SUMMARY
Endonuclease G (EndoG) is a mitochondrial protein that is released from mitochondria and relocated into the nucleus to promote chromosomal DNA fragmentation during apoptosis. Here, we show that oxidative stress causes cell death defects in C. elegans through an EndoG-mediated cell death pathway. In response to high ROS levels, homodimeric CPS-6—the C. elegans homolog of EndoG—is dissociated into monomers with diminished nuclease activity. Conversely, the nuclease activity of CPS-6 is enhanced and its dimeric structure is stabilized by its interaction with the worm AIF homolog, WAH-1, which shifts to disulfide cross-linked dimers under high ROS levels. CPS-6 thus plays an unexpected role in acting as a ROS sensor to regulate the life and death of cells. Modulation of the EndoG dimer conformation could present an avenue for prevention and treatment of diseases resulting from oxidative stress.
Keywords: Endonuclease G, apoptosis, nuclease, mitochondria dysfunction, crystal structure
INTRODUCTION
Endonuclease G (EndoG) is a mitochondrial nuclease that is released from the mitochondria during apoptosis and translocated into the nucleus to promote chromosomal DNA fragmentation (Büttner et al., 2007; Li et al., 2001; Parrish et al., 2001). In C. elegans, the EndoG homolog CPS-6 is required for apoptotic DNA degradation and normal progression of cell death (Parrish et al., 2001; Parrish and Xue, 2003, 2006). Recently, EndoG activity has been linked to neuronal cell loss in Parkinson’s disease (Büttner et al., 2013) and apoptosis in cancer cells (Basnakiana et al., 2006). In addition to its role in cell death, EndoG is also important for normal cellular proliferation (Büttner et al., 2007; Huang et al., 2006). The nuclease activity of EndoG is suggested to be involved in mitochondrial DNA biogenesis, such as DNA replication and recombination (Cote and Ruiz-Carrillo, 1993; Huang et al., 2006; Zan et al., 2011). EndoG-deficient mice have elevated levels of reactive oxygen species (ROS) due to a decreased electron transport rate and develop cardiac hypertrophy, suggesting a direct link between EndoG and ROS in mitochondrial dysfunction (McDermott-Roe et al., 2011).
In C. elegans, the WAH-1 protein—the homolog of human apoptosis-inducing factor (AIF)—interacts with CPS-6 to enhance its nuclease activity and its ability to promote apoptotic DNA fragmentation (Parrish and Xue, 2003; Wang et al., 2007; Wang et al., 2002). Similarly, human EndoG interacts with and is stimulated by human AIF, which is located in the mitochondrial intermembrane space and also translocates from the mitochondria to the nucleus during apoptosis (Kalinowska et al., 2005; Susin et al., 1999). AIF is an oxidoreductase with two different conformations: a reduced dimeric form (FAD- and NADH-bound) and an oxidized monomeric form (FAD-bound) (Ferreira, 2014; Miramar et al., 2001; Sevrioukova; Ye et al., 2002). Like EndoG, mice with reduced AIF activity have increased oxidative stress and ROS-induced damage in neurons, and mice lacking AIF in their muscle cells develop severe dilated cardiomyopathy and skeletal muscle atrophy, suggesting that AIF regulates mitochondrial respiration and homeostasis (Joza et al., 2005; Klein et al., 2002). However, unlike AIF, WAH-1 does not appear to contain any binding sites for nicotinamide, implying that WAH-1 may have a distinct redox role as an oxidoreductase.
To decipher the link between EndoG and ROS, we investigated the biochemical and structural properties of CPS-6 under different redox conditions. We found that under oxidized conditions, CPS-6 dimers dissociate into monomers with a diminished endonuclease activity. In contrast, WAH-1 is a monomer in a reduced environment and shifts to a disulfide cross-linked dimer in response to oxidized conditions. Our biochemical and functional analyses suggest that WAH-1 acts as an antioxidant and an activator to prevent dimeric CPS-6 dissociation and to preserve maximal CPS-6 activity under oxidative stress. Thus, our study provides molecular and mechanistic insights into the intricate relationships among high ROS levels and the activities of CPS-6 and WAH-1 and reveals the important roles of these two proteins in mediating appropriate life versus death responses to oxidative stress.
Results
Oxidized conditions compromise cell death in C. elegans by inhibiting the CPS-6 pathway
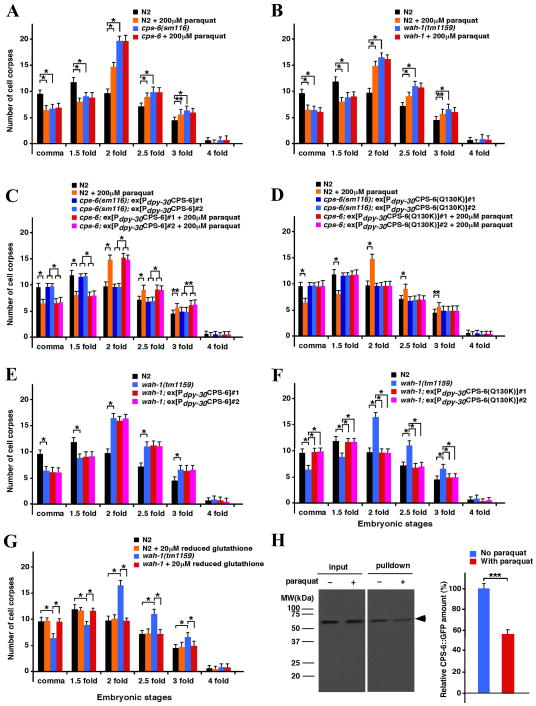
To examine the effects of oxidation on programmed cell death in C. elegans, we conducted a time-course analysis of C. elegans embryonic cell death in the presence of increasing concentrations of paraquat—a compound that generates superoxide and H2O2. Treatment of wild-type N2 animals with low concentrations of paraquat (20 μM, 2 μM and 200 nM) did not cause an obvious cell death defect, whereas treatment of N2 animals with 200 μM paraquat resulted in delayed appearance of apoptotic cell corpses (‘delay-of-cell-death defect’), similar to that observed in the cps-6(sm116) loss-of-function mutant (Figure 1A and Figure S1A). To examine if paraquat affects the activity of CPS-6, we treated cps-6(sm116) animals with 200 μM paraquat and found that, at this concentration, paraquat did not enhance the delay-of-cell-death defect of the cps-6(sm116) mutant (Figure 1A), suggesting that paraquat affects cell death by targeting either a component in the CPS-6 cell death pathway or CPS-6 itself. Consistent with the role of WAH-1 as the activator of CPS-6, a loss-of-function mutation (tm1159) in wah-1 causes a delay-of-cell-death defect similar to that of the cps-6(sm116) mutant, and interestingly, this delay-of-cell-death defect of wah-1(tm1159) was not enhanced by treatment with 200 μM paraquat (Figure 1B), confirming that paraquat affects cell death by targeting a component in the CPS-6 pathway.
Figure 1. Time-course analysis of C. elegans embryonic cell corpses under oxidative or reduced conditions.
(A–G) Embryonic cell corpses were scored from the indicated strains in the presence of the oxidative reagent paraquat or reduced glutathione. L4-stage animals were exposed to 200 μM paraquat or 20 μM reduced glutathione on NGM agar plates, and their progenies were analyzed as described. Stages of embryos examined were comma, 1.5-fold, 2-fold, 2.5-fold, 3-fold and 4-fold. The y-axis represents the mean of cell corpses scored in embryos (n=15), and error bars represent the standard deviation (SD). (H) The co-immunoprecipitation assay shows that CPS-6::GFP associated with CPS-6::FLAG was reduced following treatment with paraquat.
CPS-6 dimers dissociate into monomers in oxidized conditions
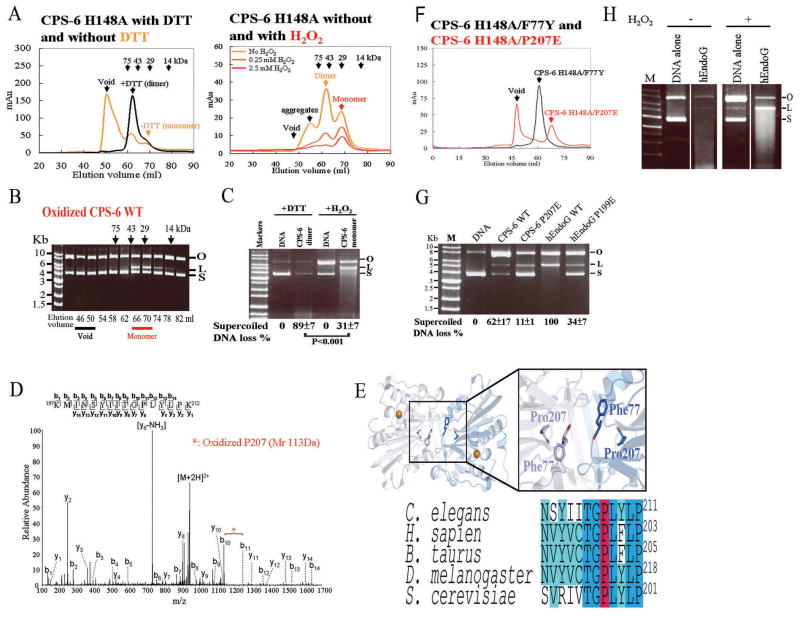
To determine if the nuclease activity of CPS-6 is affected by the redox potential, we purified from E. coli His-tagged CPS-6 protein (residues 63–305) without its N-terminal mitochondrial targeting signal. Due to the difficulty of expressing a large quantity of the wild-type CPS-6 protein, which is highly toxic to E. coli, we used the nuclease-defective CPS-6(H148A) mutant with a mutated catalytic residue for biochemical analysis. The crystal structure of CPS-6(H148A) shows that it forms a homodimer (Lin et al., 2012). Affinity-purified CPS-6(H148A) proteins with or without the reducing reagent dithiothreitol (DTT, 2.5 mM) were applied to a Superdex 75 (S-75) size exclusion column. In the presence of DTT, recombinant CPS-6(H148A) was eluted at 62 ml as a homodimer (Figure 2A, left panel). By contrast, in the absence of DTT, the CPS-6(H148A) dimers partially dissociated into monomers, eluted at 68 ml, alone with the high-order oligomeric aggregates eluted in the void peak (light orange line in Figure 2A, left).
Figure 2. Dimeric CPS-6 dissociates into monomers with diminished nuclease activity under oxidized conditions.
(A) CPS-6(H148A) was eluted as a dimer (~62 ml) by size exclusion chromatography (Superdex 75) in the presence of 2.5 mM DTT, but was eluted as monomers (~68 ml), dimers and high-order oligomers (void) in the absence of DTT for 32 hours (in yellow). In the right panel, CPS-6(H148A) was completely dissociated into monomers in the presence of 2.5 mM H2O2. (B) Oxidized monomeric CPS-6 (fractions 66–70 ml) digested plasmid supercoiled DNA. (C) Reduced dimeric CPS-6 (fractions 58-62 ml) cleaved supercoiled plasmid DNA more efficiently (89 ± 7% vs. 31 ± 7%) than the oxidized monomeric CPS-6 (fractions 66–70 ml). (D) LC-MS/MS analysis indicated that Pro207 was oxidized with an increased mass of 16 Da within the tryptic peptide CPS-6-Lys197-Lys212. (E) The structure of the CPS-6 dimer with the two highlighted interfacial residues, F77 and P207. The bottom panel shows the sequence alignment of the EndoG protein sequences. (F) CPS-6(H148A/F77Y) and CPS-6(H148A/P207E) were eluted by S-75 chromatography as dimers and monomers, respectively, in the presence of the reducing agent. (G) In the presence of H2O2, the nuclease activity of hEndoG decreased in the plasmid nicking assays. (H) Wild-type CPS-6 and hEndoG digested DNA more efficiently than their respective oxidizing mimicking monomers, CPS-6(P207E) and hEndoG(P199E).
To test the stability of the oxidized monomers under oxidative conditions, the eluted dimeric and monomeric peak fractions (fractions 60–70 ml) were re-applied successively to size exclusion chromatography (S-75 column) in three different H2O2 concentrations, 0, 0.25 mM, and 2.5 mM (Figure 2A, right). Oxidized CPS-6 remained a stable monomer in the presence of the oxidizing agent, but CPS-6 dimers completely dissociated into monomers in the presence of 2.5 mM H2O2. CPS-6 retained its integrity and was not degraded by the incubation with H2O2 as shown by the SDS-PAGE for each fraction (Figure S2A). The molecular weight of the oxidized CPS-6 was increased by 115 Dalton comparing to the reduced CPS-6 as measured by mass spectrometry (Figure S2B). The molecular masses of the reduced homodimer and the oxidized monomer were estimated to be 57 kDa and 37 kDa, respectively, according to the protein markers used for S-75 calibration (the calculated molecular mass of CPS-6 monomer is 28.6 kDa). Taken together, these results suggest that CPS-6 dimers can be oxidized and shifted monomers in oxidized conditions.
To determine if the nuclease activity of CPS-6 is affected by the redox conditions, a small amount (0.2 mg) of the recombinant wild-type CPS-6 was purified for enzymatic assays and incubated with 2.5 mM DTT or 2.5 mM H2O2 at 4°C for 16 hours to generate the reduced or oxidized forms of CPS-6, respectively. The monomer fractions (elution volume: 66–70 ml) of the oxidized wild-type CPS-6 eluted from size exclusion chromatography (S-75) could cleave supercoiled plasmid DNA, demonstrating that the monomeric CPS-6 had some nuclease activity under oxidizing conditions (Figure 2B). To compare the nuclease activity of the reduced and oxidized CPS-6, purified DTT-treated reduced dimeric CPS-6 (collected from fractions 58–62 ml) and purified H2O2-treated oxidized monomeric CPS-6 (collected from fractions 66–70 ml) were incubated with plasmid DNA. Since the plasmid DNA was partially nicked by H2O2, which converted supercoil plasmid DNA (labeled as S) to nicked open circular form (labeled as O), the DNA control lanes in the presence of H2O2 and DTT appeared differently in Figure 2C. The nuclease activity of CPS-6 was determined by deducting the background DNA cleavage in the presence of H2O2 or DTT. The dimeric CPS-6 efficiently cleaved the supercoiled plasmid DNA (89 ± 7% supercoiled DNA digested) under the reduced conditions, whereas the monomeric CPS-6 only digested 31 ± 7% supercoiled DNA (Figure 2C) under the oxidized conditions. Taken together, these results suggest that CPS-6 dimers shift to monomers in oxidized conditions and that CPS-6 monomers have a diminished nuclease activity compared with that of CPS-6 dimers.
The interfacial residues of CPS-6 dimer are oxidized, disrupting dimer formation
It is possible that the interfacial residues of CPS-6 dimers were oxidized, leading to disassociation of CPS-6 dimers. To identify the residues in CPS-6 that are oxidized upon oxidative treatments, both reduced and oxidized CPS-6 proteins were trypsin-digested and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). CPS-6 tryptic fragments located at the dimeric interfacial surface exhibited different molecular masses in oxidized and reduced conditions (Table S1), and the mass spectroscopy results are shown in Figure 2D and Figure S2C. Two dimer-interfacial residues (F77 and P207) and one non-interfacial residue (M198) were oxidized, resulting in increased molecular masses of 16 Da each for peptides His72-Arg81 and Lys197-Lys212, respectively (Table S1 and Figure 2D). These results suggest that CPS-6 is modified by oxidation at two dimer-interfacial residues, which may cause dimer dissociation (see Figure 2E).
We generated the CPS-6(H148A/F77Y) and CPS-6(H148A/P207E) mutants to mimic the oxidized states of F77 and P207 residues in CPS-6, since it has previously been shown that phenylalanine can be oxidized and converted to tyrosine (Molnar et al., 2005) and that proline can be oxidized and converted to glutamic and aminoadipic semialdehydes (Requena et al., 2001). In the absence of H2O2, CPS-6(H148A/F77Y) was eluted as dimers, whereas CPS-6(H148A/P207E) was eluted as monomers, suggesting that the P207E substitution in CPS-6(H148A/P207E) causes CPS-6 to dissociate into monomers and that an oxidized P207 residue promotes dissociation of CPS-6 dimers into monomers.
Because the interfacial proline residue is highly conserved among EndoG proteins in different species (Figure 2E), oxidative treatment of human EndoG (hEndoG) may also cause oxidation of the same proline residue (P199). We incubated the reduced and oxidized forms of hEndoG with the supercoiled plasmid DNA and found that the plasmid could be cleaved into both open circular (O) and linear (L) forms (Figure 2H). However, the endonuclease activity of H2O2-treated hEndoG was diminished significantly, suggesting that the diminished endonuclease activity under oxidized conditions is a general and conserved phenomenon for both hEndoG and CPS-6 (Figure 2H). Moreover, the nuclease activities of both CPS-6(P207E) and hEndoG(P199E) were significantly diminished compared with respective wild-type proteins (Figure 2G). These results support our hypothesis that the conserved proline residue at the dimeric interface of CPS-6 and EndoG plays an important role in modulating dimer stability and enzyme activity. We conclude that the dimeric interfacial residues of CPS-6 can be oxidized to shift CPS-6 from dimers to monomers with a diminished nuclease activity under oxidative stress.
WAH-1 monomers shift to dimers in response to oxidized conditions
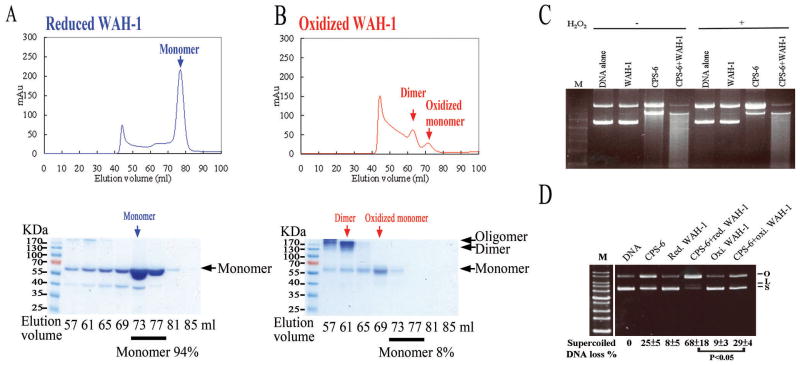
WAH-1 has been shown to enhance the nuclease activity of CPS-6 (Wang et al., 2002), but how WAH-1 stimulates CPS-6 activity is unknown. To examine how WAH-1 affects the nuclease activity of CPS-6 under different redox conditions, we purified His-tagged WAH-1 (residues 214 to 700) devoid of the N-terminal mitochondrial targeting signal and the disordered regions (residues 1 to 213). Affinity-purified WAH-1 was pre-treated with either reducing (2.5 mM DTT) or oxidizing (2.5 mM H2O2) reagents for 32 hours and applied to a Superdex 200 (S-200) size exclusion column. Following DTT (reducing) treatment, WAH-1 was eluted as a monomer with an estimated molecular mass of 60 kDa (Figure 3A), which is close to the calculated molecular mass of the WAH-1 monomer (53.6 kDa). After oxidizing treatment with 2.5 mM H2O2, WAH-1 eluted in two major peaks, corresponding to a monomeric and a dimeric species (Figure 3B). Protein samples collected from different fractions of our S-200 chromatography were further analyzed by SDS-PAGE and visualized by Coomassie blue staining in the absence of a reducing agent. Monomers comprised approximately 94% of the reduced WAH-1 samples (Figure 3A), but only 8% of the oxidized WAH-1 samples (Figure 3B). Moreover, proteins from the 57 and 61 ml fractions of oxidized WAH-1 migrated with a protein size of over 130 kDa (as determined by SDS-PAGE), indicative of the formation of covalently-linked WAH-1 dimers or oligomers.
Figure 3. WAH-1 is a monomer under reduced conditions and a dimer under oxidized conditions.
(A) WAH-1 was eluted as a monomer (60 kDa) in Superdex 200 (S-200) size exclusion chromatography in reduced conditions. The chromatographic fractions from the S-200 column were analyzed by SDS-PAGE (bottom panel). (B) WAH-1 was eluted in two peaks (oxidized dimer and monomer) in the presence of 2.5 mM H2O2. (C) The nuclease activity of CPS-6 was reduced in the oxidized condition, but was enhanced by WAH-1. (D) The reduced monomeric form of WAH-1 significantly enhanced the nuclease activity of oxidized CPS-6 in plasmid DNA nicking assays. The mean percentages with standard errors shown at the bottom of the gel were calculated from three independent experiments.
We next considered if WAH-1 stimulates the activity of CPS-6 under different redox conditions. Plasmid-nicking assays using CPS-6 were performed with or without reduced WAH-1 (incubated with 2.5 mM DTT) or oxidized WAH-1 (incubated with 2.5 mM H2O2) under oxidative conditions. The nuclease activity of CPS-6 was greatly diminished in the presence of H2O2, but addition of WAH-1 enhanced the nuclease activity of CPS-6 (Figure 3C). We then examined whether the oxidized dimeric WAH-1 (62 ml fraction; Figure 3B) or the reduced monomeric WAH-1 (77 ml fraction) could stimulate the nuclease activity of CPS-6. Plasmid DNA nicking assays demonstrated that 68% of supercoiled DNA was digested by CPS-6 in the presence of the reduced monomeric WAH-1, whereas only 29% of supercoiled DNA was digested by CPS-6 in the presence of oxidized dimeric WAH-1 (Figure 3D) under oxidative conditions. Taken together, these results suggest that under oxidative conditions, the nuclease activity of CPS-6 is diminished but can be enhanced by the reduced monomeric WAH-1.
Oxidized dimeric WAH-1 may be formed by non-covalent physical association of WAH-1 monomers or by disulfide bonds between cysteine residues of two WAH-1 monomers. To identify the cysteine residues potentially responsible for the formation of dimeric WAH-1, we digested reduced and oxidized WAH-1 proteins using thermolysin and analyzed the samples by nano liquid chromatography-nano electrospray ionization-tandem mass spectrometry (nanoLC–nanoESI-MS/MS). The MS/MS peaks were assigned by MassMatrix—an automated search and match program—which scored the overall peak assignment and mass matches with a probability-based algorithm (Xu and Freitas, 2007). The thermolysin fragments from oxidized WAH-1 containing predominantly disulfide bond-forming residues are summarized in Tables S2 and S3, and Figure S3. The spectrometry data revealed that under oxidized conditions, the C345 residues of both WAH-1 proteins in the dimer formed inter-molecular disulfide bonds. In addition, C345 formed inter- or intra-molecular disulfide bonds with either C240 or C535, suggesting the dynamic nature of the C345 residue as a key redox determinant in WAH-1. In summary, these data show that WAH-1 is mainly a monomer in reduced conditions, but shifts to a disulfide cross-linked dimer in oxidized conditions.
WAH-1 enhances the stability and nuclease activity of dimeric CPS-6
To investigate how WAH-1 affects the conformation and activity of CPS-6 in solution, S-75 size exclusion chromatography was employed to monitor the ratio of the different forms of CPS-6(H148A) proteins in the presence of H2O2 and WAH-1. Under reduced conditions, we found 31% monomer and 69% dimer of CPS-6(H148A) (Figure S4A, upper panel). Under oxidized conditions, CPS-6(H148A) was primarily in the monomeric form, with 16% dimer and 84% monomer (Figure S4A, bottom panel). Addition of reduced monomeric WAH-1 to CPS-6 in oxidized conditions substantially increased the amount of CPS-6 dimer from 16% to 40% (Figure S4B). These results suggest that reduced monomeric WAH-1 stabilizes the dimeric conformation of CPS-6, leading to enhanced endonuclease activity of CPS-6.
It has already been shown that WAH-1 directly interacts with CPS-6 to stimulate its endonuclease activity (Wang et al., 2002). CPS-6 has two intrinsic fluorophore residues, W99 and W184, that allow monitoring of protein-protein binding events. Increased fluorescence emission (λmax=347 nm) can be detected upon an interaction between CPS-6 and WAH-1. We detected an increased emission resulting from interactions between CPS-6 and WAH-1 in the fluorescence spectra compared with those of WAH-1 and CPS-6(H148A) proteins alone (Figure S4C). A dissociation constant (Kd) of 69.8 ± 2.8 nM was estimated between wild-type WAH-1 and CPS-6(H148A), and CPS-6 formed a one-to-one complex with WAH-1 as estimated from the binding curve (Figures S4D and S4E). We further noticed that CPS-6 and WAH-1 eluted as a complex in the size exclusion chromatography in the absence of DTT. This complex had a molecular mass of about 200 kDa as measured by DLS, suggesting that one CPS-6 dimer was interacting with two WAH-1 monomers (Figure S4F). Moreover, WAH-1 enhanced the nuclease activity of purified dimeric CPS-6 proteins, but not that of oxidized monomeric CPS-6 proteins (Figure S4G), indicating that WAH-1 interacts with dimeric CPS-6 to enhance its activity. Taken together, these results suggest that WAH-1 directly interacts with CPS-6 to form a hetero-tetramer that stabilizes the dimeric conformation of CPS-6 and, thus, stimulates its endonuclease activity.
A CPS-6(Q130K) mutation results in a more stable dimer with stronger nuclease activity
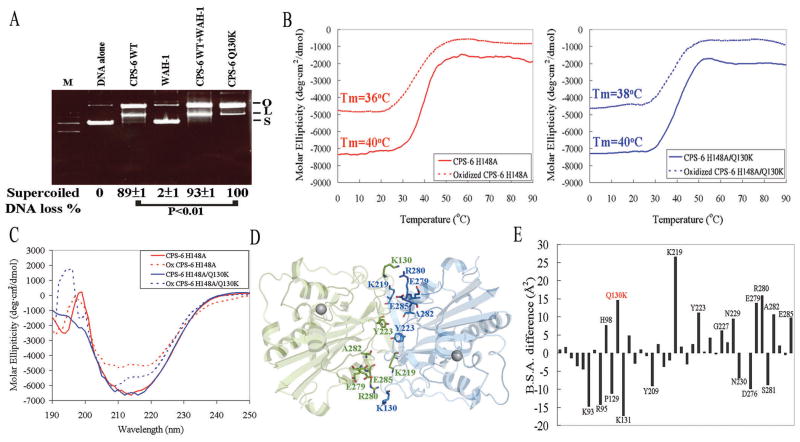
We have shown that CPS-6 was in equilibrium both in dimeric and monomeric conformations, but that dimeric CPS-6 digested DNA more efficiently. To corroborate our hypothesis that the more stable CPS-6 dimer has greater nuclease activity, we mutated one of the CPS-6 dimer interface residues, Q130K, and found that this CPS-6 mutant digested plasmid DNA more efficiently (100%) than the wild-type CPS-6 (89 ± 1%) (Figure 4A, P < 0.01). Because the Q130K mutation may stabilize CPS-6 dimers, we measured the melting point temperatures (Tm) of CPS-6(H148A) and CPS-6(H148A/Q130K) using circular dichroism (CD) at 215 nm in both the reduced and oxidized conditions. We found that although the Tm (40ºC) was identical under the reduced condition for both proteins, CPS-6(H148A/Q130K) had a slightly higher Tm of 38ºC compared to 36ºC for CPS-6(H148A) in the oxidized condition (Figure 4B). In addition, greater CD signal loss at a wavelength of 215 nm was observed for CPS-6(H148A) than for CPS-6(H148A/Q130K) following addition of the oxidative reagent (Figure 4C), suggesting that the secondary structure of CPS-6(H148A/Q130K) was more resistant to oxidants that can destabilize the dimeric protein conformation.
Figure 4. Crystal structural study reveals that CPS-6(Q130K) is a more stable dimer with greater nuclease activity.
(A) CPS-6(Q130K) has greater nuclease activity than wild-type CPS-6. (B) The thermal melting points (Tm) measured by circular dichroism (CD) at 215 nm for CPS-6(H148A) and CPS-6(H148A/Q130K) with or without 2.5 mM H2O2. (C) The CD spectra of CPS-6 mutants show a larger decrease at 215 nm for CPS-6(H148A) than for CPS-6(H148A/Q130K) with the addition of H2O2. (D) The crystal structure of CPS-6(H148A/Q130K) shows that several residues, including H98, K130, Y223, K219, E279, R280, A282 and E285 are located in the dimeric interface. (E) The X-ray crystal structure of CPS-6(H148A/Q130K) reveals a greater buried surface area (B.S.A.) for 39 interfacial residues compared to that of CPS-6(H148A) (PDB:3S5B).
To examine the molecular and structural basis by which the Q130K mutation confers increased protein stability, we determined the crystal structure of the CPS-6(H148AQ/130K) mutant at a resolution of 2.8 Å (Figure 4D). The diffraction and structural refinement statistics are listed in Table S4. The crystal structure of CPS-6(H148A/Q130K) is almost identical to that of CPS-6(H148A) (Lin et al., 2012), with an average rmsd of 0.31 Å for 452 superimposed Cα–atoms. However, closer examination revealed increased buried dimeric interface areas of 1445.8 Å2 for CPS-6(H148A/Q130K) compared to 1392.2 Å2 for the CPS-6(H148A) dimer (PDB entry: 3S5B). The buried surface areas (B.S.A.) for 39 interfacial residues in CPS-6(H148A/Q130K) were larger compared to those of CPS-6(H148A). Of these 39 interfacial residues, H98, K130 (mutated from Q130 in wild type CPS-6), K219, Y223, E279, R280, A282 and E285 each had an increase of more than 5.0 Å2 (Figure 4E). Therefore, we conclude that CPS-6(H148A/Q130K) is a more stable dimer due to an increased dimeric interface, which enhances CPS-6 nuclease activity.
The Q130K mutation in CPS-6 suppresses the oxidative effect of paraquat on C. elegans
To examine the effect of CPS-6(Q130K) mutation on the cell death in C. elegans, we performed transformation rescue assay with CPS-6(Q130K) in the oxidative conditions. We expressed wild-type CPS-6 and CPS-6(Q130K) in the cps-6(sm116) mutant under the control of the dpy-30 gene promoter, which directs ubiquitous gene expression in C. elegans, and treated transgenic animals with 200 μM paraquat. Expression of wild-type CPS-6 fully rescued the delay-of-cell-death defect of the cps-6(sm116) mutant in the absence of paraquat, but failed to rescue the defect in the presence of 200 μM paraquat (Figure 1C). In contrast, expression of CPS-6(Q130K) fully rescued the cell death defect of the cps-6(sm116) mutant regardless of the paraquat treatment (Figure 1D), indicating that CPS-6(Q130K) can overcome the oxidative effect of 200 μM paraquat to confer wild-type CPS-6 activity. These results are consistent with our in vitro data showing that CPS-6(Q130K) dimers are more stable and have a greater nuclease activity than wild-type CPS-6 and provide further supporting evidence that a more stable CPS-6 dimer is more resistant to the influence of oxidative agents in vivo.
The major role of WAH-1 in C. elegans cell death is to stabilize CPS-6
CPS-6 needs to interact with and be activated by WAH-1 to promote apoptosis, as loss of wah-1 causes a delay-of-cell-death defect similar but not greater than that of the cps-6(sm116) mutant (Wang et al., 2002). Because CPS-6(Q130K) dimers are structurally more stable and are able to retain their pro-apoptotic activities in response to oxidative conditions, we tested whether CPS-6(Q130K) is dependent on WAH-1 to promote apoptosis. As expected, expression of wild-type CPS-6 failed to rescue the delay-of-cell-death defect of wah-1(tm1159) animals (Figure 1E). Surprisingly, expression of CPS-6(Q130K) in wah-1(tm1159) animals fully rescued the cell death defect (Figure 1F). These results indicate that CPS-6(Q130K) can function independently of WAH-1 to promote apoptosis and suggest that the major function of WAH-1 is to stabilize CPS-6 dimers during apoptosis.
We have shown that the cysteine residues located in WAH-1 form disulfide bonds in the presence of H2O2 (Tables S2 and S3), which potentially can also serve as a reducing agent. Because the primary structure of reduced glutathione also features cysteine residues capable of forming disulfide bonds, we tested whether reduced glutathione can replace WAH-1 as a reducing agent to stimulate CPS-6 activity. Treatment of N2 animals with reduced glutathione at various concentrations did not appear to affect cell death (Figure S1B). As expected, the delay-of-cell-death defect of the cps-6(sm116) mutant was not rescued by 20 μM reduced glutathione (Figure S1C). However, the delay-of-cell-death phenotype of the wah-1(tm1159) mutant was fully rescued by the addition of 20 μM reduced glutathione (Figure 1G). These results indicate that the major function of WAH-1 in cell death is to provide a reduced environment that stimulates the endonuclease and pro-apoptotic activity of CPS-6.
CPS-6 dimerization is compromised under oxidative conditions in vivo
To investigate how paraquat treatment affects dimerization of CPS-6 in vivo, we performed a co-immunoprecipitation assay using animals that have a FLAG epitope knockin at the carboxyl terminus of CPS-6 in the genome (cps-6::flag) and, at the same time, express a CPS-6::GFP fusion protein under the control of the dpy-30 gene promoter, which directs a ubiquitous, low level of gene expression in C. elegans (Nakagawa et al., 2014). We incubated these animals with or without 200 μM paraquat in NGM plates for 3 days and then performed co-immunoprecipitation assays on lysates of these animals using anti-FLAG affinity beads. We found that the amount of CPS-6::GFP associated with CPS-6::FLAG was reduced following treatment with paraquat, which did not alter the expression level of CPS-6::GFP (Figure 1H, left). By measuring the signal intensities of the precipitated CPS-6::GFP bands from three independent experiments, the amount of CPS-6::GFP associated with CPS-6::FLAG in paraquat-treated animals was estimated to be 56% of that in non-treated animals (Figure 1H, right), indicating that the oxidative treatment indeed compromised the formation of CPS-6 dimers.
Discussion
In this study, we provide strong in vitro and in vivo evidence to demonstrate, for the first time, that hydrogen peroxide induces disassociation of CPS-6 dimers into monomers, resulting in reduced endonuclease activity and decreased pro-apoptotic activity. Hydrogen peroxide also oxidizes monomeric WAH-1 and produces oxidized WAH-1 dimers cross-linked by inter-disulfide bonds predominantly between two C345 residues, as well as intra- or inter-disulfide bonds among C240, C345 and C535. We suggest that cross-linking between two WAH-1 monomers provides the reduction potential that is important for stabilizing CPS-6 dimers, thereby countering the oxidative effect generated by H2O2 in mitochondria. Moreover, reduced glutathione can successfully rescue the cell death defect induced by loss of WAH-1, suggesting that WAH-1 has a glutathione-like activity, albeit not as an enzyme that can remove superoxide or convert hydrogen peroxide to water. WAH-1 thus may act as an antioxidant to protect CPS-6 from oxidation and thereby preserve its maximal endonuclease activity under oxidative stress.
In mammals, dimeric AIF might play a similar role to WAH-1 by protecting EndoG from oxidation by ROS so that EndoG can maintain its endonuclease activity for its pro-survival function in mitochondria or for its pro-apoptotic role in the nucleus during apoptosis. As the nuclease activity of EndoG decreases under high ROS levels, mitochondrial DNA biogenesis is compromised, which could lead to mitochondrial dysfunction. This is supported by observations that both EndoG-knockout (McDermott-Roe et al., 2011) and AIF-deficient (Joza et al., 2005; Klein et al., 2002) mice had elevated levels of ROS in mitochondria and developed mitochondria dysfunction-related heart diseases. As EndoG has a dual role in regulating both the life and death of cells in both yeast and mammals (Buttner et al., 2007; Huang et al., 2006; Li et al., 2001), our results suggest that oxidative stress is linked to the regulation of cell metabolism by directly modulating the activity of EndoG. In addition, our results suggest causal relationships between high ROS levels, compromised activities of EndoG and AIF, and mitochondrial dysfunction.
Increased ROS is associated with various pathologic conditions, including neurodegenerative diseases, cancers, aging and inflammatory diseases (Li et al., 2013; Schon and Przedborski, 2011; Sena and Chandel, 2012). How mitochondrial proteins respond to oxidative damages to effect cell death and cell survival remains unclear. Here, we demonstrate that the interfacial region located within human and C. elegans EndoG dimer is responsible for the regulation of the nuclease activity in a ROS-sensitive manner. We provide molecular insights into the underlying mechanism by which ROS directly oxidizes EndoG/CPS-6 and reduces its nuclease activity, leading to cell death defects. In conclusion, the endonuclease and cell-killing activity of EndoG/CPS-6 is regulated by ROS levels and the redox potential of its activator, AIF/WAH-1. Oxidative stress impairs the nuclease activity of EndoG/CPS-6 and thus compromises both the life and death of a cell. Our findings may open a new direction for preventing cell loss in aging-related diseases and for preventing the undesirable survival of cancer cells.
EXPERIMENTAL PROCEDURES
Protein expression and purification
All of the wild-type and mutant CPS-6 and WAH-1 recombinant proteins were cloned, expressed and purified according to the previously-described protocol (Lin et al., 2012).
Nuclease activity assays
The recombinant CPS-6 was incubated with 2.5 mM DTT or 2.5 mM H2O2 at 4°C for 16 hours to generate the reduced or oxidized forms of CPS-6, respectively. For the DNase activity assays shown in Figure 2 (the eluted oxidized protein fractions collected from S-75), CPS-6 enzymes were incubated with 25 ng pET28 plasmid DNA in 50 mM Tris-HCl pH7.4, 200 mM NaCl, 2 mM MgCl2 and 2.5 mM DTT at 37ºC for 60 min.
Supplementary Material
Acknowledgments
This work was supported by research grants from Academia Sinica and the Ministry of Science and Technology, Taiwan, R.O.C. to H.Y., and NIH grants R01 GM059083, R01 GM079097 and R35 GM118188 to D.X. The tandem mass data were acquired at the Academia Sinica Common Mass Spectrometry Facilities located at the Institute of Biological Chemistry. Portions of this research were carried out at the National Synchrotron Radiation Research Center; a national user facility supported by the National Science Council of Taiwan. The Synchrotron Radiation Protein Crystallography Facility is supported by the National Core Facility Program for Biotechnology. Funding for open access charge provided by Academia Sinica, Taiwan.
Footnotes
AUTHOR CONTRIBUTIONS
JLJL and WZY performed the biochemical and structural experiments. AN, RS-G, PZ, ZZ, XG and SM performed the in vivo experiments. JLJL, AN, DX and HSY designed experiments, analyzed data, and wrote the manuscript.
Accession Numbers
The atomic coordinates for the CPS-6(H148A/Q130K) crystal structure have been deposited in the RCSB Protein Data Bank with the accession number 4QN0
Supplemental Information includes Supplemental Experimental Procedures, four tables and six figures and can be found with this article online.
References
- Basnakiana AG, Apostolova EO, Yina X, Abirib SO, Stewarta AG, Singha AB, Shaha SV. Endonuclease G promotes cell death of non-invasive human breast cancer cells. Exp Cell Res. 2006;312 doi: 10.1016/j.yexcr.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner S, Habernig L, Broeskamp F, Ruli D, FNV, Vlachos M, Macchi F, VK, Carmona-Gutierrez D, Eisenberg T, et al. Endonuclease G mediates α-synuclein cytotoxicity during Parkinson’s disease. EMBO J. 2013;32:3041–3054. doi: 10.1038/emboj.2013.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büttner S, Eisenberg T, Carmona-Gutierrez D, Ruli D, Knauer H, Ruckenstuhl C, Sigrist C, Wissing S, Kollroser M, Frohlich KU, et al. Endonuclease G regulates budding yeast life and death. Mol Cell. 2007;25:233–246. doi: 10.1016/j.molcel.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Cote J, Ruiz-Carrillo A. Primers for mitochondrial DNA replication generated by Endonuclease G. Science. 1993;261:765–769. doi: 10.1126/science.7688144. [DOI] [PubMed] [Google Scholar]
- Ferreira P, Villanueva R, Martínez-Júlvez M, Herguedas B, Marcuello C, Fernandez-Silva P, Cabon L, Hermoso JA, Lostao A, Susin SA, Medina M. Structural insights into the coenzyme mediated monomer-dimer transition of the pro-apoptotic apoptosis inducing factor. Biochemistry. 2014;53:4204–4215. doi: 10.1021/bi500343r. [DOI] [PubMed] [Google Scholar]
- Huang KJ, Ku CC, Lehman IR. Endonuclease G: A role for the enzyme in recombination and cellular proliferation. Proc Natl Acad Sci USA. 2006;103:8995–9000. doi: 10.1073/pnas.0603445103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joza N, Oudit GY, Brown D, Bénit P, Kassiri Z, Vahsen N, Benoit L, Patel MM, Nowikovsky K, Vassault A, et al. Muscle-specific loss of apoptosis-inducing factor leads to mitochondrial dysfunction, skeletal muscle atrophy, and dilated cardiomyopathy. Mol Cell Biol. 2005;25:10261–10272. doi: 10.1128/MCB.25.23.10261-10272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowska M, Garncarz W, Pietrowska M, Garrard WT, Widlak P. Regulation of the human apoptotic DNase/RNase Endonuclease G: involvement of Hsp70 and ATP. Apoptosis. 2005;10:821–830. doi: 10.1007/s10495-005-0410-9. [DOI] [PubMed] [Google Scholar]
- Klein JA, Longo-Guess CM, Rossmann MP, Seburn KL, Hurd RE, Frankel WN, Bronson RT, Ackerman* SL. The harlequin mouse mutation downregulates apoptosis-inducing factor. Nature. 2002;419:367–374. doi: 10.1038/nature01034. [DOI] [PubMed] [Google Scholar]
- Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- Li X, Fang P, Mai J, Choi ET, Wang H, Yang X-F. Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J Hemat & Oncol. 2013;6:1–19. doi: 10.1186/1756-8722-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JLJ, Nakagawa A, Lin CL, Hsiao Y-Y, Yang W-Z, Wang Y-T, Doudeva LG, Skeen-Gaar RR, Xui D, Yuan HS. Structural insights into apoptotic DNA degradation by CED-3 Protease Suppressor-6 (CPS-6) from Caenorhabditis elegans. J Biol Chem. 2012;287:7110–7120. doi: 10.1074/jbc.M111.316075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott-Roe C, Ye J, Ahmed R, Sun XM, Serafín A, Ware J, Bottolo L, Muckett P, Cañas X, Zhang J, et al. Endonuclease G is a novel determinant of cardiac hypertrophy and mitochondrial function. Nature. 2011;478:114–118. doi: 10.1038/nature10490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miramar MD, Costantini P, Ravagnan L, Saraiva LM, Haouzi D, Brothers G, Penninger JM, Peleato ML, Kroemer G, Susin SA. NADH oxidase activity of mitochondrial apoptosis-inducing factor. J Biol Chem. 2001;276:16391–16398. doi: 10.1074/jbc.M010498200. [DOI] [PubMed] [Google Scholar]
- Molnar GA, Nemes V, Biro Z, Ludany A, Wagner Z, Wittmann I. Accumulation of the hydroxyl free radical markers meta-, orthotyrosine and DOPA in cataractous lenses is accompanied by a lower protein and phenylalanine content of the water-soluble phase. Free Rad Res. 2005;39:1359–1366. doi: 10.1080/10715760500307107. [DOI] [PubMed] [Google Scholar]
- Parrish J, Li L, Klotz K, Ledwich D, Wang X, Xue D. Mitochondrial endonuclease G is important for apoptosis in C. elegans. Nature. 2001;412:90–94. doi: 10.1038/35083608. [DOI] [PubMed] [Google Scholar]
- Parrish JZ, Xue D. Functional genomic analysis of apoptotic DNA degradation in C. elegans. Mol. Cell. 2003;11:987–996. doi: 10.1016/s1097-2765(03)00095-9. [DOI] [PubMed] [Google Scholar]
- Parrish JZ, Xue D. Cuts can kill: the roles of apoptotic nucleases in cell death and animal development. Chromosoma. 2006;115:89–97. doi: 10.1007/s00412-005-0038-0. [DOI] [PubMed] [Google Scholar]
- Requena JR, Chao CC, Levine RL, Stadtman ER. Glutamic and aminoadipic semialdehydes are the main carbonyl products of metal-catalyzed oxidation of proteins. Proc Natl Acad Sci USA. 2001;98:69–74. doi: 10.1073/pnas.011526698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon EA, Przedborski S. Mitochondria: The next (neurode)generation. Neuron. 2011;70:1033–1053. doi: 10.1016/j.neuron.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukova IF. Redox-linked conformational dynamics in apoptosis-inducing factor. J Mol Biol. 2009;390:924–938. doi: 10.1016/j.jmb.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M, et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang J, Gengyo-Ando K, Gu L, Sun C-L, Yang C, Shi Y, Kobayashi T, Shi Y, Mitani S, et al. C. elegans mitochondrial factor WAH-1 promotes phosphatidylserine externalization in apoptotic cells through phospholipid scramblase SCRM-1. Nat Cell Biol. 2007;9:541–549. doi: 10.1038/ncb1574. [DOI] [PubMed] [Google Scholar]
- Wang X, Yang C, Chai J, Shi Y, Xue D. Mechanisms of AIF-mediated apoptotic DNA degradation in Caenorhabditis elegans. Science. 2002;298:1587–1592. doi: 10.1126/science.1076194. [DOI] [PubMed] [Google Scholar]
- Xu H, Freitas MA. A mass accuracy sensitive probability based scoring algorithm for database searching of tandem mass spectrometry data. BMC Bioinfor. 2007;8:133. doi: 10.1186/1471-2105-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Cande C, Stephanou NC, Jiang S, Gurbuxani S, Larochette N, Daugas E, Garrido C, Kroemer G, Wu H. DNA binding is required for the apoptogenic action of apoptosis inducing factor. Nat Struct Biol. 2002;9:680–684. doi: 10.1038/nsb836. [DOI] [PubMed] [Google Scholar]
- Zan H, Zhang J, Al-Qahtani A, Pone EJ, White CA, Lee D, Yel L, Mai T, Casali P. Endonuclease G plays a role in immunoglobulin class switch DNA recombination by introducing double-strand breaks in switch regions. Mol Immun. 2011;48:610–622. doi: 10.1016/j.molimm.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.