Abstract
We have developed a baculovirus-based display system for identifying antigen mimotopes for MHC class I-specific T cells. The mouse MHC class I molecule, Dd, was displayed on baculovirus-infected insect cells with a library of 9- and 10-mer peptides tethered via a flexible linker to the N terminus of β2 microglobulin. As a test case, the library was screened by flow cytometry by using a multimeric fluorescent αβTCR from a mouse T cell specific for Dd plus an unknown self peptide. A mimotope was identified that, when bound to Dd, stimulated the T cell to secret IL-2. The sequence of the mimotope was used to identify a self peptide present in a mouse protein, Spin. The Spin peptide, when complexed with Dd, also activated the T cell. This technique should be generally useful in identifying and manipulating MHC class I peptide mimotopes and epitopes.
Keywords: T cell receptor, peptide, epitope
The identification of peptide antigen epitopes is an important step in isolating and modulating class I MHC (MHCI)-restricted T cells involved in protective and pathological immune responses. The identification of MHCI peptide antigens is difficult in autoimmunity and cancer, because, in these cases, the peptide antigen may be derived from any of the host's proteins expressed in the target tissue. As a consequence, although there are many candidate antigens, in almost no case is the T cell epitope(s) involved in human autoimmunity definitively known. Likewise, despite years of work, the list of tumor-associated antigens with potential for use in immunotherapy is still quite small and concentrated in a few types of cancer.
Many approaches have been taken to identify MHCI peptide epitopes derived from unknown self proteins. One approach has been to identify peptide mimotopes in peptide libraries. Mimotopes differ in sequence from the unknown peptide epitope, but they nevertheless bind to the appropriate MHCI molecule and are recognized by the specific CD8+ T cell (1–3). They can be used to track the T cell in vivo and for immune modulation of the T cell response. Sometimes, the sequence of the mimotope can be used to deduce the sequence of the epitope (4).
Peptide display libraries have been very powerful tools in other situations (reviewed in refs. 5–7), but they are not widely used to identify T cell mimotopes, because the peptide can be identified and used to enrich the library only when it is properly associated with the appropriate MHC molecule. We recently developed baculovirus-infected insect cells as a display platform for class II MHC (MHCII) molecules covalently bound to a library of potential peptide mimotopes (8). “Fishing” in this library with soluble fluorescent T cell receptors, we were able to identify peptide mimotope/MHC complexes that bound to the soluble receptors and stimulated T cells bearing the same receptors. In this present study, we have adapted this method for use with MHCI. In this case, we expressed the peptide library covalently bound to β2 microglobulin (β2m) paired with membrane anchored MHCI Dd heavy chain. Fishing with a fluorescent soluble αβTCR, we found a mimotope for a T cell reactive to Dd plus an unknown self peptide. We used the mimotope sequence to identify a self peptide that could also stimulate the same T cell.
Materials and Methods
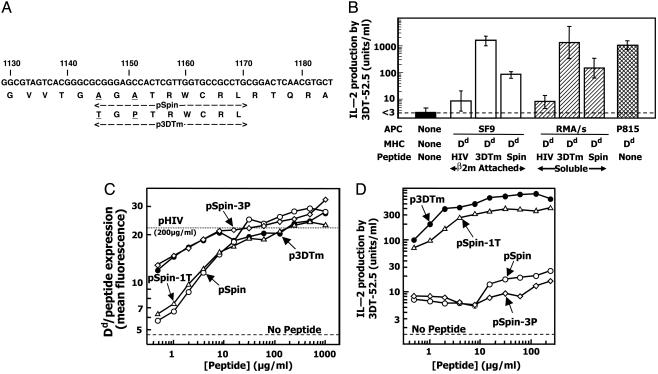
Synthetic Peptides, Oligonucleotides, and DNA Sequencing. The peptides, pHIV (RGPGRAFVTI), pSPIN (AGATRWCRL), and p3DTm (TGPTRWCRL), and variants of pSpin, pSpin-1T (TGATRWCRL), and pSpin-3P (AGPTRWCRL), were synthesized in the Molecular Resource Center of National Jewish Medical and Research Center, as were all oligonucleotides. Automated DNA sequencing was also performed in this facility.
Cell Lines and T Cell Hybridomas. SF9 and High Five were obtained from Invitrogen. An antigen-presenting cell line, SF9-ICAM/B7, was produced as described (8). The T cell hybridoma 3DT-52.5 was produced as described (9). The Dd-pHIV-specific T cell hybridoma, B4.2.3 (10), was provided by David Margulies (National Institutes of Health). Mouse mastocytoma, P815, was obtained from the American Type Culture Collection. The TAP2 mutant cell line, RMA/S-Dd (11), was provided by David Raulet (University of California, Berkeley).
Soluble αβTCR. A soluble version of the 3DT-52.5 αβTCR (AV01S1/AJ25 and BV0801/BD1/BJ2S3) was made by expression in baculovirus, as described (12, 13). The receptor was purified from the supernatant of the baculovirus-infected cells by immunoaffinity chromatography by using the 3DT-52.5 αβTCR-specific mAb, KJ12-98 (14).
Flow Cytometry. Two Dd reactive mouse mAbs (15, 16), 34-2-12 (Dd/α3-specific) and 34-5-8 (specific for Dd occupied with a peptide), were used in these experiments. In flow cytometry experiments, 34-2-12 was detected with a phycoerythrin-labeled goat anti-mouse IgG2a (Fisher), and 34-5-8 was directly fluoresceinated. A multivalent-fluorescent version of the soluble 3DT-52.5 αβTCR was made as described (8). Briefly, sparsely biotinylated (one to two biotins per molecule) ADO-304 (anti-Cα) mAb was precomplexed with Alexa Fluor 647-labeled streptavidin (Molecular Probes). The complex was purified by size-exclusion chromatography and preloaded with soluble 3DT-52.5 αβTCR just before use. Analytical flow cytometry was performed with a FACSCalibur flow cytometer (Beckton Dickinson), and data were analyzed by flowjo software (Tree Star, Ashland, OR). For sorting, a Moflo instrument was used (Dako/Cytomation).
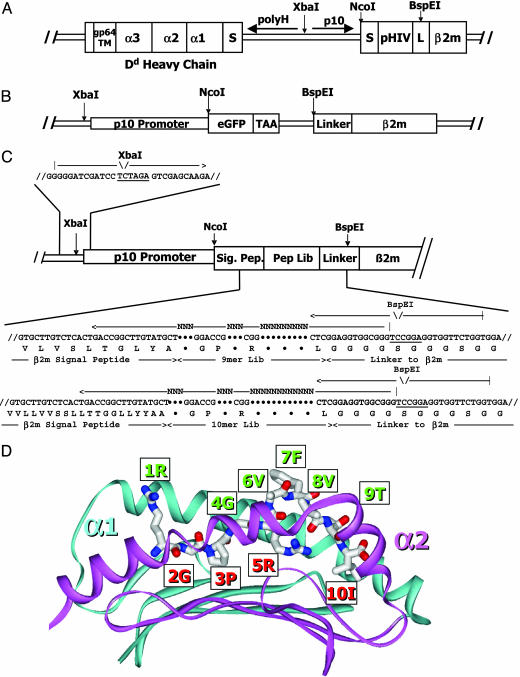
Dd, β2m, and Peptide Constructions. For displaying Dd on the surface of baculovirus-infected insect cells, modifications (Fig. 1A) were made to reported (17) constructs for preparing soluble Dd with a covalently attached peptide from HIV gp120 (pHIV). Using a two-promoter baculovirus transfer plasmid (18), the sequence encoding the extracellular portion of the mouse Dd heavy chain was cloned behind the polyhedrin promoter attached to a sequence encoding the baculovirus gp64 transmembrane domain and cytoplasmic tail. The sequence encoding mouse β2m was cloned behind the p10 promoter in the same plasmid. The sequence encoding pHIV and a flexible glycinerich linker was inserted between the β2m signal peptide and the N terminus of β2m.
Fig. 1.
Baculovirus-mediated display of Dd/β2m with an attached pHIV or a library of peptides. (A) Schematic diagram of the baculovirus construction leading to surface display on infected insect cells of Dd/β2m with pHIV attached. (B) Schematic diagram of the E. coli transfer plasmid constructed to accept PCR fragments encoding libraries of Dd-binding peptides. (C) Strategy for producing and cloning PCR fragments that encode a library of 9- and 10-mer Dd-binding peptides. (D) A ribbon representation of the α1 (cyan) and α2 (magenta) domains of Dd with a wire-frame representation of pHIV (CPK coloring) in the peptide-binding groove. The figure was derived from the crystal structure of the Dd/pHIV complex (21) by using weblab viewer pro (Molecular Simulations, Waltham, MA). The major anchor residues of pHIV are labeled in red, and the surface-exposed residues of pHIV are labeled in green.
The construction of a library of peptides tethered to Dd via the N terminus of β2m was performed in two steps. First, the β2m gene in the transfer plasmid encoding Dd-pHIV was disrupted by sequence encoding enhanced GFP with a tumor-associated antigen termination codon to prevent reading through into the β2m gene, as illustrated in Fig. 1B. When incorporated into baculovirus, cells expressing this construct produced GFP rather than a surface Dd molecule. Second, randomized PCR primers were used, as illustrated in Fig. 1C, to construct PCR fragments encoding peptides that could bind to Dd. The oligonucleotides were synthesized such that the Dd-binding anchor positions 2, 3, and 5 and the C-terminal amino acid of the peptide (Fig. 1D) were fixed as G, P, R, and L, whereas other positions were randomized. Two kinds of fragments were made, one encoding 9- and the other 10-aa peptides. The mixture of fragments was cloned into the transfer plasmid replacing GFP and restoring a functional β2m gene. The resulting library of plasmids was incorporated into baculovirus by the standard homologous recombination method by using Sapphire baculovirus DNA (Orbigen, San Diego). The estimated size of library was ≈105 independent clones.
IL-2 Assays. T cell hybridoma cells (105) were added to microtiter wells containing (i) 105 P815 cells, (ii) 105 RMA/S-Dd cells plus soluble peptide, or (iii) 5 × 104 SF9-ICAM/B7 insect cells infected 3 days previously with baculovirus encoding Dd with a covalently bound peptide. After overnight incubation, the culture supernatants were assayed for IL-2, as described (19).
Results
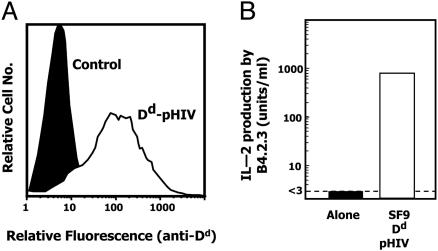
Functional Surface Expression of a Peptide Tethered to Dd Molecules on Insect Cells. We previously established methods to display functional MHCII peptides on baculovirus-infected insect cells (8). As described in Fig. 1 A and Materials and Methods, we adapted this system to display the mouse MHCI molecule, Dd, covalently bound to the peptide, pHIV. We had previously produced a secreted form of this complex in baculovirus (17) in which the pHIV was attached to the N terminus of β2m via a flexible linker. This construct was modified to tether Dd-pHIV to the infected insect cell surface via the transmembrane/cytoplasmic region of the baculovirus gp64 molecule. SF9 insect cells infected with baculovirus encoding this construct displayed Dd-pHIV at a high level on their surface (Fig. 2A) and, when coexpressing ICAM and B7.1, could stimulate IL-2 production by the Dd-pHIV-specific T cell, B4.2.3 (10) (Fig. 2B).
Fig. 2.
Functional display of baculovirus-encoded Dd-pHIV on infected insect cells. (A) SF9 cells infected 3 days previously with baculovirus encoding a surface anchored version Dd-pHIV (open histogram) or a control virus (filled histogram) were analyzed by flow cytometry by using an anti-Dd-specific mAb, 34-2-12. (B) B4.2.3 hybridoma T cells were cultured alone (filled bar) or with Dd-pHIV displaying SF9-ICAM/B7 cells (open bar). After 24 h, culture supernatants were assayed for IL-2.
Construction of a Library of Peptides Bound to Dd. To prepare a library of displayed peptides bound to Dd, we used a strategy described in Fig. 1 B and C and Materials and Methods. We disrupted the β2m gene by replacing the DNA encoding the β2m signal peptide and linked pHIV with the GFP gene (Fig. 1B). This plasmid was then used as a recipient for PCR fragments constructed with degenerate oligonucleotides that randomized certain codons encoding amino acids in the peptide. These replaced the GFP gene and restored a functional peptide-β2m gene (Fig. 1C). In the peptide, we fixed the codons at positions P2, P3, P5, and the C terminus to encode G, P, R, and L, respectively. Based on crystal structures of Dd-pHIV (20, 21) (Fig. 1D), these are the most favored anchor amino acids of peptides naturally bound to Dd (16). Codons for the other amino acids of the peptide, which are αβTCR accessible, were randomized. Because Dd has been shown to bind either 9- or 10-aa peptides, we made two types of PCR DNA fragments, with either three or four variable amino acids between p5 and the peptide C terminus. The result was a pool of baculoviruses, the majority of which expressed Dd tethered to one of the peptides encoded in the library.
Identification of a 3DT-52.5 Mimotope in the Peptide Library. Our goal was to identify a peptide mimotope in the library for the Dd-reactive T cell hybridoma, 3DT-52.5. This T cell reacts with Dd when presented on a number of different antigen-presenting cells, including the mouse Dd-bearing tumor cell line, P815, but its self-peptide specificity is unknown (9, 22). To screen this library, we prepared a multivalent fluorescent soluble version of the 3DT-52.5 αβTCR, as described in ref. 8 and in Materials and Methods. SF9 insect cells were infected with the library at a multiplicity of infection of <1. In a preliminary enrichment experiment, those infected cells expressing surface Dd, but not GFP, were isolated by flow cytometry and mixed with fresh SF9 cells to make a new library stock (data not shown). This eliminated most of the viruses in the library that had recombined with plasmids that had not replaced the GFP gene with a sequence encoding a peptide and also eliminated viruses in which the peptide sequence contained a termination codon.
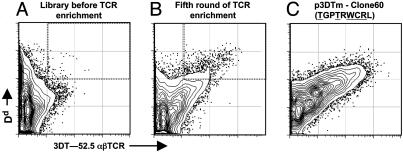
Cells infected with this preenriched library were stained simultaneously with the fluorescent 3DT-52.5 αβTCR and a Dd α3 domain-specific antibody. Those Dd-expressing cells with the brightest fluorescence with the αβTCR reagent (≈1%) were sorted and cocultured with fresh SF9 cells to create new infected cells for analysis and an enriched viral stock. The process was repeated four more times. As shown in Fig. 3A, no clearly αβTCR f luorescent population was evident among Dd-expressing cells infected with the initial viral stock. However, during five rounds of enrichment, a population emerged in which there was a direct relationship between the level of surface Dd and the level of staining with the αβTCR (Fig. 3B), indicating the enrichment of viruses expressing Dd bearing a peptide that the 3DT-52.5 receptor could recognize.
Fig. 3.
Identification of a peptide antigen mimotope for 3DT-52.5. 2D flow cytometry histograms are shown for SF9 cells infected 3 days earlier at a multiplicity of infection of ≈1 with baculovirus, each encoding Dd/β2m with an attached peptide. The infected cells were analyzed with fluorescent multimeric soluble αβTCR from 3DT-52.5 (x axis) and with anti-Dd/α3 (y axis). (A) Cells infected with the Dd-peptide library after enrichment for Dd, but not GFP, expression. Among the cells strongly positive for Dd expression, the 1% with the brightest staining with the fluorescent αβTCR (dotted region) were sorted to create a new enriched viral stock. (B) Cells infected with the Dd-peptide library that had been enriched four previous cycles as in A. Single infected cells within the dotted area were sorted into individual wells of a 96-well plate with fresh SF9 cells to prepare clonal viral stocks. (C) Cells were infected with a viral clone stock (no. 60) that encoded the peptide TGPTRWCRL (p3DTm).
During the last round of enrichment, the viruses were cloned by sorting single infected cells into culture wells containing fresh SF9 cells. Each clone was rechecked for 3DT-52.5 αβTCR binding, and its encoded peptide sequence was determined by sequencing a PCR fragment produced by using the viral DNA as template. Six clones that showed uniform Dd vs. αβTCR staining as well as an unambiguous peptide sequence all had the same nucleotide sequence encoding the same nonamer peptide: TGPTRWCRL (the underlined amino acids are the positions varied in the library). Thus, it seems most likely that a single virus was present in the enriched population. We designated the peptide mimotope, p3DTm. Fig. 3C shows the Dd vs. αβTCR staining pattern of SF9 cells infected with one of these clones. As expected for a single homogeneous Dd–peptide complex, there was a direct relationship between the level of Dd expressed and the level of fluorescent αβTCR binding.
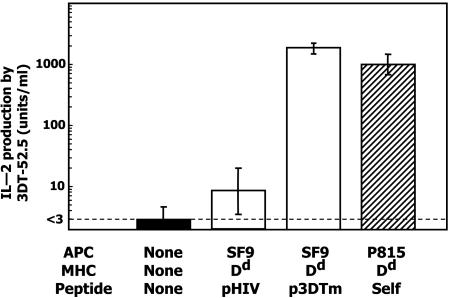
To test the function of Dd-p3DTm, we compared the ability of SF9-ICAM/B7 cells infected with the Dd-p3DTm expressing virus to that of the Dd expressing mouse cell line, P815, in stimulating IL-2 production by 3DT-52.5. As shown in Fig. 4, Dd-p3DTm specifically activated 3DT-52.5 hybridoma cells and, interestingly, the stimulation was consistently slightly better than that produced by endogenous peptide(s) presented by Dd on P815 cells.
Fig. 4.
Stimulation of 3DT-52.5 by the library peptide mimotope. 3DT52-5 hybridoma T cells were cultured alone (filled bar) or with SF9-ICAM/B7 cells (open bar) displaying Dd/β2m attached to either pHIV or p3DTm or with p815 mouse tumor cells (hatched bar). After 24 h, culture supernatants were assayed for IL-2.
Identification of a 3DT-52.5 Self-Peptide Epitope. A search of the mouse sequence database with the mimotope sequence revealed only one protein containing a similar sequence (GenBank accession no. AAH11467), a murine homolog of the Drosophila protein, spinster (23). This protein influences programmed cell death in the development of the insect neural and reproductive systems. Homologs have been identified in other species, including mice (two isoforms) and humans (three isoforms). Fig. 5A shows the portion of the mouse spinster homolog that contains a sequence identical to p3DTm at seven of nine positions and four of the five positions varied in our library. We named this peptide pSpin.
Fig. 5.
Identification of a peptide epitope for 3DT-52.5. (A) Portion of the sequence of one of the mouse spin genes (GenBank accession no. AAH11467) showing the location of the peptide (pSpin) related in sequence to p3DTm. Base numbering starts at the initiation ATG of spin. (B) 3DT-52.5 hybridoma T cells were cultured alone (filled bar); with SF9-ICAM/B7 cells (open bars) displaying Dd/β2m attached to either pHIV, p3DTm, or pSpin; or with RMA/S-Dd cells (single hatched bars) alone or in the presence of 62.5 μg/ml soluble p3DTm or pSpin or with P815 mouse tumor cells (double hatched bar). After 24 h in culture, supernatants were assayed for IL-2. (C) RMA/S-Dd cells were incubated overnight alone (dashed line), with 200 μg/ml of pHIV (dotted line), or with various concentrations of the following peptides: p3DTm (filled circles), pSpin (open circles), pSpin-1T (open triangle), or pSpin-3P (open diamonds). The RMA/S-Dd cells were analyzed by flow cytometry for surface expression of Dd/peptide by using the peptide-sensitive anti-Dd mAb, 34-5-8. (D) RMA/S-Dd cells prepared as in C were cultured overnight with 3DT-52.5 hybridoma T cells. The culture supernatants were then assayed for IL-2.
To show that pSpin is likely to be at least one of the epitopes recognized by 3DT-52.5 on Dd-bearing mouse antigen-presenting cells, we first demonstrated that the spinster homolog was expressed in the 3DT-52.5-stimulating mouse Dd-bearing cell line, P815. A DNA fragment encoding the appropriate portion of the gene was produced by using PCR with P815 cDNA as template and was sequenced to confirm the presence of the peptide sequence (data not shown). We then compared, in two ways, the responses of 3DT-52.5 to p3DTm and pSpin presented by Dd (Fig. 5B). First, SF9-ICAM/B7 cells infected with baculovirus expressing Dd tethered either to p3DTm or to pSpin, but not to pHIV, stimulated 3DT-52.5 IL-2 production. Second, the Tap-deficient Dd cell line, RAM/S-Dd (11), presenting synthetic versions of p3DTm and pSpin, but not pHIV, stimulated 3DT-52.5 IL-2 production.
The response of 3DT-52.5 to pSpin was weaker than to p3DTm, whether soluble or Dd-tethered peptide was used (Fig. 5B). This could have been due to the difference at p1 (predicted to interact with the 3DT-52.5 αβTCR), at p3 (a Dd anchor position), or at both. To test these possibilities, we synthesized pSpin-1T (p1A changed to T) and pSpin-3P (p3 A changed to P). All four peptides were compared for their ability to up-regulate the expression of Dd on the surface of RMA/S-Dd (Fig. 5C), a measure of the relative efficiency of Dd binding by the peptides (16). p3DTm was approximately five times more efficient than pSpin in up-regulating Dd expression on RMA/S-Dd cells. Changing the p3A of pSpin to P improved its ability to stabilize Dd on RMA/S-Dd cells to that of p3DTm. However, the change of the p1A to T had no effect on Dd binding. Thus, as expected, changing the p3 amino acid to a more favorable anchor residue improved peptide binding to Dd.
All four peptides were also tested for presentation by RMA/S-Dd to 3DT-52.5 (Fig. 5D). The hybridoma responded more weakly to pSpin than to p3DTm throughout the concentration range tested. Changing the p1A to T improved its ability to stimulate 3DT-52.5 nearly to that of p3DTm. The A to P change at p3 did not improve the response despite the improved ability to bind to Dd. These results indicate the interaction of the T at p1 with the 3DT-52.5 αβTCR is more important than the optimal P at p3 in determining the stronger reactivity of 3DT-52.5 to p3DTm compared to pSpin.
Because the response of 3DT-52.5 to mouse P815 cells is very strong, the weak response to pSpin may indicate that it is not the only epitope presented by these cells. Alternatively, pSpin may be the sole natural epitope for 3DT-52.5, but p815 may simply present this peptide much better than peptide-loaded RMA/S-Dd or infected SF9 cells. In this case, p3DTm acts as a “heteroclitic” mimotope, i.e., a peptide that interacts better with this TCR than the natural epitope.
Discussion
For many years, peptide mimotopes for MHCI-restricted T cells have been identified in chemically synthesized or genetic peptide libraries that have been distributed in aliquots. With this approach, the peptide sequence is obtained either by sequential reduction in the number of peptides per aliquot or by deduction based on the pattern of aliquots with stimulatory activity. A recent version of the latter method is called positional scanning (24). For each position in the peptide, 20 aliquots of the library are prepared. In each aliquot, this position is fixed as one of the 20 amino acids, but the other positions in the peptide are randomized. Therefore, for peptides of 9 aa in length, the complete library has 180 aliquots. These are tested for presentation by the appropriate MHC molecule to the T cell in question. A positive reaction indicates that the fixed amino acid in that aliquot is preferred or at least tolerated at that position. This information can be used to construct a second set of potential peptide mimotopes that are rescreened for presentation and T cell recognition. The major advantages of this approach are that the libraries contain all possible versions of the peptide and, because the number of aliquots is small, the screening of the library is very rapid. The major disadvantages of this method are its expense, the fact that the library is exhaustible, the requirement for a second round of synthesis to deconvolute the data, and, perhaps most importantly, that its success depends on T cells with high peptide sensitivity for activation but with low sensitivity to competition from nonrecognized peptides. Despite these limitations, the method has been very useful in identifying T cell mimotopes (reviewed in ref. 24).
Display libraries circumvent many of the shortcomings of distributed libraries, because all members of the library are tested independently and enriched from one aliquot without the need to screen distributed samples. Furthermore, because the screening of the library involves a binding assay, the stringency of the binding can be manipulated to select for a particular affinity/avidity of binding. Because these libraries are by definition genetically encoded, they are relatively inexpensive to construct by using various PCR and cloning strategies and, once constructed, the libraries are never exhausted. Although Escherichia coli phage has been the most widely developed platform for display libraries, its use for identifying T cell peptide antigen mimotopes has been limited because of the poor assembly and expression of MHC molecules within E. coli.
A recent advance in display libraries has been the development of yeast as a display platform (25). As a eukaryotic organism, yeast is better than E. coli at expressing complex eukaryotic proteins, such as MHC molecules, antibodies, and αβTCRs (26–29). Frequently, however, considerable engineering and mutation of these molecules have been required for efficient surface display, and the use of yeast surface MHC display for identifying TCR peptide mimotopes has not been reported.
Baculovirus has long been used to express a wide variety of eukaryotic proteins. We have developed methods in baculovirus for genetically covalently tethering antigenic peptides to MHCI and MHCII molecules (17, 18). In a previous study (8), we showed that a library encoding randomized peptides attached to the mouse MHCII molecule, IAb, could be constructed in baculovirus and displayed very efficiently on baculovirus-infected insect cells. Using flow cytometry with fluorescent multimerized αβTCRs, we isolated baculoviruses from this library encoding mimotopes that stimulated the T cells from which the αβTCRs were derived. In this study, we have extended our development of baculovirus as a display vector to peptide libraries bound to the mouse MHCI molecule, Dd. We screened this library with an αβTCR from the T cell hybridoma 3DT-52.5, specific for Dd plus an unknown self peptide(s). We identified a mimotope peptide that, when bound to Dd, activated the hybridoma. We then identified a self protein, Spin, that encoded a peptide of similar sequence that also stimulated 3DT-52.5 but less well then the mimotope. In using the mimotope sequence to find the Spin peptide in the mouse genome, we were fortunate that the Dd peptide anchor amino acids are very well defined, and that relatively unique amino acids such as cysteine and tryptophan appear to be part of the T cell recognition epitope. Identifying epitopes in databases from mimotope sequences can be expected to be much more difficult when MHC anchor amino acids are more forgiving, fewer peptide exposed amino acids are contacted by the αβTCR, and/or αβTCR contact amino acids are more common amino acids.
In our experiments described here and previously (8), the sizes of the peptide libraries were ≈105, far less than needed to have each of the 3.2–64 × 106 possible peptide sequences (assuming five or six varied positions) present in the library. Although in our studies thus far, these libraries were sufficiently large to obtain mimotopes for the αβTCRs used, it seems likely that more saturating libraries will be required for this method to be broadly useful in mimotope/epitope discovery. The size of our libraries was limited by the relatively inefficient technique of first cloning the library in an E. coli plasmid and then transferring it to baculovirus DNA by homologous recombination. Therefore, we have adapted the techniques developed by Ernst et al. (30, 31) to bypass the E. coli stage and clone PCR DNA fragments directly into baculovirus DNA.
Details of this approach will be published elsewhere (F.C., P.M., and J.W.K., unpublished results). Briefly, for both MHCI and MHCII libraries, we have introduced sites for homing nucleases into baculovirus DNA encoding the GFP-bearing recipient MHC construct. These enzymes recognize long rare DNA sequences absent in the large (136-kb) baculovirus genome. In both cases, we have introduced a site for SceI into the region between the p10 and polyhedrin promoter and placed a site for CeuI in the region encoding the linker between the peptide and the MHC molecule. Cutting the viral DNA with SceI and CeuI linearizes the viral DNA and releases the GFP gene, leaving 4-bp nonpalindromic 5′ overhangs. We have added sites for the restriction enzyme, BstXI, to the ends of the PCR DNA fragment encoding the peptide library. Because this enzyme recognizes an interrupted palindrome and leaves a 4-bp 5′ overhang, the BstXI sites were constructed such that these overhangs matched those left by the homing nucleases. Therefore, the fragment can be ligated directly into the linear viral DNA, recircularizing it to an infectious form and restoring expression of the MHC/peptide genes. In our initial experiments, transfection of insect cells with this DNA has allowed us to produce viral libraries with >3 × 107 independent clones expressing surface MHC/peptide.
Finally, we hope that a major use of this technique will be to complement other methods for the discovery of mimotopes/epitopes for T cells of unknown specificity. However, we also point out that it may prove useful in the rapid screening of variants of known antigenic peptides to increase or decrease the affinity of a MHC–peptide complex for a given αβTCR, thus providing a tool to find better antigens for cancer therapy or altered peptide ligands for suppression of autoimmunity.
Acknowledgments
We thank Amy Marrs and Randy Anselment of the National Jewish Molecular Resource Center, National Jewish Medical and Research Center, for oligonucleotide and peptide syntheses as well as automated DNA sequencing. We also thank Shirley Sobus, Josh Loomis, and Bill Townend of the National Jewish Flow Cytometry Facility, National Jewish Medical and Research Center, for help with flow cytometric analysis and sorting. This work was supported by U.S. Public Health Service Grants AI-17134, AI-18785, and AI-22295.
Author contributions: Y.W. and J.W.K. designed research; Y.W., A.R., R.H., J.W., F.C., and J.W.K. performed research; Y.W., A.R., R.H., J.W., F.C., and J.W.K. analyzed data; and Y.W., P.M., and J.W.K. wrote the paper.
Abbreviations: MHCn, class n MHC; β2m, β2 microglobulin.
References
- 1.Anderson, B., Park, B. J., Verdaguer, J., Amrani, A. & Santamaria, P. (1999) Proc. Natl. Acad. Sci. USA 96, 9311-9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinilla, C., Rubio-Godoy, V., Dutoit, V., Guillaume, P., Simon, R., Zhao, Y., Houghten, R. A., Cerottini, J. C., Romero, P. & Valmori, D. (2001) Cancer Res. 61, 5153-5160. [PubMed] [Google Scholar]
- 3.Linnemann, T., Tumenjargal, S., Gellrich, S., Wiesmuller, K., Kaltoft, K., Sterry, W. & Walden, P. (2001) Eur. J. Immunol. 31, 156-165. [DOI] [PubMed] [Google Scholar]
- 4.Lieberman, S. M., Evans, A. M., Han, B., Takaki, T., Vinnitskaya, Y., Caldwell, J. A., Serreze, D. V., Shabanowitz, J., Hunt, D. F., Nathenson, S. G., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 8384-8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Partidos, C. D. (2000) Curr. Opin. Mol. Ther. 2, 74-79. [PubMed] [Google Scholar]
- 6.Lee, S. Y., Choi, J. H. & Xu, Z. (2003) Trends Biotechnol. 21, 45-52. [DOI] [PubMed] [Google Scholar]
- 7.Szardenings, M. (2003) J. Recept. Signal Transduct. Res. 23, 307-349. [DOI] [PubMed] [Google Scholar]
- 8.Crawford, F., Huseby, E., White, J., Marrack, P. & Kappler, J. W. (2004) PLoS Biol. 2, 523-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Endres, R. O., Marrack, P. & Kappler, J. W. (1983) J. Immunol. 131, 1656-1662. [PubMed] [Google Scholar]
- 10.Kozlowski, S., Takeshita, T., Boehncke, W. H., Takahashi, H., Boyd, L. F., Germain, R. N., Berzofsky, J. A. & Margulies, D. H. (1991) Nature 349, 74-77. [DOI] [PubMed] [Google Scholar]
- 11.Correa, I. & Raulet, D. H. (1995) Immunity 2, 61-71. [DOI] [PubMed] [Google Scholar]
- 12.Kappler, J., White, J., Kozono, H., Clements, J. & Marrack, P. (1994) Proc. Natl. Acad. Sci. USA 91, 8462-8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, C. P., Crawford, F., Marrack, P. & Kappler, J. (1998) Proc. Natl. Acad. Sci. USA 95, 4522-4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kappler, J., Kubo, R., Haskins, K., White, J. & Marrack, P. (1983) Cell 34, 727-737. [DOI] [PubMed] [Google Scholar]
- 15.Ozato, K., Mayer, N. M. & Sachs, D. H. (1982) Transplantation 34, 113-120. [DOI] [PubMed] [Google Scholar]
- 16.Corr, M., Boyd, L. F., Padlan, E. A. & Margulies, D. H. (1993) J. Exp. Med. 178, 1877-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White, J., Crawford, F., Fremont, D., Marrack, P. & Kappler, J. (1999) J. Immunol. 162, 2671-2676. [PubMed] [Google Scholar]
- 18.Kozono, H., White, J., Clements, J., Marrack, P. & Kappler, J. (1994) Nature 369, 151-154. [DOI] [PubMed] [Google Scholar]
- 19.White, J., Kappler, J. & Marrack, P. (2000) Methods Mol. Biol. 134, 185-193. [DOI] [PubMed] [Google Scholar]
- 20.Li, H., Natarajan, K., Malchiodi, E. L., Margulies, D. H. & Mariuzza, R. A. (1998) J. Mol. Biol. 283, 179-191. [DOI] [PubMed] [Google Scholar]
- 21.Achour, A., Persson, K., Harris, R. A., Sundback, J., Sentman, C. L., Lindqvist, Y., Schneider, G. & Karre, K. (1998) Immunity 9, 199-208. [DOI] [PubMed] [Google Scholar]
- 22.Gay, D., Maddon, P., Sekaly, R., Talle, M. A., Godfrey, M., Long, E., Goldstein, G., Chess, L., Axel, R., Kappler, J., et al. (1987) Nature 328, 626-629. [DOI] [PubMed] [Google Scholar]
- 23.Sanyal, S. & Ramaswami, M. (2002) Neuron 36, 335-338. [DOI] [PubMed] [Google Scholar]
- 24.Nino-Vasquez, J. J., Allicotti, G., Borras, E., Wilson, D. B., Valmori, D., Simon, R., Martin, R. & Pinilla, C. (2004) Mol. Immunol. 40, 1063-1074. [DOI] [PubMed] [Google Scholar]
- 25.Boder, E. T. & Wittrup, K. D. (1997) Nat. Biotechnol. 15, 553-557. [DOI] [PubMed] [Google Scholar]
- 26.Kieke, M. C., Cho, B. K., Boder, E. T., Kranz, D. M. & Wittrup, K. D. (1997) Protein Eng. 10, 1303-1310. [DOI] [PubMed] [Google Scholar]
- 27.VanAntwerp, J. J. & Wittrup, K. D. (1998) J. Mol. Recognit. 11, 10-13. [DOI] [PubMed] [Google Scholar]
- 28.Starwalt, S. E., Masteller, E. L., Bluestone, J. A. & Kranz, D. M. (2003) Protein Eng. 16, 147-156. [DOI] [PubMed] [Google Scholar]
- 29.Brophy, S. E., Holler, P. D. & Kranz, D. M. (2003) J. Immunol. Methods 272, 235-246. [DOI] [PubMed] [Google Scholar]
- 30.Ernst, W., Grabherr, R., Wegner, D., Borth, N., Grassauer, A. & Katinger, H. (1998) Nucleic Acids Res. 26, 1718-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ernst, W. J., Grabherr, R. M. & Katinger, H. W. (1994) Nucleic Acids Res. 22, 2855-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]