. doi: 10.1056/NEJMe030041. [DOI] [PubMed] [Google Scholar]
- 20.Cookson WO, Moffatt MF. Genetics of asthma and allergic disease. Hum. Mol. Genet. 2000;9:2359–2364. doi: 10.1093/hmg/9.16.2359. [DOI] [PubMed] [Google Scholar]
- 21.Vercelli D. Genetic polymorphism in allergy and asthma. Curr. Opin. Immunol. 2003;15:609–613. doi: 10.1016/j.coi.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 22.Graves PE, et al. A cluster of seven tightly linked polymorphisms in the IL-13 gene is associated with total serum IgE levels in three populations of white children. J. Allergy Clin. Immunol. 2000;105:506–513. doi: 10.1067/mai.2000.104940. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, et al. An IL13 coding region variant is associated with a high total serum IgE level and atopic dermatitis in the German multicenter atopy study (MAS-90) J. Allergy Clin. Immunol. 2000;106:167–170. doi: 10.1067/mai.2000.107935. [DOI] [PubMed] [Google Scholar]
- 24.Wang M, et al. A common IL-13 Arg130Gln single nucleotide polymorphism among Chinese atopy patients with allergic rhinitis. Hum. Genet. 2003;113:387–390. doi: 10.1007/s00439-003-1001-x. [DOI] [PubMed] [Google Scholar]
- 25.Heinzmann A, et al. Association study of the IL13 variant Arg110Gln in atopic diseases and juvenile idiopathic arthritis. J. Allergy Clin. Immunol. 2003;112:735–739. doi: 10.1016/s0091-6749(03)01887-6. [DOI] [PubMed] [Google Scholar]
- 26.Heinzmann A, et al. Genetic variants of IL-13 signalling and human asthma and atopy. Hum. Mol. Genet. 2000;9:549–559. doi: 10.1093/hmg/9.4.549. [DOI] [PubMed] [Google Scholar]
- 27.He JQ, et al. Genetic variants of the IL13 and IL4 genes and atopic diseases in at-risk children. Genes Immun. 2003;4:385–389. doi: 10.1038/sj.gene.6363985. [DOI] [PubMed] [Google Scholar]
- 28.Tsunemi Y, et al. Interleukin-13 gene polymorphism G4257A is associated with atopic dermatitis in Japanese patients. J. Dermatol. Sci. 2002;30:100–107. doi: 10.1016/s0923-1811(02)00065-8. [DOI] [PubMed] [Google Scholar]
- 29.DeMeo D, et al. Univariate and multivariate family-based association analysis of the IL-13 ARG130GLN polymorphism in the Childhood Asthma Management Program. Genet. Epidemiol. 2002;23:335–348. doi: 10.1002/gepi.10182. [DOI] [PubMed] [Google Scholar]
- 30.van der Pouw Kraan TC, et al. An IL-13 promoter polymorphism associated with increased risk of allergic asthma. Genes Immun. 1999;1:61–65. doi: 10.1038/sj.gene.6363630. [DOI] [PubMed] [Google Scholar]
- 31.Howard TD, et al. Identification and association of polymorphisms in the Interleukin-13 gene with asthma and atopy in a Dutch population. Am. J. Respir. Cell. Mol. Biol. 2001;25:377–384. doi: 10.1165/ajrcmb.25.3.4483. [DOI] [PubMed] [Google Scholar]
- 32.Liu X, et al. Associations between total serum IgE levels and the 6 potentially functional variants within the genes IL4, IL13, and IL4RA in German children: the German Multicenter Atopy Study. J. Allergy Clin. Immunol. 2003;112:382–388. doi: 10.1067/mai.2003.1635. [DOI] [PubMed] [Google Scholar]
- 33.Hummelshoj T, et al. Association between an interleukin-13 promoter polymorphism and atopy. Eur. J. Immunogenet. 2003;30:355–359. doi: 10.1046/j.1365-2370.2003.00416.x. [DOI] [PubMed] [Google Scholar]
- 34.Howard TD, et al. Gene-gene interaction in asthma: IL4RA and IL13 in a Dutch population with asthma. Am. J. Hum. Genet. 2002;70:230–236. doi: 10.1086/338242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffjan S, Nicolae D, Ober C. Association studies for asthma and atopic diseases: a comprehensive review of the literature. Respir. Res. [serial online] 2003;4:14. http://respiratory–research. doi: 10.1186/1465-9921-4-14. com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nat. Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 37.Arima K, et al. Upregulation of IL-13 concentration in vivo by the IL13 variant associated with bronchial asthma. J. Allergy Clin. Immunol. 2002;109:980–987. doi: 10.1067/mai.2002.124656. [DOI] [PubMed] [Google Scholar]
- 38.Chen W, Ericksen MB, Levin LS, Khurana Hershey GK. Functional effect of the R110Q IL13 genetic variant alone and in combination with IL4RA genetic variants. J. Allergy Clin. Immunol. 2004;114:553–560. doi: 10.1016/j.jaci.2004.04.044. [DOI] [PubMed] [Google Scholar]
- 39.Madhankumar AB, Mintz A, Debinski W. Alanine-scanning mutagenesis of alpha-helix D segment of interleukin-13 reveals new functionally important residues of the cytokine. J. Biol. Chem. 2002;277:43194–43205. doi: 10.1074/jbc.M205047200. [DOI] [PubMed] [Google Scholar]
- 40.Minty A, et al. Interleukin-13 is a new human lymphokine regulating inflammatory and immune responses. Nature. 1993;362:248–250. doi: 10.1038/362248a0. [DOI] [PubMed] [Google Scholar]
- 41.Takeda K, Kamanaka M, Tanaka T, Kishimoto T, Akira S. Impaired IL-13-mediated functions of macrophages in STAT6-deficient mice. J. Immunol. 1996;157:3220–3222. [PubMed] [Google Scholar]
- 42.Brown, H., and Prescott, R. 1999. Applied mixed models in medicine. John Wiley & Sons Ltd. Chichester, United Kingdom/New York, New York, USA. 442 pp.
- 43.Vercelli D, et al. Human recombinant interleukin 4 induces FcεR2/CD23 on normal human monocytes. J. Exp. Med. 1988;167:1406–1416. doi: 10.1084/jem.167.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Köhler I, Rieber EP. Allergy-associated Iε and Fcε receptor II (CD23b) genes activated via binding of an interleukin-4-induced transcription factor to a novel responsive element. Eur. J. Immunol. 1993;23:3066–3071. doi: 10.1002/eji.1830231204. [DOI] [PubMed] [Google Scholar]
- 45.Vercelli D. IgE and its regulators. Curr. Opin. Allergy Clin. Immunol. 2001;1:61–65. doi: 10.1097/01.all.0000010986.58020.8d. [DOI] [PubMed] [Google Scholar]
- 46.Jabara HH, Brodeur SR, Geha RS. Glucocorticoids upregulate CD40 ligand expression and induce CD40L-dependent immunoglobulin isotype switching. J. Clin. Invest. 2001;107:371–378. doi: 10.1172/JCI10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hershey GK. IL-13 receptors and signaling pathways: an evolving web [review] J. Allergy Clin. Immunol. 2003;111:677–690. doi: 10.1067/mai.2003.1333. [DOI] [PubMed] [Google Scholar]
- 48.Kioi M, Kawakami K, Puri RK. Mechanism of action of interleukin-13 antagonist (IL-13E13K) in cells expressing various types of IL-4R. Cell. Immunol. 2004;229:41–51. doi: 10.1016/j.cellimm.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 49.Eisenmesser EZ, Horita DA, Altieri AS, Byrd RA. Solution structure of interleukin-13 and insights into receptor engagement. J. Mol. Biol. 2001;310:231–241. doi: 10.1006/jmbi.2001.4765. [DOI] [PubMed] [Google Scholar]
- 50.Carlson CS, Eberle MA, Kruglyak L, Nickerson DA. Mapping complex disease loci in whole-genome association studies. Nature. 2004;429:446–452. doi: 10.1038/nature02623. [DOI] [PubMed] [Google Scholar]
- 51.Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease [review] Nat. Genet. 2003;33(Suppl.):228–237. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 52.Donaldson DD, et al. The murine IL-13 receptor α2: Molecular cloning, characterization and comparison with murine IL-13 receptor α1. J. Immunol. 1998;161:2317–2324. [PubMed] [Google Scholar]
- 53.Wood N, et al. Enhanced Interleukin-13 responses in mice lacking IL-13 receptor α2. J. Exp. Med. 2003;197:703–709. doi: 10.1084/jem.20020906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng T, et al. Cytokine regulation of IL-13Rα2 and IL-13Rα1 in vivo and in vitro. J. Allergy Clin. Immunol. 2003;111:720–728. doi: 10.1067/mai.2003.1383. [DOI] [PubMed] [Google Scholar]
- 55.Oshima Y, Joshi BH, Puri RK. Conversion of interleukin-13 into a high affinity agonist by a single amino acid substitution. J. Biol. Chem. 2000;275:14375–14380. doi: 10.1074/jbc.275.19.14375. [DOI] [PubMed] [Google Scholar]
- 56.LeVan TD, et al. A common single nucleotide polymorphism in the CD14 promoter decreases the affinity of Sp protein binding and enhances transcriptional activity. J. Immunol. 2001;167:5838–5844. doi: 10.4049/jimmunol.167.10.5838. [DOI] [PubMed] [Google Scholar]
- 57.Schweiger A, et al. Differences in proliferation of the hematopoietic cell line TF-1 and cytokine production by peripheral blood leukocytes induced by 2 naturally occurring forms of human IL-3. J. Allergy Clin. Immunol. 2001;107:505–510. doi: 10.1067/mai.2001.113239. [DOI] [PubMed] [Google Scholar]
- 58.Tokuhiro S, et al. An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat. Genet. 2003;35:341–348. doi: 10.1038/ng1267. [DOI] [PubMed] [Google Scholar]
- 59.Silverman ES, et al. Transforming growth factor-β1 promoter polymorphism C-509T is associated with asthma. Am. J. Respir. Crit. Care Med. 2004;169:214–219. doi: 10.1164/rccm.200307-973OC. [DOI] [PubMed] [Google Scholar]
- 60.Ozaki K, et al. Functional variation in LGALS2 confers risk of myocardial infarction and regulates lymphotoxin-α secretion in vitro. Nature. 2004;429:72–75. doi: 10.1038/nature02502. [DOI] [PubMed] [Google Scholar]
- 61.Knight JC, Keating BJ, Kwiatkowski DP. Allele-specific repression of lymphotoxin-α by activated B cell factor-1. Nat. Genet. 2004;36:394–399. doi: 10.1038/ng1331. [DOI] [PubMed] [Google Scholar]
- 62.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat. Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 63.Tepper RI, et al. IL-4 induces allergic-like inflammatory disease and alters T cell development in transgenic mice. Cell. 1990;62:457–467. doi: 10.1016/0092-8674(90)90011-3. [DOI] [PubMed] [Google Scholar]
- 64.Dent LA, Strath M, Mellor AL, Sanderson CJ. Eosinophilia in transgenic mice expressing interleukin 5. J. Exp. Med. 1990;172:1425–1431. doi: 10.1084/jem.172.5.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jabara HH, Ahern DJ, Vercelli D, Geha RS. Hydrocortisone and IL-4 induce IgE isotype switching in human B cells. J. Immunol. 1991;147:1557–1560. [PubMed] [Google Scholar]
- 66.Moy FJ, Diblasio E, Wilhelm J, Powers R. Solution structure of human IL-13 and implication for receptor binding. J. Mol. Biol. 2001;310:219–230. doi: 10.1006/jmbi.2001.4764. [DOI] [PubMed] [Google Scholar]
- 67.Monticelli S, Ghittoni R, Kabesch M, Vercelli D. Myb proteins repress human ε germline transcription by inhibiting STAT6-dependent promoter activation. Mol. Immunol. 2002;38:1129–1138. doi: 10.1016/s0161-5890(02)00037-8. [DOI] [PubMed] [Google Scholar]
- 68.Albesiano E, et al. Activation-induced cytidine deaminase in chronic lymphocytic leukemia B cells: expression as multiple forms in a dynamic, variably sized fraction of the clone. Blood. 2003;102:3333–3339. doi: 10.1182/blood-2003-05-1585. [DOI] [PubMed] [Google Scholar]
- 69.Jabara HH, Fu SM, Geha RS, Vercelli D. CD40 and IgE: Synergism between anti-CD40 mAb and IL-4 in the induction of IgE synthesis by highly purified human B cells. J. Exp. Med. 1990;172:1861–1864. doi: 10.1084/jem.172.6.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.