Abstract
Species evolutionary relationships have traditionally been defined by sequence similarities of phylogenetic marker molecules, recently followed by whole-genome phylogenies based on gene order, average ortholog similarity or gene content. Here, we introduce genome conservation—a novel metric of evolutionary distances between species that simultaneously takes into account, both gene content and sequence similarity at the whole-genome level. Genome conservation represents a robust distance measure, as demonstrated by accurate phylogenetic reconstructions. The genome conservation matrix for all presently sequenced organisms exhibits a remarkable ability to define evolutionary relationships across all taxonomic ranges. An assessment of taxonomic ranks with genome conservation shows that certain ranks are inadequately described and raises the possibility for a more precise and quantitative taxonomy in the future. All phylogenetic reconstructions are available at the genome phylogeny server: <http://maine.ebi.ac.uk:8000/cgi-bin/gps/GPS.pl>.
INTRODUCTION
Prior to the genome era, compositional signatures (1) or sequence alignments (2) were used to delineate the phylogenetic patterns across organisms. The availability of entire genome sequences has sparked a further development of methods for phylogenetic reconstructions on a genome-wide scale. Following this long tradition in molecular evolution, similar methods were expanded to encompass complete genome information, based on either compositional patterns (3,4) or concatenated alignments of orthologs (5,6).
More recently, methods that exploit the entire gene complement of completely sequenced genomes have been developed. These include phylogenies based on patterns of conservation of gene order (7), gene fusion events (8), gene content (7,9–13), protein folds (12), and average ortholog similarity (14). Only a few of these approaches were successful in resolving difficult cases, such as correctly grouping pathogens having highly reduced genomes with their free-living relatives and clustering Proteobacteria into a monophyletic group (7,9,14). In addition, most of the above mentioned methods are (i) not scalable to hundreds of species (e.g. concatenated alignments), (ii) unable to place correctly species with reduced genomes (13) and (iii) strongly affected by the number and phylogenetic proximity of species (e.g. gene order) (15).
Herein, we define a new composite measure termed ‘genome conservation’, which expresses both the conservation of sequence and gene content between two genomes. This value is derived from the sum of alignment scores between all proteins for every pair of organisms. Larger genomes tend to share more genes, irrespective of their phylogenetic distance (9). Thus, a higher conservation score can result from a higher number of shared genes, rather than from phylogenetic proximity. To counterbalance this effect, the results were normalized before tree reconstruction (see Materials and Methods).
The genome conservation method is naturally adjusted for missing genes and for gene lengths. If a gene is absent in one of the compared genomes, its contribution to the similarity is zero. However, if this absence is a direct result of reductive evolution and difference in genome sizes, absence of this gene would be calibrated by the normalization scheme described below. The gene length is taken into account in the summation of the BLAST scores: longer genes generate greater alignment scores and thus, would be more important contributors than shorter genes. Thus, the final similarity is based on both gene content and average sequence similarity of genes, with the adjustment for gene length.
MATERIALS AND METHODS
To obtain a similarity measure between any pair of genomes, we have compared all proteins using BlastP (16), with an e-value cut-off of e-10, and used the ‘bit-score’ as the measure of similarities between two sequences. The bit-score has the advantages of being independent of the searched database size. Moreover, it does not calibrate for protein length, thus, longer proteins have a greater impact than shorter ones. To eliminate noise created by paralogy in the cases when multiple hits were observed, only the best hit was used (i.e. the most significant sequence similarity). The total number of sequence similarities thus obtained exceeds 25 million pairs for 153 genomes. We will denote the sum of all best hits between genomes A and B as Σ(A,B).
The usage of best BlastP hits, when compared with orthologs, abolishes the issue of ortholog identification, which is still an unresolved problem for datasets of this size. However, it results in non-reciprocal genomic similarities, that is Σ(A,B) ≠ Σ(B,A). To calculate the conservation between genome A and genome B, we used the minimum of the two values, i.e. min(Σ(A,B), Σ(B,A)). To normalize for differences in genome sizes, we calculated the genome conservation distance measure D in two ways (D1 and D2): D1 = 1-S/min(Σ(A,A), Σ(B,B)), or D2 = −ln(S/(√2 * Σ(A,A) * Σ(B,B)/√((Σ(A,A))2 + (Σ(B,B))2))). D1 (9) and D2 (7) correspond to strategies for the transformation and normalization of self-similarity and adjustment for genome sizes, proposed previously. The tree discussed in the text was produced using D2, for consistency with gene content trees, reported to be optimal with this normalization (7). The phylogeny generated using D1 (Supplement 4) produces equally good results.
Gene content trees were generated as described elsewhere (7), using identical data as for the genome conservation tree. Average ortholog similarity was computed as the pairwise score divided by the smallest number of hits between the genomes, divided by 1000 as a scaling factor, or Σ(A,B)/(min(N(A,B), N(B,A)) * 1000), where N(A,B) is the number of hits between genomes A and B. These values were also normalized between 0 and 100, for comparison with other measures (Figure 2).
Figure 2.
Similarity matrices across all completely sequenced organisms, derived from genome conservation (A), gene content (B) and average ortholog similarity (C). Each matrix element represents a pairwise comparison of the corresponding genomes. Genome conservation and gene content were computed using D1 normalization (see Materials and Methods). Species are ordered consistently across the different matrices, sorted according to their position on the genome conservation tree (Supplement 1), and major clades are indicated in (A). The conservation levels in percentages are color-coded, and the values for individual pairwise scores for genome conservation are available (Supplement 3). It is evident that there are three fields of values, seen as lighter blue sub-matrices representing Eukarya, Archaea and Bacteria, from top left to bottom right in (A). The diagonal values of 100% represent self-similarity. Highly similar groups are evident, for instance Escherichia coli strains (red or yellow) and enterobacteria (green), both within γ-proteobacteria. For the comparison of the matrices, see text.
The values of pairwise distances were used to construct a distance matrix; trees were calculated using QuickTree (17). Bootstrap values were generated by resampling the pool of alignment scores between pairs of genomes for 1000 times. This procedure was not applied to the gene content tree, as it requires a jack knife approach rather than genuine bootstrapping (7).
In order to evaluate the conservation within taxonomic units, we computed an average of the conservation values, while eliminating taxonomic over-representation. Within each species, we averaged the pairwise conservation values of its strains. For higher taxonomic ranks (e.g. genus, family), the conservation between each pair of its sub-ranking taxa was averaged, thereby avoiding bias of unequal taxonomic sampling of sequenced species.
RESULTS AND DISCUSSION
Ideally, a species distance metric should enable the reliable inference of phylogeny across variable evolutionary time scales and taxonomic ranges. To assess the performance of genome conservation, we used standard neighbor-joining procedures (17,18) and produced a phylogenetic tree for 153 species with known genomes (19). The obtained tree (Supplement 1) clusters all the major clades consistently with current taxonomic knowledge (20) and is similar to the gene content-based tree (7,9), with a number of important exceptions, described below. Overall, compared to gene content phylogeny (7), a previously proposed and widely accepted method for whole-genome-based phylogenetic reconstruction, genome conservation produces significantly improved results (Supplement 2).
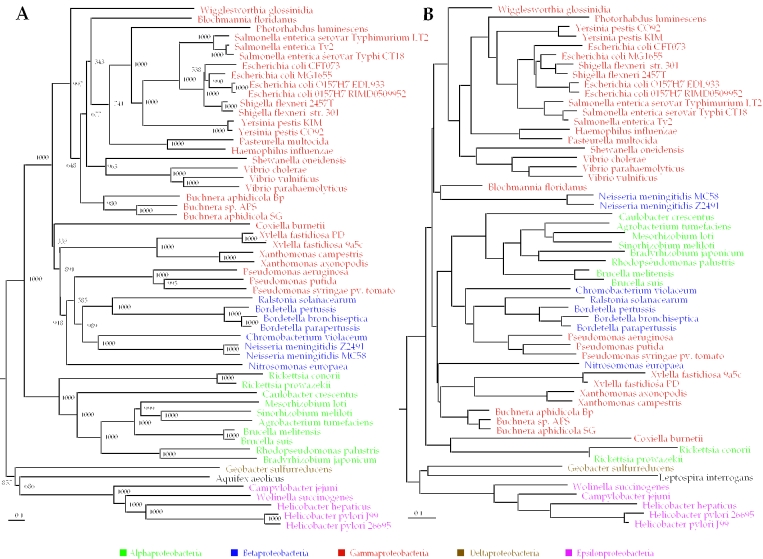
In the genome conservation tree, the alpha-, gamma-, delta- and epsilon proteobacteria form highly supported clades and consistently form a monophyletic proteobacterial clade (Figure 1A). Beta proteobacteria form a clade inside gamma proteobacteria, and there are two inconsistencies with the accepted taxonomy. First, Aquifex aeolicus is placed within Proteobacteria. This placement of Aquifex is also suggested by an independent study (21). Second, the Pseudomonas clade and Nitrosomonas europaea seem to be flipped with respect to each other. Both are coupled to tree nodes with low bootstrap values (686 and 585 out of 1000, respectively). In comparison, the gene content method fails to produce a plausible phylogenetic reconstruction of Proteobacteria (Figure 1B), or other taxa (Supplement 2). Furthermore, the average ortholog similarity approach generates a plausible scenario for evolution of Proteobacteria in the shallow branches of the tree, but fails to group both delta and epsilon subdivisions with other Proteobacteria, defining them as separate deeply branching groups (data not shown).
Figure 1.
Part of the complete tree of life containing the Proteobacteria generated by genome conservation (A) and gene content (B) methods. Classes are color-coded, and the Spirochaetum Leptospira interrogans and deeply branching Aquifex aeolicus are shown in black. Trees were generated using D2 normalization as described in Materials and Methods; the complete tree is available in Supplement 1.
The recognition of abundance of horizontal gene transfer (HGT) among Archaea and Bacteria led to the questioning whether a reliable bacterial phylogeny can possibly be reconstructed (22). Yet, the overall agreement of whole-genome based methods, such as genome conservation, average pairwise sequence similarity, gene content and 16S rRNA phylogenies clearly demonstrates the existence of a consistent bacterial phylogeny (23).
In comparative genomics, it is crucial to accurately measure the evolutionary distance between organisms. Levels of conservation of 16S rRNA sequence may not be sufficient to estimate the evolutionary distance and guarantee species identity, especially in the case of recently divergent organisms (24). Presently, species distances are often estimated in millions of years of divergence (25–27). However, the time of divergence estimated from the ‘molecular clock’ is extremely imprecise (28) and only organisms with fossil records can be dated with some accuracy (29). Another popular form of estimating evolutionary distance is measuring the number of neutral substitutions per site. This technique is appropriate for higher eukaryotes or closely related bacteria; however, saturation in mutations hinders reliable estimations for highly divergent bacterial species (30).
We propose the use of pairwise genome conservation metric as a stable whole-genome based evolutionary measurement to assess conservation between organisms. A pictorial representation of the genome conservation matrix across all presently sequenced organisms readily demonstrates the ability of this species distance metric to define evolutionary relationships at variable taxonomic ranges, from strain variants up to the three domains of life (Figure 2A; the complete set of values is available as Supplement 3).
We have compared the similarity matrices derived from the three principal genome-based phylogeny methods, namely genome conservation, gene content and average ortholog similarity. All matrices evidently contain a strong phylogenetic signal, represented both by the diagonal (self-hits) and various groupings of related taxa (Figure 2). All matrices are also able to clearly separate the three domains of life and delineate closely related groups. These similarity matrices are transformed to distance matrices for the construction of phylogenetic trees (see Materials and Methods), which produce different results.
Massive gene loss in some intracellular parasites, such as Buchnera and Wolbachia, creates an effect where these species share their entire gene content with multiple, distantly related lineages. The similarity estimated from gene content, normalized by the size of the minimal genome, fails to accurately estimate species distances (Figure 2B). The genome conservation approach also suffers from the same effect, however on a significantly lower level (Figure 2A). Finally, average ortholog similarity (Figure 2C) is independent of genome size, and thus resistant to the problem of drastically reduced genome sizes.
However, it is evident that genome conservation allows the detection of phylogenetic groupings at variable taxonomy ranges, from stains up to entire domains of life (Figure 2A). Despite the fact that some of these patterns are also present in the gene content and average ortholog similarity matrices (Figure 2B and C, respectively), their resolution is less pronounced, reflected by a blurred distribution of color-encoded similarity across taxa.
Having demonstrated that the genome conservation metric reflects meaningful evolutionary relationships, we subsequently explored its ability to resolve long-standing arguments in defining the concept of bacterial taxa (31). Using genome conservation as a measure of evolutionary divergence, we investigated how the levels of taxonomical classification in Bacteria correspond to evolutionary distances (Figure 3). Overall, there is a clear decrease in genome conservation at the higher taxonomic ranks. However, the definition of some taxonomic units is not precise, and the ranges of genome conservation for various ranks often overlap (Figure 3). The other two measures, namely, gene content and average ortholog similarity, also exhibit a gradual reduction across increasing taxonomic distances, yet within a narrower value range, which renders them less effective in detecting taxonomic ranks (data not shown). It is worth noting that in all cases, the ranks of genus and family are the least well-defined, according to any genomic similarity measure.
Figure 3.
Genome conservation within bacterial taxonomic ranks. Error bars mark standard deviations. See text for discussion, genome conservation computed using D1 normalization (see Materials and Methods).
In particular, we found that the broadest distribution of genome conservation scores is observed within the genus rank. The similarity between species belonging to the same genus can vary considerably. For example, Mycobacterium tuberculosis and M.bovis are 96% similar whereas Mycoplasma gallisepticum, M.pneumoniae and M.penetrans are only 16% similar. Another example of questionable classification involves Prochlorococcus marinus strains MIT9313 and MED4, which present a challenging case for rRNA-based taxonomy. These two strains are 97% similar in their 16S rRNA sequence (32), while representing distinct genetic populations, with a 2-fold difference in genome size and content (33), as well as specific phenotypic properties. Genome conservation between these strains is only 49%, which is more typical for distances between genera within a family, rather than strains within a species. The genome conservation measurement of classification provides a possibility of a precise and quantitative definition of each taxonomic unit and a guide for future taxonomic classification.
In summary, the genome conservation method uses a genomic perspective of gene content and couples it with sequence divergence at the whole-genome level. Despite the limitation that an entire genome is required in order to place a species in its taxonomic context, this approach can delineate poorly resolved taxa and potentially be coupled with local rRNA-based phylogenies. However, with the new approaches to whole-genome sequencing in environmental genomics (34), the genome conservation approach can provide an unambiguous and consistent classification system for the newly discovered species. The proposed species distance metric provides a clear measure based on sequence divergence for use in comparative genomics and taxonomy.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We would like to thank Russell F. Doolittle (UCSD) for his comments. We also thank Lee Bofkin (EBI) and members of the Computational Genomics Group for valuable discussions. C.A.O. acknowledges additional support from IBM Research. All computations were carried out on the IBM 200-CPU cluster at the EBI. Funding to pay the Open Access publication charges for this article was provided by EMBL.
REFERENCES
- 1.Fox G.E., Stackebrandt E., Hespell R.B., Gibson J., Maniloff J., Dyer T.A., Wolfe R.S., Balch W.E., Tanner R.S., Magrum L.J., et al. The phylogeny of prokaryotes. Science. 1980;209:457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- 2.Doolittle R.F. Similar amino acid sequences: chance or common ancestry? Science. 1981;214:149–159. doi: 10.1126/science.7280687. [DOI] [PubMed] [Google Scholar]
- 3.Kreil D.P., Ouzounis C.A. Identification of thermophilic species by the amino acid compositions deduced from their genomes. Nucleic Acids Res. 2001;29:1608–1615. doi: 10.1093/nar/29.7.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qi J., Wang B., Hao B.I. Whole proteome prokaryote phylogeny without sequence alignment: a K-string composition approach. J. Mol. Evol. 2004;58:1–11. doi: 10.1007/s00239-003-2493-7. [DOI] [PubMed] [Google Scholar]
- 5.Brown J.R., Douady C.J., Italia M.J., Marshall W.E., Stanhope M.J. Universal trees based on large combined protein sequence data sets. Nature Genet. 2001;28:281–285. doi: 10.1038/90129. [DOI] [PubMed] [Google Scholar]
- 6.Rokas A., Williams B.L., King N., Carroll S.B. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature. 2003;425:798–804. doi: 10.1038/nature02053. [DOI] [PubMed] [Google Scholar]
- 7.Korbel J.O., Snel B., Huynen M.A., Bork P. SHOT: a web server for the construction of genome phylogenies. Trends Genet. 2002;18:158–162. doi: 10.1016/s0168-9525(01)02597-5. [DOI] [PubMed] [Google Scholar]
- 8.Enright A.J., Ouzounis C.A. Functional associations of proteins in entire genomes by means of exhaustive detection of gene fusions. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-9-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snel B., Bork P., Huynen M.A. Genome phylogeny based on gene content. Nature Genet. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- 10.Fitz-Gibbon S.T., House C.H. Whole genome-based phylogenetic analysis of free-living microorganisms. Nucleic Acids Res. 1999;27:4218–4222. doi: 10.1093/nar/27.21.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tekaia F., Lazcano A., Dujon B. The genomic tree as revealed from whole proteome comparisons. Genome Res. 1999;9:550–557. [PMC free article] [PubMed] [Google Scholar]
- 12.Lin J., Gerstein M. Whole-genome trees based on the occurrence of folds and orthologs: implications for comparing genomes on different levels. Genome Res. 2000;10:808–818. doi: 10.1101/gr.10.6.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.House C.H., Fitz-Gibbon S.T. Using homolog groups to create a whole-genomic tree of free-living organisms: an update. J. Mol. Evol. 2002;54:539–547. doi: 10.1007/s00239-001-0054-5. [DOI] [PubMed] [Google Scholar]
- 14.Clarke G.D., Beiko R.G., Ragan M.A., Charlebois R.L. Inferring genome trees by using a filter to eliminate phylogenetically discordant sequences and a distance matrix based on mean normalized BLASTP scores. J. Bacteriol. 2002;184:2072–2080. doi: 10.1128/JB.184.8.2072-2080.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolf Y.I., Rogozin I.B., Grishin N.V., Koonin E.V. Genome trees and the tree of life. Trends Genet. 2002;18:472–479. doi: 10.1016/s0168-9525(02)02744-0. [DOI] [PubMed] [Google Scholar]
- 16.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howe K., Bateman A., Durbin R. QuickTree: building huge Neighbour-Joining trees of protein sequences. Bioinformatics. 2002;18:1546–1547. doi: 10.1093/bioinformatics/18.11.1546. [DOI] [PubMed] [Google Scholar]
- 18.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 19.Janssen P., Enright A.J., Audit B., Cases I., Goldovsky L., Harte N., Kunin V., Ouzounis C.A. COmplete GENome Tracking (COGENT): a flexible data environment for computational genomics. Bioinformatics. 2003;19:1451–1452. doi: 10.1093/bioinformatics/btg161. [DOI] [PubMed] [Google Scholar]
- 20.Wheeler D.L., Church D.M., Edgar R., Federhen S., Helmberg W., Madden T.L., Pontius J.U., Schuler G.D., Schriml L.M., Sequeira E., et al. Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res. 2004;32:D35–D40. doi: 10.1093/nar/gkh073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dutilh B.E., Huynen M.A., Bruno W.J., Snel B. The consistent phylogenetic signal in genome trees revealed by reducing the impact of noise. J. Mol. Evol. 2004;58:527–539. doi: 10.1007/s00239-003-2575-6. [DOI] [PubMed] [Google Scholar]
- 22.Doolittle W.F. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 23.Kurland C.G., Canback B., Berg O.G. Horizontal gene transfer: a critical view. Proc. Natl Acad. Sci. USA. 2003;100:9658–9662. doi: 10.1073/pnas.1632870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fox G.E., Wisotzkey J.D., Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 25.Hedges S.B. The origin and evolution of model organisms. Nature Rev. Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- 26.Hedges S.B., Parker P.H., Sibley C.G., Kumar S. Continental breakup and the ordinal diversification of birds and mammals. Nature. 1996;381:226–229. doi: 10.1038/381226a0. [DOI] [PubMed] [Google Scholar]
- 27.Kumar S., Hedges S.B. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 28.Graur D., Martin W. Reading the entrails of chickens: molecular timescales of evolution and the illusion of precision. Trends Genet. 2004;20:80–86. doi: 10.1016/j.tig.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Benton M.J., Ayala F.J. Dating the tree of life. Science. 2003;300:1698–1700. doi: 10.1126/science.1077795. [DOI] [PubMed] [Google Scholar]
- 30.Gribaldo S., Philippe H. Ancient phylogenetic relationships. Theor. Popul. Biol. 2002;61:391–408. doi: 10.1006/tpbi.2002.1593. [DOI] [PubMed] [Google Scholar]
- 31.Cohan F.M. What are bacterial species? Annu. Rev. Microbiol. 2002;56:457–487. doi: 10.1146/annurev.micro.56.012302.160634. [DOI] [PubMed] [Google Scholar]
- 32.Moore L.R., Rocap G., Chisholm S.W. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature. 1998;393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 33.Rocap G., Larimer F.W., Lamerdin J., Malfatti S., Chain P., Ahlgren N.A., Arellano A., Coleman M., Hauser L., Hess W.R., et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 34.Venter J.C., Remington K., Heidelberg J.F., Halpern A.L., Rusch D., Eisen J.A., Wu D., Paulsen I., Nelson K.E., Nelson W., et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.