Abstract
We describe the cloning and characterization of a human homolog of the yeast transcription/RNA-processing factor Ssu72, following a yeast two-hybrid screen for pRb-binding factors in the prostate gland. Interaction between hSsu72 and pRb was observed in transfected mammalian cells and involved multiple domains in pRb; however, so far, mutual effects of these two factors could not be demonstrated. Like the yeast counterpart, mammalian Ssu72 associates with TFIIB and the yeast cleavage/polyadenylation factor Pta1, and exhibits intrinsic phosphatase activity. Mammals contain a single ssu72 gene and a few pseudogenes. During mouse embryogenesis, ssu72 was highly expressed in the nervous system and intestine; high expression in the nervous system persisted in adult mice and was also readily observed in multiple human tumor cell lines. Both endogenous and ectopically expressed mammalian Ssu72 proteins resided primarily in the cytoplasm and only partly in the nucleus. Interestingly, fusion to a strong nuclear localization signal conferred nuclear localization only in a fraction of transfected cells, suggesting active tethering in the cytoplasm. Suppression of ssu72 expression in mammalian cells by siRNA did not reduce proliferation/survival, and its over-expression did not affect transcription of candidate genes in transient reporter assays. Despite high conservation, hssu72 was unable to rescue an ssu72 lethal mutation in yeast. Together, our results highlight conserved and mammalian specific characteristics of mammalian ssu72.
INTRODUCTION
Transcription by RNA polymerase II is coupled to RNA processing, including capping, splicing and cleavage/polyadenylation. The C-terminal repeat (CTD) of RNA pol II orchestrates both processes by recruiting RNA processing factors (1–4). Indeed, CTD directly binds polyadenylation factors and its truncation inhibits transcript cleavage in vivo (5,6). For instance, the cleavage-polyadenylation-specificity-factor (CPSF) is recruited to the promoter by interaction with TFIID. After transcription starts, CPSF dissociates from TFIID and becomes associated with CTD (7). The CTD undergoes cyclic phosphorylation and dephosphorylation allowing transcription initiation and re-initiation by RNA pol II (8). Recent evidence implicates a novel yeast factor, Ssu72, in both CTD dephosphorylation and RNA processing/termination.
Yeast Ssu72 was initially identified by Hampsey's group (9) in a screen for suppressors of sua7-1, a cold-sensitive mutation in yeast TFIIB, hence Ssu72 (Suppressor of sua7-1 clone 2). The sua7-1 mutation shifts transcription start site selection downstream at the ADH1 promoter; the ssu72 mutation combined with sua7-1 induces further shift downstream. A null mutation in ssu72 is lethal. A screen for suppressors of a temperature-sensitive ssu72 strain identified Rpb2, the second largest subunit of RNA pol II (10). Accordingly, biochemical analysis revealed direct Ssu72–Rbp2 and Ssu72–TFIIB physical interactions (10,11). Mutation in ssu72 disrupts the expression of a subset of yeast genes, such as CYC1 (10). Similar repression was observed when the CYC1 promoter, fused to a reporter gene, was expressed in mutant ssu72 cells in transient transfection experiments, indicating that Ssu72 affects promoter strength directly (10).
A genetic screen for factors that can rescue a temperature-sensitive ssu72 strain identified Fcp1, the only known RNA pol II CTD phosphatase, and consistent with this, Ssu72 harbors intrinsic phosphatase activity (12,13). Recent analysis revealed that Ssu72 dephosphorylates Ser-5 in the CTD of RNA pol II, and together with Fcp1, which targets Ser-2, regenerates initiation competent hypo-phosphorylated RNA pol II (14). The yeast Ssu72 also has a phosphatase-independent function. Large-scale analysis of protein complexes in yeast identified Ssu72 as a component of a cleavage and polyadenylation factor (CPF) complex (15). Ssu72 interacts directly with the Pta1 subunit of CPF and is implicated in transcript cleavage and termination (16–19).
The retinoblastoma tumor suppressor, pRb, controls cell proliferation, survival and differentiation and its inactivation contributes to the progression of most human malignancies (20). pRb exerts diverse biological effects by interacting with over 100 proteins, most of which are involved in transcriptional regulation (21). So far, pRb was documented to employ three mechanisms of transcription repression. First, pRb directly interacts and quenches the trans-activation domain of transcription activators, such as E2Fs (22). Second, once bound to an activator on promoter DNA, pRb recruits chromatin-modifying enzymes, such as histone deacetylases (HDACs), via LxCxE motifs in these proteins, and further silences gene expression (23). Third, pRb binds components of the pre-initiation complex, such as TAFII250, and may directly affect its assembly (24). In addition to transcriptional repression, pRb also binds tissue-specific differentiation factors, such as myogenin, C/EBP and CBFA1, and positively participates in the transcriptional activation of lineage-specific markers (25–27). In a screen for mammary- and prostate gland-specific cDNA libraries for pRb-binding factors, we here describe the identification and characterization of hSsu72, a human homolog of yeast Ssu72.
MATERIALS AND METHODS
Plasmids
Details on the construction of the following plasmids are available upon request. For yeast two-hybrid screens, we generated pAS1-CYH2-Rb, RbΔK11 and Δ22. For yeast complementation studies: pRS316-yssu72, pYES-yssu72, pYES-Myc-yssu72, pYES-hssu72 and pYES-Myc-hssu72. GST-Rb was used to create GST-RbΔA, -RbΔC, -RbΔ22ΔC and -RbΔLxCxE [derived from a plasmid kindly obtained from F. Dick (28)]. GST-Pta1 was a gift from X. He and C. L. Moore. Mutant Rb and ssu72 alleles were transferred into pHAT vectors (Clontech). Human ssu72-NI-1 pseudogene was cloned by PCR from human genomic DNA. Mouse ssu72 was cloned into (pBS) by RT–PCR. Human ssu72 was sub-cloned into pTAG-2HA or a pcDNA3 vector into which Myc or Flag epitopes were added in frame to the N-terminus.
SV40 TAg-NLS
P P K K K R K V
CCC CCG AAG AAA AAG CGG AAG GTC
was added between the Myc and hssu72 sequence. pcDNA3-Myc and Myc-NLS-hssu72 deletion mutants were generated by cloning linkers between restriction enzyme sites in hssu72. For tethering to promoters, hSsu72 was fused in frame with the DNA-binding domain of Lex. pLex, GAL-myc, GAL-Rb and the reporter X4G2-luciferase [4 GAL4 and 2 LexA DNA-binding elements fused upstream of the hsp70 promoter (29)] were gifts from R. Bremner.
Yeast interactive screen and complementation analysis
Yeast two-hybrid screens for pRb interacting factors were performed following the Clontech Matchmaker GAL4 two-hybrid user manual. Prior to library screening, conditions were optimized using the E1A-activation domain as prey. A human prostate Matchmaker cDNA library (catalog no. HL4037AH) in pACT2 vector was purchased from Clontech and screened as specified by the manufacturer. Plasmid DNA was recovered from putative yeast clones using the following protocol. Fresh (2–4 day old) yeast colonies were inoculated into 5 ml of SD/-Trp-Leu liquid medium, vortex vigorously to resuspend the cells and rotated 230–250 r.p.m. at 30°C overnight. The yeast cells were centrifuged at 14 000 r.p.m. for 5 min. The supernatant was decanted and the pellets were resuspended in the same buffer (∼50 μl). An aliquot of 10 μl lyticase solution was added and the samples were rotated at 200–250 r.p.m. for 30–60 min at 37°C. An aliquot of 20 μl of 10% SDS was added followed by vigorous vortexing for 1 min. The samples were subject to at least one cycle of freeze/thaw (at −20°C) and vortexed again to ensure complete lysis of the cells. The volume of the samples was adjusted to 200 μl in TE buffer (pH 7.0), and 200 μl of phenol:chloroform:isoamyl alcohol (25:24:1) was added. After high-speed vortexing for 5 min, the samples were spun down at 14 000 r.p.m. for 10 min. The supernatant was collected, 8 μl of 10 M ammonium acetate was added and the DNA samples were further purified on Qiagen mini-prep columns. The purified plasmid DNA was transformed and amplified in competent DH5α bacteria. Plasmid DNA was analyzed using PCR, restriction enzyme digestions and sequencing, and subsequently re-transformed and tested in yeast Y190 containing the different Rb baits.
For complementation analysis, ssu72(+/−) diploid yeast strain [ATCC-4021990; (30)] was transformed with human or yeast ssu72 expression vectors using lithium acetate. Transformants were selected on SD-URA(−) medium supplemented with glucose (2%). Ectopic protein expression was assessed by the SDS–UREA method from liquid cultures grown in YEPD then induced with YEP supplemented with galactose (2%) overnight. Sporulation was induced in 2% potassium acetate. Tetrads were isolated and dissected on SD supplemented with galactose (2%) and all amino acids. Dissected cultures were replica plated onto YEPD- G418 or SD-URA(−) medium to assess growth and genotype.
Antibodies
Rabbit polyclonal antibodies to hSsu72 were generated by Exalpha Biologicals, Inc. (Boston MA), following immunization with purified eluted GST-hSsu72 that was extensively dialyzed against phosphate-buffered saline (PBS). These anti-hSsu72 antibodies are now available (catalog no. X1562P). Monoclonal α-Flag and α-Myc (Cell Signaling Technology) were used at a dilution of 1:1500 for western blotting and 2.5 μg per sample for immunoprecipitation. Monoclonal anti-α-tubulin, Sigma catalog no. T-5168, was used at 1:1000 dilution for western blotting.
Western blot and co-immunoprecipitation analysis
Cells were transfected with the indicated plasmids using polyethylenimine (Aldrich catalog no. 40,8727, protocol available upon request). Three days later, plates were placed on ice and washed twice with ice-cold PBS. Cells were scrapped in PBS, transferred to 1.5 ml microfuge tubes and centrifuged at 3000–4000 r.p.m. for 5 min at 4°C. Cell pellets were resuspended in 1 ml NTEN buffer (100 mM NaCl, 0.5% NP-40, 50 mM Tris–HCl, pH 7.6) plus proteinase inhibitors and phenylmethlysulfonyl fluoride (PMSF), and disrupted by drawing through a needle 5–10 times on ice, followed by additional incubation on ice for 15 min. The lysates were cleared by centrifugation at 13 000 r.p.m. for 20 min and the supernatants were transferred to new tubes. Aliquots of 40 μl were withdrawn for straight western blots. To the rest of the lysates, α-Myc antibodies (2.5 μg) were added and placed on a rocking platform at 4°C overnight. Pre-washed (3–4 times NTEN) 100 μl secondary antibody-agarose was added for 30–60 min. Protein complexes on beads were washed six times in NTEN plus 0.1% SDS and immunoblotted as described previously (31).
Immunostaining
COS7 cells were seeded on coverslips in DMEM medium supplemented with fetal bovine serum (FBS) (10%), then transfected with 1–2 μg of recombinant or control pcDNA3 vector DNA. Twenty-four hours following transfection, coverslips were rinsed with PBS, fixed in ice-cold methanol (30 min), then washed in PBS (3 × 10 min). After blocking in 1% BSA/PBS for 1 h, α-Myc or α-hSsu72 primary antibodies were added in blocking solution for 1 h. Coverslips were rinsed with PBS (3 × 10 min) and incubated with fluorescein isothiocyanate (FITC) or Rhodamine-conjugated secondary antibodies (1:200 dilution, 1% BSA/PBS) in addition to DAPI (4′,6′-diamidino-2-phenylindole, dihydrochloride). Cover slips were rinsed with PBS (3 × 10 min) and finally mounted using Dako fluorescent mounting medium (S3023). Leptomycin B (Sigma catalog no. L2913), was added to a final concentration of 10 nM for a duration of 2 h prior to immunostaining.
Purification of GST- and Hat-fusion proteins and GST-pull down
Hat-tagged proteins were purified on Talon Metal Affinity Resins according to the manufacturer's recommendations (Clontech). For GST-fusion proteins, overnight bacterial cultures were diluted 1:10 in 2–3 l of 2YT (Ampicillin, 100 μg/ml), and cultures were grown for 1.5 h at 37°C. Isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 0.1 mM and cultures were shaken for another 2.5 h at 30°C/280 r.p.m. Cultures were spun down at 5000 r.p.m. for 15 min at 4°C and resuspended in 3 ml NNTE buffer (500 mM NaCl, 0.5% NP-40, 50 mM Tris–HCl, pH 7.6, 5 mM EDTA) containing PMSF. The resuspended bacteria were transferred to 30 ml spin tubes at 4°C, sonicated 3 × 20 s on ice and spun at 10 000 r.p.m. for 20 min at 4°C. The supernatant containing the fusion proteins were mixed with glutathione Sepharose™ 4B beads pre-washed three times in 0.5 ml ice-cold NNTE buffer in 15 ml FALCON tubes and rocked for 30–60 min at 4°C. The beads-GST fusion proteins were then washed in NNTE buffer and three times in NTEN buffer at 4°C and finally resuspended in 500 μl NTEN:glycerol (50:50) for storage for up to 10 days at −20°C. For pull-down experiments, GST-beads (amounts assessed from Coomassie blue staining of protein gels with BSA as control) were washed in NTEN three times, rocked with protein lysates or Hat-tagged proteins (Hat-Ssu72) for 2 h at 4°C and washed with NTEN buffer six times.
Phosphatase assay
Phosphatase activity was performed by monitoring the production of p-nitrophenylate from p-nitrophenylphosphate (pNPP). For phosphatase inhibition assay, excess GST-proteins on beads were mixed with minimal amounts of purified Hat-hSsu72 protein, which produced phosphatase activity within 30 min, in a final volume of 98 μl NTEN buffer in ELISA plates (MICROTEST™96, FALCON). After agitation at 37°C for 30 min at 100 r.p.m., 2 μl of 1 M pNPP was added and incubation continued for additional 30–60 min. The reaction was stopped by the addition of 3.8 vol of 0.1 M NaOH and optical density was measured at 405 nm with Vmax ELISA reader (Molecular Devices Inc.).
Small interfering RNA (SiRNA) and MTT analysis
Custom sense and anti-sense RNA oligonucleotides targeting the sequence AAC AGG GAC TCA CGT GAA GCT or AAG ACC TGT TTG ATC TGA TCC of hssu72 were purchased from Dharmacon Research Inc. and annealed according to the manufacturer's recommendations. COS7 cells were seeded at 50% confluency in DMEM medium supplemented with 10% FBS and transfected for 2 or 3 successive days using 0.2 nM of either annealed duplex targeting hssu72 or two non-complementary RNA oligonucleotides (control), using the Oligofectamine reagent (Invitrogen). The day following the last transfection, siRNA-treated and controls cells were harvested by trypsinization and the total number of cells was determined by Trypan blue exclusion staining. A portion of the harvested cells was seeded in triplicates onto 96 well plates at 2 × 103 cells/well. About 6 or 18 h later, cultures were treated with MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium-bromide; Sigma M2128] at a final concentration of 0.8 μg/ml in DMEM (10% FBS) for 1 h. Cultures were lysed overnight, and absorbency was determined at 570 nm using an ELISA plate reader. The remaining harvested cells were lysed using a Triton-X based protein extraction buffer [50 mM Tris, pH 7.5, 150 mM NaCl, 10% (v/v) Glycerol, 1% (v/v) Triton X-100 and 1 mM EGTA] for western analysis to confirm the effect of siRNA treatment on hssu72 expression.
Northern blot analysis
Total RNA from cultured cells or fresh mouse tissues was prepared using the Trizol reagent (Invitrogen). RNA samples were separated electrophoretically on formaldehyde denaturing agarose gels, transferred onto GeneScreen-Plus nylon membranes and hybridized with 32P-labeled hssu72, mssu72 or gapdh radioactive probes under conditions specified by the manufacturer.
Nuclear/cytoplasmic fractionation
COS7 cultures were transiently transfected with pcDNA3-Myc-Stra13 (nuclear marker) and pcDNA3-Myc-luciferase (cytosol marker) and harvested by trypsinization 36–48 h later. Cells were resuspended in swelling buffer (10 mM Tris–HCl, pH 7.5, 10 mM KCl and 2 mM MgCl2) and the plasma membrane was solubilized by the addition of NP-40 to a final concentration of 0.6%. Nuclei were collected by centrifugation at 1000 g. The supernatant/cytoplasmic fraction was adjusted to 10% glycerol, 150 mM NaCl. Nuclei were rinsed once in swelling buffer then lysed in Triton-X (1%) extraction buffer. The amount of protein lysates used from each fraction was normalized for analysis by western blotting.
Luciferase reporter assays
COS7 or C33A cells were transiently transfected with a pGL3-based luciferase reporter vectors, Cyclin E, p107, pG4X2 (100 ng); class B E-box elements (CACGTG) (100 ng), the SV40 immediate early promoter (10 ng) or the Rb promoter (100 ng). Expression vectors pcDNA3-Myc-hSsu72, pcDNA3-Myc-NLS-hSsu72, pcDNA3-Lex-Myc-NLS-hSsu72, Gal-Rb or Gal-myc were co-transfected together with 250 ng CMV-lacZ as internal control. Luciferase analysis and data analysis was performed using standard methods as described previously (32,33).
In situ hybridization
33P-UTP labeled sense and anti-sense riboprobes for mouse ssu72 were generated from linearized DNA templates using T7 or T3 RNA polymerase (Fermentas). An aliquot of 8 μm sections from paraffin-embedded embryonic tissues were processed, hybridized (1.5 × 105 c.p.m./μl), washed and counter-stained as described previously (34,35). Images were captured using an AxioCam MRc digital camera (Zeiss). For the dark field phase, identical exposure time and digital processing were used for sense and anti-sense probes.
RESULTS
Identification of human Ssu72 following a yeast interactive screen with a pRb bait
In an effort to identify mammary and prostate gland-specific pRb-binding factors, we performed yeast two-hybrid screens with pRb as bait. Previous analysis had shown that when ectopically expressed in yeast, human pRb is highly phosphorylated by cyclin-dependent kinases (36). We therefore used as bait a constitutively active pRb allele, RbΔK11, in which phosphorylation is inhibited by mutations in 11 of the 16 Ser/Thr phospho-acceptor sites in pRb (37). This has allowed us to perform yeast two-hybrid screens at relatively high concentrations of the His3 antagonist, 3-AT. The initial screen was carried out in the presence of 20 mM 3-AT. A total of 200 clones isolated after 6 days of incubation were collected and re-examined on plates containing 35 mM 3-AT. Approximately 40% survived and were selected for DNA isolation and shuttling into bacteria. Plasmid DNA from unique clones, identified by PCR analysis and restriction enzyme digestion, was purified and subjected to a second round of screens with RbΔK11 and RbΔ22, a non-functional allele of pRb, as baits. In the second screen, we used both the His3 and LacZ reporters to score for positive clones.
Our initial screen of a mammary gland library identified multiple factors that were all previously shown to interact with pRb, including HSP70, Id2 and cyclin D1 (data not shown). Screening of a prostate library identified additional factors, four of which represented full- or near full-length human homologs of the yeast ssu72 gene (Figure 1A and B). When re-transformed into yeast cells and tested on His- plates or by the lacZ reporter assay, the human ssu72 cDNA clones interacted with pRbΔK11 as well as wild-type pRb, but only weakly with the pocket-mutant pRbΔ22 (data not shown).
Figure 1.
Molecular cloning of hSsu72 following yeast two-hybrid screen with RbΔK11 as a bait. (A) Schematic representation of hSsu72 clones recovered from the yeast two-hybrid screen with RbΔK11 as bait. The approximate size of clone no. 113 was deduced from fragment size; the exact size of the other clones was determined by DNA sequencing. (B) Sequence alignment and amino acid conservation in human, mouse and yeast Ssu72 proteins. The human and mouse proteins are almost identical with the exception of the two indicated conserved substitutions. Red denotes identical residues; orange, similar; black, non-conserved. Note that the N-terminus is generally more conserved than the C-terminus. The PPase domain and the putative LxCxE are indicated.
The interaction between pRb and hSsu72 was confirmed in vitro and in transfected cells in vivo (Figure 2A and B). Interestingly, human Ssu72 contains an LxCxE motif frequently found in pRb-binding factors (Figure 1A and B). However, interaction between the two factors was independent of the LxCxE motif in hSsu72 or the LxCxE-binding motif in pRb but rather involved multiple domains in pRb (Figure 2B–E). Despite numerous experiments, we have been so far unable to identify a context in which a mutual effect between pRb and hSsu72 could be demonstrated (see below and Discussion).
Figure 2.
Interaction of pRb and hSsu72 in vitro and in vivo. (A) COS7 cells were transfected with plasmids expressing Myc-tagged pRb, Myc-RbΔ22ΔC and/or Flag-tagged Ssu72 as indicated. Four percent of protein lysates were analyzed directly by western blotting (lanes 1–5). The remaining lysates were immunoprecipitated with anti-Myc antibodies, washed extensively and immunoblotted with anti-Flag antibodies to reveal co-immunoprecipitated Flag-Ssu72. Ssu72 was co-sedimented only in the presence of pRb (lane 9) but not RbΔΔ22Δc (lane 10) or in the absence of Rb (lane 8). Asterisk in both panels denotes uncharacterized proteins. IP, immunoprecipitation; IB, immunoblot. (B–C) GST-pull down experiments demonstrating that interaction of pRb and hSsu72 is not mediated by the LxCxE-binding site in pRb (B–C) or the LxCxE motif in Ssu72 (C). In GST-RbLxCxEmut, the LxCxE-binding domain is disrupted by I753A, N757A and M761A substitutions (28,61). hSsu72 LxCxE contains a LTCEE to LTGEE substitution. Similar substitution of the central cystein in the LxCxE motifs in adenovirus E1A and human HSP75 completely disrupted the interaction of these proteins with pRb (62). The indicated proteins were expressed in COS7 cells by transfecting the corresponding expression vectors. Lysates were mixed with the indicated GST-fusion proteins, extensively washed and immunoblotted with antibodies reactive to HA, Flag or large Tag, which is constitutively expressed in COS7 cells. Note that interaction of pRb with HDAC and Tag but not Ssu72 is disrupted by mutations in the LxCxE-binding domain in pRb. (D) Schematic representation of the GST-Rb fusion protein and various deletion mutants used in the following panel. (E) GST-pull down experiments of the indicated GST-Rb fusion proteins with transfected HA-hSsu72 or endogenous Tag in COS7 cells. Deletions in the A or B regions (GST-RbΔA and (GST-RbΔ22) or C-terminal domain (GST-RbΔC), reduce but did not completely inhibit binding to hSsu72; double deletions of exon 22 and in the C-terminus (GST-RbΔCΔ22) abrogated pRb- hSsu72 interaction. Coomassie staining of the GST-fusion proteins is shown. Inputs in all panels were 4%.
Mammalian hSsu72 associates with TFIIB and yeast Pta1
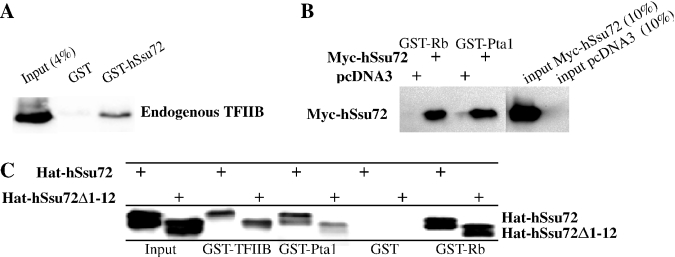
We first tested whether hSsu72 is biochemically conserved by investigating its ability to interact with known binding partners of its yeast counterpart. Yeast Ssu72 was initially identified via its genetic interaction with yTFIIB; it was subsequently shown to associate with GST-yTFIIB in pull-down experiments with in vitro labeled Ssu72 (9,11). As shown in Figure 3A, GST-pull down assays with total protein lysates of COS7 cells revealed that endogenous mammalian TFIIB was co-precipitated in the presence of GST-hSsu72 but not GST alone.
Figure 3.
Human hSsu72 interacts with TFIIB and yeast Pta1. (A) Interaction of hSsu72 with TFIIB. COS7 lysates were mixed with GST-hSsu72 or GST proteins immobilized on beads, washed extensively and immunoblotted with antibodies reactive to TFIIB. GST-hSsu72 but not GST efficiently precipitated endogenous TFIIB. (B) Interaction of hSsu72 with yeast Pta1. COS7 cells transfected with pcDNA3-Myc-Ssu72 or vector alone were lysed and mixed with GST-Rb or GST-Pta1 proteins, washed extensively and immunoblotted with antibodies reactive to a Myc-specific epitope. Both human Rb and yeast Pta1 efficiently interacted with human Ssu72. (C) Hat-hSsu72 or Hat-hSsu72Δ1-12, containing an in-frame deletion that removes the PPase domain, was purified on Talon resins, mixed with the indicated GST-fusion proteins, washed extensively and immunoblotted with polyclonal antibodies reactive to hSsu72. Pta1, pRb and TFIIB interacted with both Hat-hSsu72 and Hat-hSsu72Δ1-12.
A second major binding partner for Ssu72 in yeast is Pta1, a subunit of the yeast cleavage and polyadenylation complex (16,17,38). Myc-hSsu72 was ectopically expressed in mammalian cells and lysates were mixed with GST-Pta1 or GST alone. As shown in Figure 3B, Myc-hSsu72 co-sedimented in the presence of GST-Pta1 but not GST. The interaction of hSsu72 with GST-Pta1 appeared to be as efficient as the interaction with GST-pRb (Figure 3B). Thus, the phylogenetic conservation of Ssu72 is sufficient to allow not only intra-species binding to TFIIB but also inter-species interaction with the yeast factor Pta1.
Interaction with pRb, TFIIB or yeast Pta1 does not alter hSsu72 intrinsic phosphatase activity
To confirm that human Ssu72 exhibits protein phosphatase (PPase) activity, we tested purified GST- and Hat-tagged-hSsu72 fusion proteins in phosphatase assays using pNPP as a substrate. Phosphatase activity was observed within 20–30 min under standard phosphatase conditions (100 mM citrate, pH 6.5) as well as binding conditions (containing 0.5% NP-40) (see Supplementary Material, Figure SD.1A; data not shown). A single point mutation, converting Cystein 12 to Serine [GST-hSsu72:C12S (12,13)] in the phosphatase domain (VCSSNQNRS), or deletion of the entire domain (GST-hSsu72:113–194) completely abolished phosphatase activity (Figure SD.1A). Addition of GST-hTFIIB, GST-Pta1 or GST-Rb did not affect hSsu72 phosphatase activity compared with GST alone (Supplementary Material, Figure SD.1B). Consistent with this, deletion of the phosphatase domain (amino acids 1–12) in Hat-hSsu72 did not abolish interaction with TFIIB, Pta1 or pRb (Figure 3C), indicating that these factors do not interact with hSsu72 via its PPase domain.
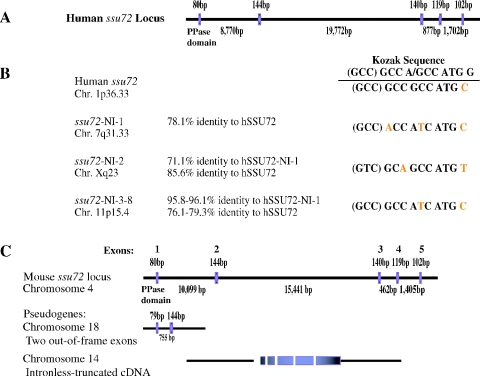
Human and mouse genomes encode single ssu72 genes
BLAST searches of mammalian NCBI EST databases revealed a single expressed ssu72 cDNA sequence per species (data not shown). Accordingly, only a single locus for ssu72 was found in the human genome (Figure 4A). Located on chromosome 1p36, it spans approximately 32 kb and its coding region is divided into five exons. We also identified eight intronless ssu72-related sequences, which likely represent pseudogenes (Figure 4B). Typically, these pseudogenes display 75–85% identity with human ssu72, have different Kozak translation initiation sequence and can be divided into three groups according to location and similarity to one another (Figure 4B). In silico analysis of expressed sequence tag databases failed to detect ssu72-pseudogene sequences, suggesting that none of them is expressed.
Figure 4.
Structural organization of mammalian ssu72 genes. (A) Structural organization of human ssu72 on chromosome 1. The intron/exon boundaries were deduced from the genomic sequence. Exon 1 denotes only the coding region (transcription start site is unknown). (B) Location, conservation and sequences around the translation initiation sites in ssu72 and its intronless pseudogenes. (C) Structural organization of mouse ssu72 on chromosome 4 and two related pseudogenes, which are likely non-functional due to internal deletions/mutations.
BLAT searches of mouse genomic sequences also revealed a single functional ssu72 gene, located on chromosome 4 (Figure 4C). The overall organization into five exons and the intron/exon boundaries in the human and mouse ssu72 genes are conserved (Figure 4C; data not shown). Two partial sequences with homology to ssu72 are also present in the mouse genome. These sequences are clearly non-functional as they are severely truncated/mutated (Figure 4C). Thus, the intronless homologs of ssu72 likely represent non-functional pseudogenes; each mammalian genome, just like yeast, appears to contain a single functional ssu72.
Identification of the ssu72 transcripts and Ssu72 protein
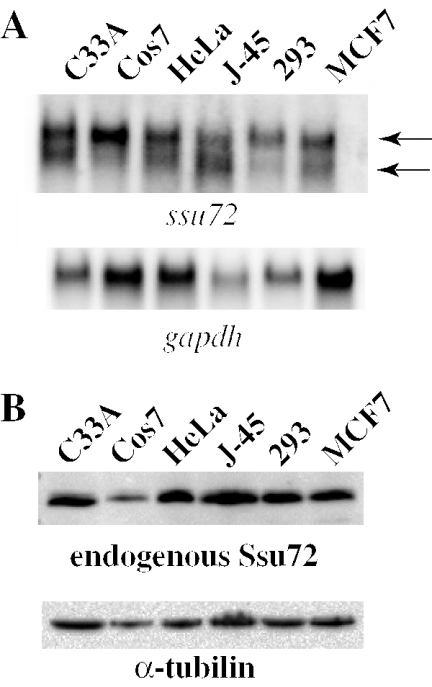
To assess the expression of human ssu72, we performed northern blot analysis on RNA extracted from multiple mammalian tumor cell lines. These included human cervical carcinoma cells (C33A), SV40 transformed monkey kidney cells (COS7); human cervical epithelial adenocarcinoma (HeLa); human T lymphocytes (J-45); human transformed kidney cells (293); and human breast adenocarcinoma (MCF7). A doublet of different relative intensity was detected in all cell lines (Figure 5A). The lower transcript was predominant in the J-45 lymphocytes, whereas the upper transcript was more abundant in the other cell types, in particularly the two kidney-derived cells 293 and COS7. The nature of these different transcripts, which may originate from alternative splicing or promoter usage, is unknown. Northern blots performed in parallel with the hssu72-NI-1 pseudogene probe produced only low levels of signals possibly due to cross hybridization with hssu72 (data not shown).
Figure 5.
Identification of ssu72 transcripts and Ssu72 protein. (A) Aliquots containing 50 μg of total RNA from the indicated cell lines were separated by electrophoresis through a 1% agarose/formaldehyde gels, northern blotted and probed with 32P-radiolabeled human ssu72 cDNA as a probe. Note a doublet with variable intensity in the different cell types. Gapdh was used as internal RNA loading control. (B) Western blot analysis of indicated cell lines with rabbit polyclonal antibodies raised against GST-hSsu72. Note a single band of ∼30 kDa in all lines. α-tubulin was used as internal protein loading control.
To identify endogenous hSsu72, we raised rabbit polyclonal antibodies against full-length GST-hSsu72 (see Materials and Methods). Specificity of the hSsu72 sera was verified in a series of competition experiments with transfected 2HA-tagged hSsu72, which migrates slower than endogenous Ssu72 on denaturing polyacrylamide gels, in the presence or absence of GST or GST-hSsu72 as competitors. In addition, as described below (Figure SD-2), ssu72 siRNA specifically knocked down the expression of Ssu72 as determined by immunoblotting with these antibodies. To assess the expression of Ssu72, we performed western blot analysis on the same cell panel described above. A single band of ∼30 kDa corresponding to endogenous Ssu72 was observed in all the cell lines (Figure 4B).
Ssu72 resides primarily in the cytoplasm and partly in nucleus
To determine the subcellular localization of Ssu72, we performed nuclear-cytoplasmic fractionation of COS7 cells followed by western blotting. As control, we transfected the cells with Myc-luciferase as a cytoplasmic marker and Myc-Stra13 as a nuclear marker. While the two markers localized exclusively in the cytoplasm and nuclear fractions, respectively, endogenous Ssu72 was found in both fractions but predominantly in the cytosol (Figure 6A). Ectopically expressed untagged and Myc-tagged hSsu72 isoforms also localized primarily in the cytoplasm of transfected cells, as was determined by immunofluorescence microscopy (Figure 6B). Notably, the yeast proteomic nuclear localization project also assigned cytoplasmic > nuclear localization to the yeast Ssu72 (39).
Figure 6.
Regulation of mammalian Ssu72 subcellular localization. (A) Shown are duplicate COS7 cultures transiently transfected with a nuclear marker, Myc-Stra13 and a cytoplasmic marker, Myc-luciferase. Nuclear and cytoplasmic fractions were prepared, separated on polyacrylamide–SDS gels and immunoblotted with anti-Myc antibody to determine the efficiency of the fractionation procedure and anti-hSsu72 antibody to reveal subcellular localization of endogenous Ssu72. (B) Immunofluorescent localization of ectopically expressed untagged or Myc-tagged hSsu72 proteins using anti-hSsu72 or anti-Myc specific antibodies, respectively. Nuclei were labeled by DAPI staining. (C) Schematic structure of Myc- hSsu72 and Myc-NLS-hSsu72 deletion mutants. (D) Cytoplasmic localization of two representative Myc-hSsu72 deletion mutants, Δ16-40 and Δ64-119. All other Myc-hSsu72 deletion mutants shown in (C) also resided in the cytoplasm (data not shown). (E) Immunofluorescent analysis of COS7 cells ectopically expressing Myc-NLS-hSsu72 in which hSsu72 is fused to the SV40 Tag NLS. About 45% of transfected cells exhibited complete nuclear localization (top), 45% cytoplasmic localization (bottom); the rest (10%) were found throughout the cell (data not shown). (F) Immunofluorescent analysis of COS7 cells ectopically expressing the same two deletion mutants, Δ16-40 and Δ64-119 (D), fused to SV40 TAg NLS (Myc-NLS-hSsu72). Both deletion mutants were completely nuclear. All other Myc-NLS-hSsu72 deletion mutants shown in (C) also exhibited exclusive nuclear localization (data not shown). Secondary fluorescent antibodies used were Rhodamine-conjugated secondary anti-rabbit antibody (A, top) and FITC-conjugated secondary anti-mouse antibody (all other panels).
In an attempt to identify a discrete region in hSsu72 involved in the regulation of subcellular localization, we created a series of in-frame deletions spanning the entire hSsu72 polypeptide (Figure 6C). All the deletion mutants localized primarily in the cytoplasm like full-length hSsu72 (Figure 6D). Also, Leptomycin B, an inhibitor of the nuclear export receptor, CRM1, did not induce obvious nuclear accumulation of transfected Myc-hSsu72 (data not shown).
Subcellular localization of hSsu72 fused to a strong nuclear localization signal
We next asked whether fusing a strong NLS such as the SV40 large Tag, PPKKKRKV (40), to the N-terminus of hSsu72 would confer nuclear localization. The resulting protein, NLS-hSsu72, was localized exclusively in the nucleus in ∼45%, exclusively in the cytoplasm in ∼45%, and throughout the cell in ∼10% of transfected cells (Figure 6E; data not shown). Remarkably, when the NLS was fused in frame to the series of internal deletion mutants, it invariably induced complete nuclear localization in all the transfected cells (Figure 6F). Thus, regulation of hSsu72 subcellular localization requires the integrity of the protein entire amino acid sequence and can be conferred at least in part by fusion to a strong NLS.
Mouse ssu72 is dynamically regulated during embryonic development
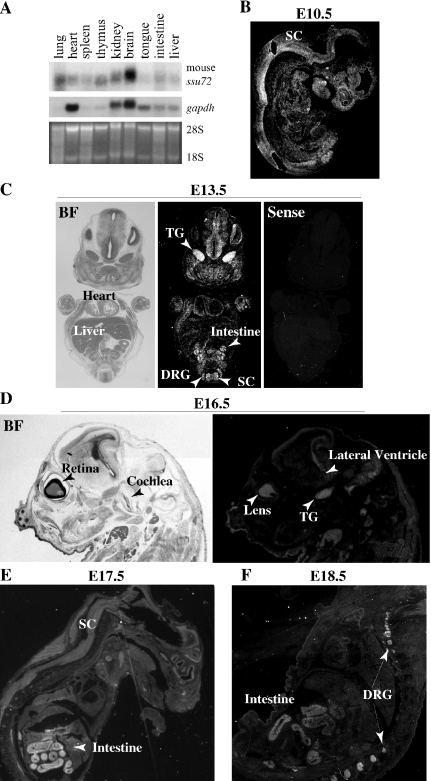
The widespread expression of ssu72 in multiple cell lines, our findings that it appears to represent a unique gene in mammals and the known function of the yeast Ssu72 in transcription/RNA processing suggest that it is likely a ‘housekeeping’ gene invariably expressed in all tissues. To directly examine this conjecture, we cloned the mouse ssu72 gene and performed northern blot analysis on multiple adult tissues. A single hybridizing band was detected in the various tissues (Figure 7A). Strikingly, mssu72 was expressed at low level in most tissues but highly in the brain, suggesting that the gene may be dynamically regulated.
Figure 7.
Dynamic regulation of mouse ssu72 expression during development. (A) Aliquots containing 20 μg of total RNA from the indicated mouse adult tissues were subjected to northern blot analysis with 32P-labeled mssu72 cDNA probe. As loading control, the blots were re-hybridized with gapdh and the levels of 28S and 18S ribosomal RNAs revealed by EtBr staining. (B–F) Radioactive section in situ hybridization analysis of ssu72 during mouse embryogenesis. (B) Sagittal section of an E10.5 embryo showing widespread expression most prevalent in the nervous system. (C) Bright field (BF) and dark field images of coronal sections of E13.5 embryos hybridized with sense or anti-sense mouse ssu72 as indicated. At this stage, ssu72 expression becomes restricted to the developing gut and nervous system, with high expression in TG, DRG and spinal cord. (D) Bright and dark field images of sagittal sections of E13.5 embryos with high expression in lens, TG and the CNS around the lateral ventricle. (E and F) Sagittal sections of E17.5 and E18.5 fetuses showing exclusive and high expression in the nervous system, primarily in DRG of the PNS, and intestine. Other tissues including heart, liver, lung and muscles do not express significant levels of mouse ssu72.
To further pursue mouse ssu72 gene expression, we performed radioactive in situ hybridization analysis during embryogenesis. At embryonic (E) day 10.5, low level of mouse ssu72 transcripts was detected throughout the embryo with relative accumulation in spinal cord and brain folds (Figure 7B). Thereafter, mouse ssu72 expression was further induced in the developing nervous system and gut. At E13.5, high expression was observed throughout the CNS both in the ventricular (mitotic) and marginal (post mitotic) zones, in the PNS in dorsal root (DRG) and trigeminal (TG) ganglia, and the developing gut (Figure 7C). During fetal development, expression in the CNS and PNS persisted, and expression in the intestine was further induced (Figure 7D–F). Expression in the intestine was observed throughout the mucosal villi, which contains epithelial cells and other cell types. Thus, in both the nervous system and gut, ssu72 appears to be expressed in both proliferating and non-proliferating cells. High ssu72 expression was also detected in the lens (Figure 7D). In contrast, other tissues such as liver, lung, bone, cardiac and skeletal muscles exhibited no detectable expression of ssu72. These results strongly implicate mouse ssu72 in the development of a subset of tissues including the nervous system and intestine.
Ssu72 is dispensable for growth and proliferation in COS7 cells
As ssu72 loss-of-function mutations result in lethality in Saccharomyces cerevisiae (9), we next wished to determine whether the gene is essential for growth or viability in mammals. We employed siRNA technology to knockdown ssu72 expression in mammalian cells (41,42). A dramatic inhibition of Ssu72 expression in COS7 cells was attained with two different siRNA pairs targeted to distinct regions in the N-terminus of the Ssu72 coding region (see Supplementary Material, Figure SD-2). Inhibition was often observed after two successive transfections, but maximal and reliable effects were obtained after three transfections. Less efficient inhibition of Ssu72 expression was obtained in HeLa cells, probably reflecting reduced transfection efficiency (data not shown), hence subsequent analysis was performed only on COS7 cells. Despite the dramatic inhibition of Ssu72 protein expression, the morphology and total number of cells were unaffected as determined by microscopic examination and trypan blue exclusion assays (Figure SD-2; data not shown) (43). The levels of dead cells were low and similar in both siRNA-treated and control cultures (data not shown). Furthermore, no difference in proliferation or viability was observed by the MTT assay, performed 6 and 18 h post siRNA treatment (Figure SD-2). We conclude that Ssu72 is dispensable at least in certain mammalian cells in culture or that dramatically reduced levels of Ssu72 suffice for cell survival/proliferation.
Over-expression of native, nuclear- or DNA-targeted hSsu72 exerts minimal effects on transcriptional activity of candidate promoters
To test whether hSsu72 can alter gene expression in mammalian cells, we performed transient luciferase (luc) reporter assays in the absence or presence of transfected hSsu72. As we have not yet identified transcriptional targets regulated by Ssu72, several candidate promoters were tested. Over-expression of hSsu72 or NLS-hSsu72 had no effect on human RbP.luc (32) and (CACGTG)2SV40.luc (33) (Supplementary Material, Figure SD-3). Three other promoters: cyclinEP.luc, p107P.luc and DHFRP.luc were also not responsive (data not shown). We also created a translational fusion of hSsu72 with the DNA-binding domain of Lex (Lex-Myc-NLS-hSsu72) and tested its effect on X4G2.luciferase, a synthetic minimal promoter containing binding sites for GAL4 and Lex. Again, no significant effect was observed when Lex-Myc-NLS-hSsu72 was transfected in the presence or absence of GAL-Rb (repressor) or GAL-myc (activator) (data not shown). These results suggest that like in yeast, hSsu72 may target specific promoters, yet to be defined in mammals, or that the high expression of the endogenous protein renders the transfection of additional nuclear hSsu72 inconsequential.
Mammalian ssu72 is unable to functionally complement its homolog in S.cerevisiae
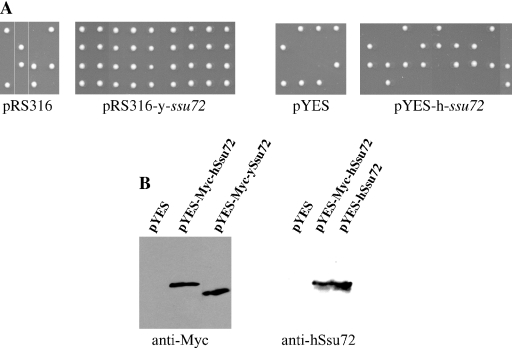
The conservation between human and yeast Ssu72 in primary sequence, binding partners, phosphatase activity and subcellular localization on the one hand, but dynamic regulation and non-essential function in COS7 cells on the other, prompted us to ask whether hssu72 could functionally complement an ssu72 mutation in yeast. To test this possibility, we used yeast RF79977, a diploid strain with one ssu72 allele inactivated by insertion of a neoR marker (30). Sporulation of RF79977 transformants for pRS316-yssu72, a CEN+ vector carrying the yeast ssu72 genomic locus spanning the promoter and coding regions, resulted in the recovery of four viable spores (Figure 8A). Upon replica plating into G418 containing plates to select for the endogenous ssu72−:neoR allele, two spores survived (data not shown), indicating complementation of the ssu72 defect. In contrast, transformants for the human ssu72 gene induced from a galactose responsive promoter (pYES-hssu72) or the parental vectors pYES and pRS316 gave rise to only two viable spores per tetrad (Figure 8A). None of these clones survived when replica plated onto G418-plates, indicating that they contained the yeast wt ssu72 allele. Western blot analysis confirmed that Myc-tagged and untagged human ssu72 alleles were highly expressed in the RF79977 yeast transformants (Figure 8B). Thus, under these conditions, human ssu72 was unable to complement the loss of its homolog in yeast.
Figure 8.
Human ssu72 does not rescue ssu72-lethal mutation in yeast. (A) A ssu72(+/−) diploid yeast (RF79977) transformed with the yeast ssu72 locus (including promoter and coding region in pRS316 vector), the human ssu72 gene under control of a galactose-inducible promoter (pYES) or parental vectors were induced to sporulate. Upon sporulation, ssu72− spores died, giving rise to two ssu72+ colonies. Expression of yeast Ssu72 rescued the ssu72 lethal defect resulting in four colonies per tetrad, two of which contained the ssu72− neoR allele as revealed by replica plating onto G418-plates (data not shown). In contrast, hSsu72 expressing and the parental vectors failed to rescue, yielding only two clones per tetrad, which were both G418-sensitive (i.e. expressing wild-type endogenous yeast ssu72 allele). (B) Western blot analysis of RF79977 yeast ssu72(+/−) transformants grown in the presence of galactose showing high expression of the human Ssu72 in yeast.
DISCUSSION
We here describe the molecular identification of a human homolog of Ssu72 as a novel partner for pRb. We also show that hSsu72 interacts with TFIIB and yeast Pta1. The interaction of pRb with hSsu72 may represent a novel mechanism of transcriptional regulation. Notably, BRCA1 associates with the polyadenylation factor CstF-50 (44,45), suggesting common regulation of RNA processing by tumor suppressors. Interaction of pRb with hSsu72 involves multiple domains but is independent of the LxCxE-binding domain. This suggests a model in which pRb may concomitantly repress certain transcriptional activators by masking their trans-activation domain, recruit chromatin modifying enzymes via the LxCxE-binding domain and sequester hSsu72, thereby inhibiting spurious transcription activation/RNA processing. Interestingly, the B subdomain of pRb shares sequences similarities with the C-terminal domain of TFIIB (46). Thus, pRb may mimic TFIIB and inhibit its interaction with hSsu72. However, in multiple assays including reporter assays and co-localization in co-transfection experiments, Saos-2 cell suppression assays, phosphatase inhibition assays and protein-interference experiments, we were unable to demonstrate any effect of pRb on hSsu72 activity or vice versa. Identification of hSsu72 responsive genes may allow us to test the effect of pRb on this protein.
An insight into where Ssu72 may be critical has emerged from our in situ hybridization analysis. During embryogenesis, mouse ssu72 is highly expressed only during neurogenesis and gut development (Figure 7). Notably, the expression pattern of mouse ssu72 in the nervous system overlaps the expression pattern of Rb during embryogenesis (35,47). Inactivation of several PIC-associated factors, including mTAFII105, dTAFII80, TAFII30 and mTRF2 results in tissue-specific defects (48–51). For example, gene-disruption reveals that TAFII105 is specifically required in ovarian development (52). The generation of a mouse knockout model for ssu72 may identify tissues such as the nervous system and intestine where Ssu72 may play a non-redundant role. Such knockout mice may facilitate the identification of target genes for Ssu72 and allow further investigation into the combined effects of pRb and Ssu72 on transcription and RNA processing.
The phylogenetic conservation among Ssu72 gene products is remarkable (Figure 1). Nonetheless, whereas Ssu72 is essential in yeast, siRNA mediated suppression of its expression in COS7 cells resulted in no obvious effect on proliferation or viability under our experimental conditions (Figures SD-2). Inactivation of mammalian Ssu72 may inhibit the expression of only a subset of genes without compromising cell growth. Alternatively, it may play redundant roles with functionally, though not structurally, related genes. This possibility is supported by the observation that the Fcp1 phosphatase can rescue the ssu72 lethal mutation in yeast (12). In addition, a recently identified novel RNA polymerase II C-terminal Ser-5 phosphatase, SCP (53), may also play a redundant role with Ssu72. In certain cell types, Fcp1, SCP or other phosphatases may be expressed at sufficient levels to substitute for Ssu72 when the latter is absent or experimentally knockdown as in our siRNA experiments.
Despite the conservation in sequence, biological partners and biochemical functions, the human Ssu72 failed to complement an ssu72 mutation in yeast (Figure 8). Perhaps, only certain regions of hSsu72 cannot function in yeast. Indeed, while the first two-thirds of Ssu72 are highly conserved, the C-terminus is diverged across species. In particular, there are eight amino acids in the C-terminus of S.cerevisiae, which are not found in Schizosaccharomyces pombe, Drosophila melanogaster or Homo sapiens (Figure 1B; data not shown). Swapping of the human 30 amino acid at the C-terminus with the yeast C-terminal 45 amino acid or a series of N-to-C and C-to-N human-yeast Ssu72 hybrids may identify functionally conserved/non-conserved regions across these species. Interestingly, human TFIIB, which is also phylogenetically conserved, fails to complement yeast TFIIB mutant; swapping analysis revealed only a small non-conserved subdomain (54).
We found that both endogenous and ectopically expressed mammalian Ssu72 resided primarily in the cytoplasm and to a lesser extent in the nucleus (Figure 6). Since there is no obvious NLS in Ssu72, it is possible that it translocates into the nucleus by a ‘piggyback’ mechanism. However, in preliminary results, we have found that co-transfection with pRb or Myc-Pta1 did not confer obvious nuclear localization upon hSsu72 (data not shown). Alternatively, due to its small size, Ssu72 may freely diffuse into the nucleus and be actively exported into the cytoplasm. Although we found no change in subcellular localization of hSsu72 in response to the Crm1 inhibitor Leptomycin B (data not shown), we cannot rule out the possibility that Ssu72 is regulated by nuclear export via other exportins. Indeed, CRM1-independent nuclear export signals have been mapped in various proteins, such as VHL and the RNA processing factors hnRNPA1, hnRNP K and Hur (55). Usually, cytoplasmic proteins readily translocate into the nucleus when fused to strong NLSs (56,57). In contrast, fusion of hSsu72 to a strong NLS induced nuclear localization in 45% of transfected cells, whereas in the rest NLS-hSsu72 localized in the cytoplasm or throughout the cell (Figure 6E). The retention of NLS-hSsu72 in the cytoplasm of 45–55% of transfected cells suggests the existence of an active mechanism that tethers hSsu72 in the cytosol.
The partial nuclear localization of Ssu72 combined with its interaction with nuclear factors, such as pRb and TFIIB, suggest a nuclear function to this protein. Certain signals, yet to be discovered, may induce complete translocation of Ssu72 into the nucleus and lead to robust activation or repression of gene expression. Alternatively, Ssu72 may exert two independent functions: transcription/RNA processing in the nucleus and as yet to be defined activity in the cytoplasm. In this regards, it is interesting to note that the closest mammalian homolog of Pta1, albeit with very weak similarity, is symplekin, a dual residence protein that localizes in the nucleus as well as in tight junctions, adherens junctions or desmosomes (58). In the nucleus, it is involved in 3′ end processing, bringing CPSF and CStF to the same complex (59,60). Further studies will determine whether Ssu72 has a ‘cytoplasmic function’ and whether its activities in both compartments are orchestrated in the course of specific biological processes, such as neuronal and gut development.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We are grateful to Dr Charlie Boone for advice with the yeast complementation analysis. We thank Drs Rod Bremner, Fred Dick, Xiaoyuan He, Claire L. Moore and Pat O'Connor for reagents and Dr Yaacov Ben-David for comments on the manuscript. L.C.K. was supported in part by a student fellowship from OSTF; E.Z. by a CRS/CIHR New Investigator Scholarship. This work was supported by research grants from the National Cancer Institute of Canada, Canadian Institute for Health Research, and a Breast Cancer Program grant from the Terry Fox Foundation to EZ. Funding to pay the open access publication charges for this article was provided by the Terry Fox Foundation.
REFERENCES
- 1.McCracken S., Fong N., Yankulov K., Ballantyne S., Pan G., Greenblatt J., Patterson S.D., Wickens M., Bentley D.L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 2.Birse C.E., Minvielle-Sebastia L., Lee B.A., Keller W., Proudfoot N.J. Coupling termination of transcription to messenger RNA maturation in yeast. Science. 1998;280:298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- 3.Ryan K., Murthy K.G., Kaneko S., Manley J.L. Requirements of the RNA polymerase II C-terminal domain for reconstituting pre-mRNA 3′ cleavage. Mol. Cell. Biol. 2002;22:1684–1692. doi: 10.1128/MCB.22.6.1684-1692.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentley D. The mRNA assembly line: transcription and processing machines in the same factory. Curr. Opin. Cell Biol. 2002;14:336–342. doi: 10.1016/s0955-0674(02)00333-2. [DOI] [PubMed] [Google Scholar]
- 5.Fong N., Bentley D.L. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev. 2001;15:1783–1795. doi: 10.1101/gad.889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Licatalosi D.D., Geiger G., Minet M., Schroeder S., Cilli K., McNeil J.B., Bentley D.L. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol. Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- 7.Dantonel J.C., Murthy K.G., Manley J.L., Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 8.Kobor M.S., Greenblatt J. Regulation of transcription elongation by phosphorylation. Biochim. Biophys. Acta. 2002;1577:261–275. doi: 10.1016/s0167-4781(02)00457-8. [DOI] [PubMed] [Google Scholar]
- 9.Sun Z.W., Hampsey M. Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential yeast gene encoding a novel protein that affects transcription start site selection in vivo. Mol. Cell. Biol. 1996;16:1557–1566. doi: 10.1128/mcb.16.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pappas D.L., Jr, Hampsey M. Functional interaction between Ssu72 and the Rpb2 subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Cell. Biol. 2000;20:8343–8351. doi: 10.1128/mcb.20.22.8343-8351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu W.H., Pinto I., Chen B.S., Hampsey M. Mutational analysis of yeast TFIIB. A functional relationship between Ssu72 and Sub1/Tsp1 defined by allele-specific interactions with TFIIB. Genetics. 1999;153:643–652. doi: 10.1093/genetics/153.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganem C., Devaux F., Torchet C., Jacq C., Quevillon-Cheruel S., Labesse G., Facca C., Faye G. Ssu72 is a phosphatase essential for transcription termination of snoRNAs and specific mRNAs in yeast. EMBO J. 2003;22:1588–1598. doi: 10.1093/emboj/cdg141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meinhart A., Silberzahn T., Cramer P. The mRNA transcription/processing factor Ssu72 is a potential tyrosine phosphatase. J. Biol. Chem. 2003;278:15917–15921. doi: 10.1074/jbc.M301643200. [DOI] [PubMed] [Google Scholar]
- 14.Krishnamurthy S., He X., Reyes-Reyes M., Moore C., Hampsey M. Ssu72 Is an RNA polymerase II CTD phosphatase. Mol. Cell. 2004;14:387–394. doi: 10.1016/s1097-2765(04)00235-7. [DOI] [PubMed] [Google Scholar]
- 15.Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 16.He X., Khan A.U., Cheng H., Pappas D.L., Jr, Hampsey M., Moore C.L. Functional interactions between the transcription and mRNA 3′ end processing machineries mediated by Ssu72 and Sub1. Genes Dev. 2003;17:1030–1042. doi: 10.1101/gad.1075203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nedea E., He X., Kim M., Pootoolal J., Zhong G., Canadien V., Hughes T., Buratowski S., Moore C.L., Greenblatt J. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J. Biol. Chem. 2003;278:33000–33010. doi: 10.1074/jbc.M304454200. [DOI] [PubMed] [Google Scholar]
- 18.Dichtl B., Blank D., Ohnacker M., Friedlein A., Roeder D., Langen H., Keller W. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell. 2002;10:1139–1150. doi: 10.1016/s1097-2765(02)00707-4. [DOI] [PubMed] [Google Scholar]
- 19.Steinmetz E.J., Brow D.A. Ssu72 protein mediates both poly(A)-coupled and poly(A)-independent termination of RNA polymerase II transcription. Mol. Cell. Biol. 2003;23:6339–6349. doi: 10.1128/MCB.23.18.6339-6349.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H., Dibling B., Spike B., Dirlam A., Macleod K. New roles for the RB tumor suppressor protein. Curr. Opin. Genet. Dev. 2004;14:55–64. doi: 10.1016/j.gde.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 21.Morris E.J., Dyson N.J. Retinoblastoma protein partners. Adv. Cancer Res. 2001;82:1–54. doi: 10.1016/s0065-230x(01)82001-7. [DOI] [PubMed] [Google Scholar]
- 22.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 23.Harbour J.W., Dean D.C. Chromatin remodeling and Rb activity. Curr. Opin. Cell. Biol. 2000;12:685–689. doi: 10.1016/s0955-0674(00)00152-6. [DOI] [PubMed] [Google Scholar]
- 24.Shao Z., Ruppert S., Robbins P.D. The retinoblastoma-susceptibility gene product binds directly to the human TATA-binding protein-associated factor TAFII250. Proc. Natl Acad. Sci. USA. 1995;92:3115–3119. doi: 10.1073/pnas.92.8.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu W., Schneider J.W., Condorelli G., Kaushal S., Mahdavi V., Nadal-Ginard B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 26.Chen P.L., Riley D.J., Chen Y., Lee W.-H. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 27.Thomas D.M., Carty S.A., Piscopo D.M., Lee J.S., Wang W.F., Forrester W.C., Hinds P.W. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol. Cell. 2001;8:303–316. doi: 10.1016/s1097-2765(01)00327-6. [DOI] [PubMed] [Google Scholar]
- 28.Dick F.A., Sailhamer E., Dyson N.J. Mutagenesis of the pRB pocket reveals that cell cycle arrest functions are separable from binding to viral oncoproteins. Mol. Cell. Biol. 2000;20:3715–3727. doi: 10.1128/mcb.20.10.3715-3727.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bunker C.A., Kingston R.E. Transcriptional repression by Drosophila and mammalian Polycomb group proteins in transfected mammalian cells. Mol. Cell. Biol. 1994;14:1721–1732. doi: 10.1128/mcb.14.3.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winzeler E.A., Shoemaker D.D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J.D., Bussey H., et al. Functional characterization of the S.cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 31.Ho A., Li H., Hakem R., Mak T.W., Zacksenhaus E. Coupling of Caspase-9 to Apaf-1 in response to loss of pRb or cytotoxic drugs is cell type dependent. EMBO J. 2004;23:460–472. doi: 10.1038/sj.emboj.7600039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gill R.M., Hamel P.A., Jiang Z., Zacksenhaus E., Gallie B.L., Phillips R.A. Characterization of the human RB1 promoter and of elements involved in transcriptional regulation. Cell Growth Differ. 1994;5:467–474. [PubMed] [Google Scholar]
- 33.St-Pierre B., Flock G., Zacksenhaus E., Egan S.E. Stra13 homodimers repress transcription through class B E-box elements. J. Biol. Chem. 2002;277:46544–46551. doi: 10.1074/jbc.M111652200. [DOI] [PubMed] [Google Scholar]
- 34.Jiang Z., Zacksenhaus E. Coordinated expression of Rb gene family in the mammary gland. Mech. Dev. 2002;119:S39–S42. doi: 10.1016/s0925-4773(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 35.Jiang Z., Zacksenhaus E., Gallie B.L., Phillips R.A. The retinoblastoma gene family is differentially expressed during embryogenesis. Oncogene. 1997;14:1789–1797. doi: 10.1038/sj.onc.1201014. [DOI] [PubMed] [Google Scholar]
- 36.Hatakeyama M., Brill J.A., Fink G.R., Weinberg R.A. Collaboration of G1 cyclins in the functional inactivation of the retinoblastoma protein. Genes Dev. 1994;8:1759–1771. doi: 10.1101/gad.8.15.1759. [DOI] [PubMed] [Google Scholar]
- 37.Jiang Z., Zacksenhaus E. Activation of retinoblastoma protein in mammary gland leads to ductal growth suppression, precocious differentiation, and adenocarcinoma. J. Cell. Biol. 2002;156:185–198. doi: 10.1083/jcb.200106084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao J., Kessler M., Helmling S., O'Connor J.P., Moore C. Pta1, a component of yeast CF II, is required for both cleavage and poly(A) addition of mRNA precursor. Mol. Cell. Biol. 1999;19:7733–7740. doi: 10.1128/mcb.19.11.7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coelho P.S., Bryan A.C., Kumar A., Shadel G.S., Snyder M., Agarwal S., Heyman J.A., Matson S., Heidtman M., Piccirillo S., et al. A novel mitochondrial protein, Tar1p, is encoded on the antisense strand of the nuclear 25S rDNA subcellular localization of the yeast proteome. Genes Dev. 2002;16:2755–2760. doi: 10.1101/gad.1035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalderon D., Richardson W.D., Markham A.F., Smith A.E. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 41.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 42.McManus M.T., Sharp P.A. Gene silencing in mammals by small interfering RNAs. Nature Rev. Genet. 2002;3:737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 43.Harborth J., Elbashir S.M., Bechert K., Tuschl T., Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell. Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- 44.Kleiman F.E., Manley J.L. Functional interaction of BRCA1-associated BARD1 with polyadenylation factor CstF-50. Science. 1999;285:1576–1579. doi: 10.1126/science.285.5433.1576. [DOI] [PubMed] [Google Scholar]
- 45.Kleiman F.E., Manley J.L. The BARD1-CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell. 2001;104:743–753. doi: 10.1016/s0092-8674(01)00270-7. [DOI] [PubMed] [Google Scholar]
- 46.Hagemeier C., Bannister A.J., Cook A., Kouzarides T. The activation domain of transcription factor PU.1 binds the retinoblastoma (RB) protein and the transcription factor TFIID in vitro: RB shows sequence similarity to TFIID and TFIIB. Proc. Natl Acad. Sci. USA. 1993;90:1580–1584. doi: 10.1073/pnas.90.4.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zacksenhaus E., Jiang Z., Chung D., Marth J., Phillips R.A., Gallie B.L. pRb controls cell proliferation, differentiation and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev. 1996;10:3051–3064. doi: 10.1101/gad.10.23.3051. [DOI] [PubMed] [Google Scholar]
- 48.Dunah A.W., Jeong H., Griffin A., Kim Y.M., Standaert D.G., Hersch S.M., Mouradian M.M., Young A.B., Tanese N., Krainc D. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 49.Hiller M.A., Lin T.Y., Wood C., Fuller M.T. Developmental regulation of transcription by a tissue-specific TAF homolog. Genes Dev. 2001;15:1021–1030. doi: 10.1101/gad.869101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martianov I., Fimia G.M., Dierich A., Parvinen M., Sassone-Corsi P., Davidson I. Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol. Cell. 2001;7:509–515. doi: 10.1016/s1097-2765(01)00198-8. [DOI] [PubMed] [Google Scholar]
- 51.Zhang D., Penttila T.L., Morris P.L., Teichmann M., Roeder R.G. Spermiogenesis deficiency in mice lacking the Trf2 gene. Science. 2001;292:1153–1155. doi: 10.1126/science.1059188. [DOI] [PubMed] [Google Scholar]
- 52.Freiman R.N., Albright S.R., Zheng S., Sha W.C., Hammer R.E., Tjian R. Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science. 2001;293:2084–2087. doi: 10.1126/science.1061935. [DOI] [PubMed] [Google Scholar]
- 53.Yeo M., Lin P.S., Dahmus M.E., Gill G.N. A novel RNA polymerase II C-terminal domain phosphatase that preferentially dephosphorylates serine 5. J. Biol. Chem. 2003;278:26078–26085. doi: 10.1074/jbc.M301791200. [DOI] [PubMed] [Google Scholar]
- 54.Shaw S.P., Wingfield J., Dorsey M.J., Ma J. Identifying a species-specific region of yeast TF11B in vivo. Mol. Cell. Biol. 1996;16:3651–3657. doi: 10.1128/mcb.16.7.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fabbro M., Henderson B.R. Regulation of tumor suppressors by nuclear-cytoplasmic shuttling. Exp. Cell Res. 2003;282:59–69. doi: 10.1016/s0014-4827(02)00019-8. [DOI] [PubMed] [Google Scholar]
- 56.Zacksenhaus E., Bremner R., Phillips R.A., Gallie B.L. A bipartite nuclear localization signal in the retinoblastoma gene product and its importance for biological activity. Mol. Cell. Biol. 1993;13:4588–4599. doi: 10.1128/mcb.13.8.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zacksenhaus E., Jiang Z., Hei Y., Phillips R.A., Gallie B.L. Nuclear localization conferred by the pocket domain of the retinoblastoma gene product. Biochim. Biophys. Acta. 1999;1451:288–296. doi: 10.1016/s0167-4889(99)00103-2. [DOI] [PubMed] [Google Scholar]
- 58.Keon B.H., Schafer S., Kuhn C., Grund C., Franke W.W. Symplekin, a novel type of tight junction plaque protein. J. Cell. Biol. 1996;134:1003–1018. doi: 10.1083/jcb.134.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takagaki Y., Manley J.L. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xing H., Mayhew C.N., Cullen K.E., Park-Sarge O.K., Sarge K.D. HSF1 modulation of Hsp70 mRNA polyadenylation via interaction with symplekin. J. Biol. Chem. 2004;279:10551–10555. doi: 10.1074/jbc.M311719200. [DOI] [PubMed] [Google Scholar]
- 61.Lee J.O., Russo A.A., Pavletich N.P. Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7. Nature. 1998;391:859–865. doi: 10.1038/36038. [DOI] [PubMed] [Google Scholar]
- 62.Chen C.F., Chen Y., Dai K., Chen P.L., Riley D.J., Lee W.H. A new member of the hsp90 family of molecular chaperones interacts with the retinoblastoma protein during mitosis and after heat shock. Mol. Cell. Biol. 1996;16:4691–4699. doi: 10.1128/mcb.16.9.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.