Abstract
In vitro selection was used to isolate five classes of allosteric hammerhead ribozymes that are triggered by binding to certain divalent metal ion effectors. Each of these ribozyme classes are similarly activated by Mn2+, Fe2+, Co2+, Ni2+, Zn2+ and Cd2+, but their allosteric binding sites reject other divalent metals such as Mg2+, Ca2+ and Sr2+. Through a more comprehensive survey of cations, it was determined that some metal ions (Be2+, Fe3+, Al3+, Ru2+ and Dy2+) are extraordinarily disruptive to the RNA structure and function. Two classes of RNAs examined in greater detail make use of conserved nucleotides within the large internal bulges to form critical structures for allosteric function. One of these classes exhibits a metal-dependent increase in rate constant that indicates a requirement for the binding of two cation effectors. Additional findings suggest that, although complex allosteric functions can be exhibited by small RNAs, larger RNA molecules will probably be required to form binding pockets that are uniquely selective for individual cation effectors.
INTRODUCTION
In the last decade, numerous RNA motifs that bind small ligands have been created by in vitro selection (1–5) or have been found to reside naturally in messenger RNAs (6–10). One of these ligand-binding genetic elements has been shown to be a new class of ribozyme, adding another regulatory mechanism to the repertoire of RNA (11). These findings provide a support for the hypothesis that RNA has sufficient structural diversity, such that highly specific binding pockets can be formed for a variety of ligands. There has been a long-standing interest in creating novel RNA–ligand interactions and in understanding how RNA can fold to form precision binding pockets for these ligands. The importance of understanding these interactions and their bounds has been underscored with the evidence that these interactions are possible in the cellular environment and where they play a role in regulating gene expression.
Various in vitro selection strategies have been applied to generate novel RNA aptamers. One common approach involves immobilizing the target compound on a solid support, then exposing this ligand-decorated surface to populations of random-sequence RNAs. Those RNA sequences that bind to the surface are selectively eluted, amplified, and further enriched by similar iterations (4,12,13). Another strategy, termed allosteric selection, is carried out by appending a randomized RNA domain to a pre-existing ribozyme domain in such a way that binding of the target compound triggers RNA self-processing. Thus, allosteric activation of RNA self-modification is exploited to distinguish ligand-binding RNAs from those that do not bind or remain inactive upon ligand binding (3,14,15). This method is dependent upon a ligand-induced conformational change, causing the catalytic domain of the enzyme to form and activate. Allosteric selection provides the advantage of having the target free in solution with no chemical modifications or associated tethers, allowing the RNA, full spatial access. While this may or may not be important for some targets, it certainly allows for the RNAs to successfully exploit more shape-space, which increases the probability of isolating an RNA receptor with the desired characteristics. Because this approach leads to the simultaneous creation of a binding motif and an allosteric ribozyme, where a cleavage event is a direct read out of binding, it provides a very convenient platform for the study of RNA–small molecule interactions.
The key parameters that define the basic functions of an allosteric ribozyme include the kinetics and thermodynamics of folding, the dynamic range (DR) of the catalytic rate constants exhibited, the DR of the concentrations of ligand triggering ribozyme modulation, and binding specificity (16). Each of these parameters can be manipulated by tailoring the conditions used for selection. Therefore, ribozymes with specific characteristics or those functioning under atypical reaction conditions can be created and studied. While modern organisms provide us with many natural enzymes to study and characterize, in vitro selection provides us with novel enzymes—free from billions of years of biological constraints—that may be quite different from each other in their structure and function. In addition, these in vitro selected molecules tend to be simpler than natural enzymes and receptors, and thus, can serve as model compounds that can better reflect individual biochemical features of interest (17).
In this study, we set out to generate allosteric ribozymes whose activities can be controlled by the selective binding of certain mono- and divalent metal cations. Natural RNAs and proteins rely extensively on metal ions for stability and function, thus, a systematic approach to the study of metal ion interactions with biomolecules could provide much insight into these interactions. While metal ions are important for many nucleic acid enzymes (as cofactors or as a requirement for proper folding) (18), there have been few opportunities to study a discrete metal-binding domain (19). Herein, we describe the use of in vitro selection to generate a series of novel metal-binding allosteric ribozymes and their subsequent characterization.
MATERIALS AND METHODS
DNA preparations
Synthetic DNAs were prepared by standard solid-phase methods (HHMI Biopolymer/Keck Foundation Biotechnology Resource Laboratory, Yale University) and were purified by denaturing (8 M urea) PAGE. Nucleic acids were recovered from the gel by elution with 10 mM Tris–HCl (pH 7.5 at 23°C), 200 mM NaCl, and 1 mM EDTA followed by precipitation with ethanol and resuspension in deionized water. Primer 1 (5′-TAATACGACTCACTATAGGATAATAGCCGTAGGTTGCGAAAGCGACCCTGATGAG-3′) and primer 2 (5′-GAGCTCTCGCTACCGTTTCG-3′) were used to amplify DNA products generated during each round of allosteric selection.
The template DNAs used to prepare the initial population of RNAs (G0 RNAs) by in vitro transcription were generated by a primer extension reaction (100 μl total volume) containing 166 pmol primer 1, 166 pmol primer 3 (5′-GAGCTCTCGCTACCGTTTCG(N)40CTCATCAGGGTCGCTTTCGCAACCTACGGC-3′), 1 mM each of the four deoxyribonucleoside 5′-triphosphates (dNTPs), 50 mM Tris–HCl (pH 7.5 at 23°C), 75 mM KCl, 3 mM MgCl2, 10 mM DTT, and 800 U SuperScript IV reverse transcriptase (SS IV RT, Gibco). Before the addition of enzyme, primers were preannealed by heating to 90°C for 1 min followed by passive cooling to room temperature. Upon addition of enzyme, the extension reaction was incubated at 37°C for 1 h. DNA was precipitated with ethanol and resuspended in deionized water. DNA template of 80 pmol corresponding to ∼5 × 1013 distinct sequence variants, were used to prepare G0 RNA.
RNA preparations
RNAs for each round of allosteric selection were generated by in vitro transcription in a total volume of 50 μl containing 20 pmol of template DNA, 50 mM Tris–HCl (pH 7.5 at 23°C), 10 mM MgCl2, 50 mM DTT, 20 mM spermidine, 2 mM each of the four ribonucleoside 5′-triphosphates (NTPs) and 35 U per μl of T7 RNA polymerase (T7 RNAP) by incubation at 37°C for 2 h. Internally 32P-labeled RNA was prepared by the reaction described above with the addition of trace amounts of [α-32P]UTP. The resulting RNA products were purified by denaturing 10% PAGE and isolated from the gel by elution as described above. The recovered RNA was precipitated with ethanol, resuspended in dH2O, and stored at −20°C until use. G0 RNAs were prepared from the original DNA template as described above, except that the transcription reaction was scaled 4 times larger. Templates for class II and class IV molecules were generated by PCR amplification using plasmid DNAs and primers 1 and 2.
Radiolabeled RNAs carrying a 5′-32P were generated by first removing the 5′-triphosphate moiety of purified transcripts using alkaline phosphatase (AP; Boehringer Mannheim, Indianapolis, IN) and then radiolabeling using T4 polynucleotide kinase (T4 PNK; New England Biolabs, Beverly, MA) and [γ-32P]ATP, both according to the manufacturer's directions. RNA was purified by denaturing 10% PAGE and isolated from the gel and stored as described above.
Allosteric selection
Negative selection reactions were conducted with <10 μM RNA (Figure 1A) in standard buffer containing 50 mM Tris–HCl (pH 7.5 at 23°C) and 20 mM MgCl2. Positive selection reactions were conducted in standard buffer supplemented with 100 μM each of KCl, LiCl, NaCl, RbCl, CaCl2, SrCl2, CdCl2, CoCl2, MnCl2, NiCl2 and ZnCl2. Negative selections were conducted at 23°C for 30 min, except for the first two rounds, which were incubated for ∼12 h. Incubations for positive selection began at 30 min and were reduced as the generations progressed.
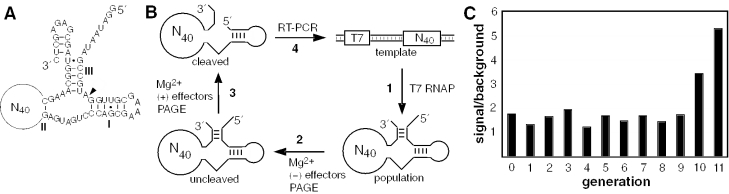
Figure 1.
Allosteric selection of allosteric hammerhead ribozymes that respond to cations. (A) RNA construct used for in vitro selection of allosteric ribozymes. The hammerhead ribozyme core is identical to that described previously (21). A 40 nt random-sequence domain (N40) replaces the majority of stem II of the hammerhead ribozyme core. The site of ribozyme cleavage is indicated by the arrowhead. (B) Scheme for the isolation of cation-dependent ribozymes by allosteric selection. Each population of ribozyme variants is prepared by transcription in vitro (1), subjected to negative (2) and positive (3) selections, and the cleaved RNAs that are enriched for allosteric function are amplified by RT–PCR (4). (C) Progress of the allosteric selection process. Values for signal and background reflect the fraction of RNA cleaved at step 3 in the presence or absence of the cation effector mixture, respectively. A ratio of 1 is expected if the addition of the cation mixture has no effect on ribozyme activity.
Reaction products were separated by 10% denaturing PAGE and both imaged and quantitated by using a PhosphorImager (Molecular Dynamics). In step 2 (Figure 1B), the uncleaved precursor RNAs were recovered from the gel and subjected to a second incubation for positive selection. In step 3, the 3′ cleavage products carrying the majority of the hammerhead ribozyme and the random-sequence domain were isolated from the gel, and the recovered sequences were amplified by reverse transcription followed by RT–PCR. Reverse transcription reactions of 50 μl of total volume contained the 3′ cleavage product recovered from the positive selection reactions, 50 pmol primer 2, 1 mM each of the four dNTPs, 50 mM Tris–HCl (pH 7.5 at 23°C), 75 mM KCl, 3 mM MgCl2, 10 mM DTT and 800 U of SS IV RT, and were incubated at 37°C for 1 h. Products of reverse transcription were precipitated with ethanol and resuspended in a PCR reaction containing 100 pmol each of primers 1 and 2, 0.2 mM dNTPs, 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris–HCl (pH 8.3 at 23°C), 0.01% gelatin and 0.05 U/μl of Taq DNA polymerase. Reactions were subjected to thermocycling until the yield of full-length DNA was determined to be near maximal by using agarose gel electrophoresis, ethidium bromide staining and UV translumination. The population of molecules at G11 was examined in greater detail by cloning and sequencing as described previously (20).
Allosteric ribozyme assays
Allosteric ribozyme assays were conducted by incubating either trace amounts of 5′-32P-labeled RNA or internally 32P-labeled RNA under standard reaction conditions. Reactions were terminated by the addition of an equal volume of gel loading buffer (87 mM Tris base, 89 mM boric acid, 20% sucrose, 0.05% bromophenol blue, 0.05% xylene cyanol, 0.1% SDS and 7.3 M urea) supplemented with EDTA to a final concentration of 40 mM. Reaction products were separated by denaturing 10% PAGE, and were imaged and quantitated as described above.
Rate constants (kobs) were derived by plotting the natural logarithm of the fraction of the total amount of RNA that remains uncleaved versus time and establishing the negative slope of the resulting line. In certain cases, data derived at later times during the reaction were not linear with data collected early in the reaction, most probably because of the presence of a certain fraction of misfolded or otherwise uncleavable RNA (typically <25%). In this event, data derived in the early phase of the reaction time course was used to establish the initial rate constant for RNA cleavage.
RESULTS AND DISCUSSION
Construct design and allosteric selection strategy
Stem II of the hammerhead ribozyme has been shown previously to be a suitable position for aptamer domains that facilitate allosteric regulation (20–22). Specifically, the formation of a G-C base pair in stem II proximal to the catalytic core of the hammerhead ribozyme [nucleotides 10.1 and 11 (23)] is typically required for the ribozyme to exhibit maximal self-cleavage activity. To select for ribozymes that are allosterically activated by metal ions, an initial population was created by largely replacing stem II with a domain of 40 randomized nucleotides (Figure 1A). This design was expected to provide ample length to the random-sequence domain such that novel cationic metal-binding pockets could be formed. Furthermore, the size and the location of the random-sequence domain should permit certain variants in the population to harness shape changes brought about by ligand binding to control the formation of stem II and thus, regulate ribozyme activity.
Iterative rounds of selection were conducted to recover molecules that cleave in the presence of a mixture of monovalent and divalent metal cations (100 μM each of K+, Li+, Na+, Rb+, Ca2+, Sr2+, Cd2+, Co2+, Mn2+, Ni2+ and Zn2+), including 20 mM MgCl2. To avoid propagating molecules that cleave independently of the candidate effectors, a ‘negative selection’ step was added where molecules that cleave in the presence of MgCl2 alone are discarded before the ‘positive selection’ step (Figure 1B). MgCl2 is included in both the negative and positive selection because Mg2+ ions (or certain other select divalent metal ions) are required for the hammerhead ribozyme to exhibit high catalytic activity under low ionic strength conditions (24). Negative selection in the presence of MgCl2 is expected to induce RNA cleavage for those variants wherein the ribozyme domain is folded properly without allosteric induction. In addition, allosteric ribozymes whose effector binding site cannot discriminate between Mg2+ and the potential cation effectors will also be induced to cleave during this step. Thus, the variants that remain uncleaved during the negative selection and undergo cleavage during the positive selection step are amplified by RT–PCR and used to generate the subsequent RNA population by in vitro transcription (Figure 1B).
Each population exhibits a slightly higher level of ribozyme activity in the presence of the cation mixture (Figure 1C). This is most probably due to a general enhancement of hammerhead ribozyme function by increased ionic strength or by the involvement of divalent metal ions other than Mg2+ in structure formation or catalysis. However, the RNA populations at G10 and G11 show a far greater level of activation in response to the addition of the cationic metal mixture. In addition, the fraction of RNA cleaved when the cation mixture is added, substantially increases over that exhibited by the RNA populations from earlier generations. These results suggest that the RNA population at G11 is enriched for ribozymes that respond allosterically to one or more of the metal ion effectors presented during positive selection.
Analysis of cation-dependent allosteric ribozymes
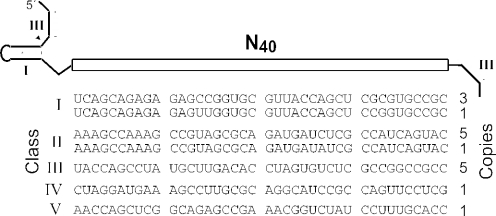
To examine the properties of individual cation-responsive ribozymes, the DNA sequences that gave rise to G11 ribozymes were cloned and sequenced. Five different classes of ribozymes were identified on examining the sequences of nucleotides corresponding to the original N40 domain (Figure 2). Initial tests for activity show that each of these classes exhibits RNA cleavage activity that increases significantly when the transition metals Cd2+, Co2+, Mn2+, Ni2+ or Zn2+ are added to the reaction (data not shown). None of the ribozymes analyzed respond to Ca2+, Sr2+ or to any of the monovalent ions included in the selection. These results suggest that the RNA structures that selectively coordinate transition metals and reject Mg2+ are more common in sequence-space relative to RNA folds that can recognize monovalent ions. Furthermore, it also appears to be challenging for RNA to form binding pockets for Ca2+ or Sr2+ that can also exclude Mg2+.
Figure 2.
Five distinct classes of allosteric ribozymes from G11 that respond to specific divalent metal ions. Clones are classified by the similarity in nucleotide sequence.
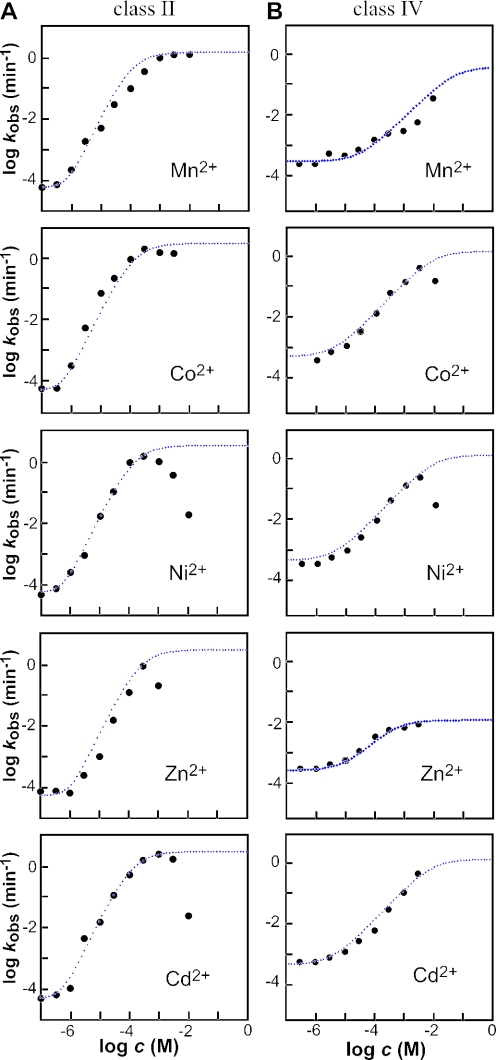
Representatives of classes II and IV were further examined to determine their kinetic characteristics in greater detail. In the absence of any of the five transition metals, the class II ribozyme exhibits its lowest rate constant (kmin) of ∼5 × 10−5 min−1 (Figure 3A). In contrast, the addition of near saturating amounts of any of the five divalent metal effectors induces self-cleavage activity by ∼50 000 fold to yield a maximum rate constant (kmax) of ∼3 min−1. Thus, allosteric activation of this class II RNA permits the ribozyme to attain a rate constant that corresponds well with that expected for the otherwise unaltered hammerhead construct (25,26). In the case of Cd2+ and Ni2+, it is clear that these metals have an inhibitory effect at concentrations in excess of 1 mM. This is most probably due to the non-specific binding of metal ions to RNA that cause disruption of RNA structure and function. This mechanism for inhibition would be different that that observed for Tb3+, which inhibits the hammerhead ribozyme by presumably replacing a catalytically important metal ion (27).
Figure 3.
The dependence of ribozyme activity on the concentration of divalent metal ion effector. Plots of the logarithm of the rate constant for ribozyme cleavage versus the logarithm of the concentration of divalent metal effector for the class II (A) and class IV (B) allosteric ribozymes. Each point represents a kobs value calculated by conducting a time course for ribozyme activity and determining the negative slope of the line from a plot of the natural logarithm of the fraction of RNA remaining uncleaved versus time (min−1). For the class II ribozyme, the dotted lines in the case of Cd2+, Co2+ and Ni2+ represent the expected plot for an ideal ribozyme with a kmax of 3 min−1, a kmin of 4.8 × 10−5 min−1 and two critical metal-binding sites each with an apparent KD of 130 μM, as determined by a variation of the Henderson–Hasselbach equation. The apparent KD, kmax and kmin parameters plotted for Mn2+ are 100 μM, 1.5 min−1 and 6.7 × 10−5 min−1 and for Zn2+ are 220 μM, 3 min−1and 4.8 × 10−5 min−1, respectively. For the class IV ribozyme, dotted lines in the case of Cd2+, Co2+ and Ni2+ represent the expected plots for and ideal ribozyme with an apparent KD of 1 mM, a kmax of 1 min−1 and one critical metal-binding site. The apparent KD, kmax and kmin parameters plotted for Zn2+ are 5 × 10−4, 0.01 min−1 and 2 × 10−4 min−1 and for Mn2+ are 0.07, 0.7 min−1 and 3.5 × 10−4 min−1, respectively.
Each of the five metal ions induces measurable allosteric activation beginning at a concentration of ∼1 μM. The concentration of metals needed to bring about half-maximal ribozyme activity is ∼100 μM in each case. However, it is important to note that these RNAs are not necessarily in thermodynamic equilibrium (see discussion on rapid switching below), which would be needed to establish the true KD values for the metals. Therefore, we define the metal ion concentration needed to induce half-maximal activation as the apparent KD value, implying that the true KD is no poorer than ∼100 μM.
Interestingly, plots of the logarithm of the apparent kobs versus the logarithm of the metal ion concentration all have a slope of ∼2. In other words, for every 10-fold increase in divalent metal ion concentration, there is ∼100-fold increase in the activity of the ribozyme. The simplest explanation for this response to increasing metal ion concentration is that there are two rate-limiting metal-binding sites that must be occupied for activation of the ribozyme. Furthermore, there is no indication from the shape of the plots that these two putative metal-binding sites are significantly different in binding affinity. If the binding affinities were significantly different, a biphasic plot would be expected, wherein two rate-limiting metal-binding events would transition to one rate-limiting metal-binding event. Two metal-binding sites in an allosteric domain of class II ribozymes could have approximately the same binding affinity, or the two metal ions might bind cooperatively. Precedence for this latter effect has recently been established for a natural glycine-binding riboswitch (28).
In contrast, the related plots for the class IV enzyme exhibit a slope of 1, indicating that only one metal-ion binding event is rate limiting. Furthermore, the apparent KD values for most effector metals with this class IV ribozyme appear to be poorer than that for the class II ribozyme. There is no indication upon examining the plots that the allosteric metal-binding site is near saturation. However, if the fully activated ribozyme can approach the maximum rate constant measured for class II ribozymes, then the apparent KD for most effector metals is likely to be no greater than 10 mM. The measured dynamic range of ribozyme activation for class IV ribozymes is ∼1000-fold. Again, certain metals (in this case Co2+ and Ni2+) appear to cause inhibition at high concentrations, and so the actual dynamic range for the ribozyme is likely to be somewhat greater.
It is important to note that the observed rate enhancements for both class II and class IV ribozymes upon addition of the divalent metal effectors is due to an interaction with the allosteric binding domain, and not due to the participation of the metals at the active site of the hammerhead. We have determined that the effects of these metals on the rate constant for a wild-type hammerhead enzyme is negligible up to 100 μM in the presence of 20 mM MgCl2 (data not shown). Thus, the overall enhancement in rate constant that occurs upon saturation with each divalent effector reflects the DR for allosteric activation, and not by replacement of effector metals in the active site of the hammerhead ribozyme.
The class II ribozyme has nearly the best possible DR, considering the constraints of the hammerhead ribozyme and the constraints of the selection conditions used. The maximum rate constant for ribozyme cleavage when fully saturated with divalent effector approaches that for an unmodified hammerhead ribozyme with this simple configuration. The lower boundary of ∼4 × 10−5 min−1 is near the limit of what would be set by trace metals that are likely to be contaminating the MgCl2 solutions used in this study. Considering that commercially available MgCl2 has up to 5 p.p.m. of each of Fe2+ and Mn2+, a 20 mM MgCl2 solution may have up to 200 nM of the metals, which are the activators of this ribozyme. This class II ribozyme typically exhibits measurable allosteric activation at effector cation concentrations of ∼1 μM. If a higher affinity metal-binding domain were to be the aim of a similar allosteric selection, a significantly lower concentration of magnesium would need to be used to reduce the level of trace effector metals in the negative selection reaction. That lower magnesium concentration would translate into a reduced rate for hammerhead self-cleavage, but in theory, should not preclude such a selection from being successful. Alternatively, one might consider beginning with a ribozyme that did not require divalent metal ions for either folding or catalysis.
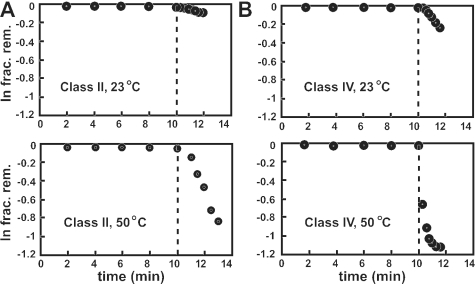
‘Rapid switching’ is the ability of an allosteric ribozyme to remain inactive in a reaction mixture in the absence of an effector, and then rapidly switch into the active conformation upon introduction of its corresponding effector. The representative class II ribozyme is only poorly activated when 1 mM Co2+ is added to an ongoing ribozyme assay that is incubated at 23°C (Figure 4A). The kobs exhibited by the class II ribozyme when Co2+ is added after a 10 min incubation under standard buffer conditions is ∼0.03 min−1, which is ∼70-fold lower than that observed when Mg2+ and the effector metal are added simultaneously. In contrast, the same ribozyme exhibits a more dramatic activation when Co2+ is added to an ongoing ribozyme assay that is incubated at 50°C. At 50°C, the kobs is ∼0.36 min−1, which more closely approximates that observed when the effector metal are added simultaneously at 23°C. Similar results are observed with the representative class IV ribozyme.
Figure 4.
Rapid switching properties of cation-dependent allosteric ribozymes. Class II (A) and class IV (B) ribozymes were assayed for allosteric function at 23 and 50°C. The fraction of precursor RNAs remaining uncleaved at various times is plotted, wherein 1 mM CoCl2 was added to the assay after 10 min (dashed line).
These findings suggest that the inactive state of the enzymes is relatively stable at room temperature, and that most of the molecules are still trapped in one or more inactive structures, even after the addition of an effector. However, when the reaction temperature is elevated, the ribozymes most probably sample alternative structures, including the active state, more rapidly. Although the rate constant for the chemical step of ribozyme cleavage probably changes somewhat with increasing temperature, the primary contributing factor for restoration of ribozyme action is most probably RNA folding. When properly folded, the chemical step for ribozyme cleavage is far faster than the interconversion of misfolded to properly folded states, and the latter rate constant for refolding should then be the main contributor to kobs. The Mg2+ cofactor for the hammerhead ribozyme and the potential cation effectors were added simultaneously in the positive selection step throughout the in vitro selection process. Therefore, the selection was conducted in a way that did not favor the isolation of RNAs, which were capable of rapid switching at room temperature. Thus, it is not surprising that the ribozymes isolated herein do not become fully activated at 23°C when pre-equilibrated with Mg2+.
Structural models for class II and IV ribozymes
An artificial phylogeny was created by performing mutagenesis and additional rounds of selection to help determine the secondary structure and consensus sequence for class II ribozymes. Three rounds of selection were conducted on a population that was generated by introducing mutations into the N40 domain with a degeneracy (29) of 0.18 per position. A number of variant class II RNAs were identified by cloning and sequencing of the resulting population (Figure 5A). All retain 40 nt in the original random-sequence domain, but most acquire mutations spanning positions 1 through 4 and positions 11 through 19. The mutations in these two regions do not co-vary, as would be expected if they participated in forming a base-paired element that was essential for allosteric function. Therefore, these regions are likely to be unstructured and not important for forming the divalent metal-binding domain.
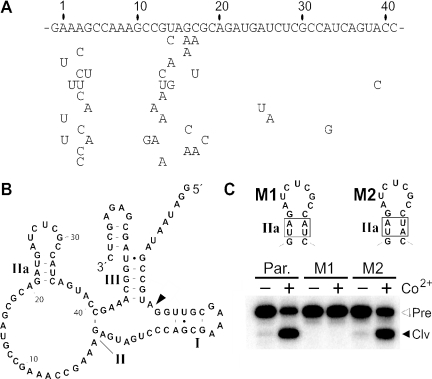
Figure 5.
Class II ribozyme artificial phylogeny and predicted secondary structure. (A) Nucleotide sequence of the N40 domain of the class II ribozyme parent is shown along with specific mutations exhibited by 12 variants recovered after additional mutagenesis and selection was completed. (B) Sequence and secondary structure model of the parental class II ribozyme. (C) Ribozyme assays conducted under standard conditions with 1 mM CoCl2 at 23°C for 5 min with the parental (Par.) class II ribozyme, or on mutants carrying either disruptive (M1) or compensatory (M2) nucleotide changes (boxed). Open and filled arrowheads identify bands corresponding to 5′-32P-labeled full-length precursor (Pre) or the 5′-cleavage product (Clv), respectively.
Secondary structure predictions (data not shown) using the mfold algorithm (30) suggest that the class II ribozyme has only a small stem-loop structure (Figure 5B; stem IIa) and no other obvious base-paired elements within the original N40 domain. Although only weakly supportive, three mutations present at positions 25, 26 and 33 are consistent with the formation of this stem-loop structure. Specifically, mutations at positions 25 and 26 are likely to be tolerated if the loop of the proposed hairpin is not critical for allosteric function. In addition, the A to G mutation at position 33 would permit retention of a G-U wobble pair within the putative stem.
To examine the proposed secondary structure element, class II ribozyme variants were created with disruptive mutations (M1) and corresponding compensatory mutations (M2), which were then tested for activity in response to added cation effector (Figure 5C). Construct M1 that carries a disrupted stem IIa fails to be activated upon the addition of 1 mM Co2+. In contrast, the M2 construct that carries four mutations, but retains base pairing potential, exhibits allosteric activity that is similar to the WT construct.
Similar results were obtained for the class IV ribozyme. The analysis of eight clones isolated after mutagenesis and an additional three rounds of selection provided an artificial phylogeny (Figure 6A). The mutations accrued by these variants are indicative of substantial regions whose sequence composition is not important, while other mutations reflect the existence of critical sequence elements or base-paired structures. For example, several variants carry mutations that strongly implicate base pairing between nucleotide positions 3 though 6 and positions 35 through 38 (Figure 5B). This putative base-paired element would extend the single G-C base pair remaining from stem II of the original hammerhead construct, and therefore might serve as a communication module-like sequence, wherein the binding of metal may serve to stabilize the potential base-paired region and hold the ribozyme in its active conformation (20,22).
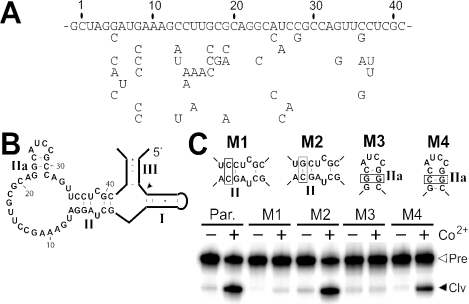
Figure 6.
Class IV ribozyme artificial phylogeny and predicted secondary structure. (A) Primary sequence of the metal-binding domain is shown for ribozymes isolated from reselection on initial populations where 18% degeneracy was introduced into the metal-binding domain. (B) Sequence and secondary structure model of the parental class IV ribozyme. (C) Standard ribozyme assays conducted on parental (Par.) or mutant (M1, M2) ribozymes containing either disruptive (M1, M3) or compensatory (M2, M4) mutations (boxed). Open and closed arrowheads indicated trace radiolabeled full-length precursor or cleaved product, respectively.
This putative base-paired element and another involving nucleotides 22 through 24 and nucleotides 29 through 31 were tested by creating disruptive (M1 and M3) and compensatory (M2 and M4) mutants (Figure 6C). Indeed, the activities of these variants upon the addition of 1 mM Co2+ are consistent with the formation of these predicted secondary structures. These base-paired elements are probably important for the activated form of the ribozyme.
Additional experiments would be needed to determine whether each ribozyme makes use of a single alternative structure to remain inactive when cation effectors are absent, or whether the RNAs remain inactive by sampling a variety of alternative structures. Under closer examination of the binding domain of the class II ribozyme, we noted the possibility to form an alternate base-paired structure where 6 bases from the binding domain (32 through 37) complement the CUGAUG consensus sequence (26) of the hammerhead ribozyme (Figure 5B). This relatively strong pairing interaction might explain the low kmin value of the ribozyme, and its poor rapid switching properties at 23°C (Figure 4A).
Selectivity for cation effectors
In addition to the class II and class IV ribozymes, the representative ribozymes from the remaining three classes show near identical activities with the same five divalent metal ions (Mn2+, Co2+, Co2+, Ni2+ and Zn2+), but remain inactive with the remaining mono- and divalent metals used in the selection (data not shown). Therefore, it appears that RNA might most commonly form binding pockets that are selective for this subset of divalent cations, suggesting that there might be some shared characteristics to this group.
To explore this issue further, we conducted a more complete survey of the effects that other cations have on the class II ribozyme (Figure 7). Two reactions were performed for each metal under otherwise standard assay conditions. One reaction was incubated with 100 μM of the metal being tested, to determine whether the test cation was able to induce the allosteric activation of ribozyme function. A second reaction was conducted with the test metal along with 100 μM Co2+ to determine if the metal were inhibitory to ribozyme function when supplemented with a known allosteric effector. For each cation tested, a concentration of 100 μM was chosen because it is near the apparent KD for the activating metals and should allow for metals with affinities similar to that of the known cation effectors to allosterically induce ribozyme cleavage.
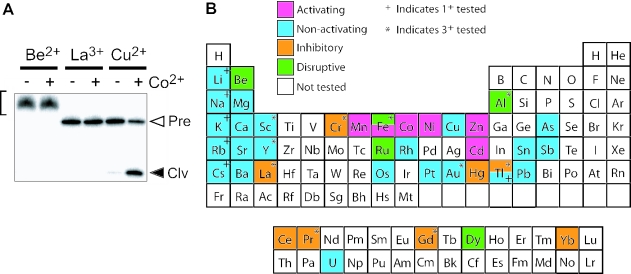
Figure 7.
The effects of various metal cations on class II ribozyme function. (A) Representative PAGE (denaturing 10%) separation of RNAs resulting from the incubation of trace amounts of 5′-32P-labeled class II ribozymes in the presence of 100 μM Be2+, La2+ or Cu2+ under standard reaction conditions at 23°C for 5 min. Reactions were conducted in the absence (−) or presence (+) of 100 μM CoCl2. Open and filled arrowheads identify bands corresponding to 5′-32P-labeled full-length precursor (Pre) or the 5′-cleavage product (Clv), respectively. Bracket identifies bands corresponding to uncleaved precursor RNAs whose gel mobility is altered, presumably due to strong interactions between RNA and the cation being tested. The results for Be2+, La2+ or Cu2+ are typical of that for cations that are disruptive, inhibitory and non-activating, respectively. (B) Summary of the various metal cations tested and their effects on class II ribozyme activity in the absence and presence of 100 μM CoCl2. Results from metal assays with Class II ribozyme are shown. Unless otherwise indicated, the metal form tested is the 2+ ionic species.
There were four outcomes observed from this assay. Metals could (i) activate, (ii) not activate, (iii) inhibit, or (iv) disruptively inhibit the ribozyme. This last effect was determined by examining ribozyme assays by PAGE and identifying those metals (such as Be2+) that did not produce distinct cleavage products, and those that also resulted in anomalous gel mobility for the uncleaved precursor RNAs (Figure 7A). Five of the metals tested (Be2+, Al3+, Fe3+, Ru2+ and Dy2+) disruptively inhibit ribozyme function as determined by the abnormal migration of the RNA during PAGE. This effect is most probably due to an interaction of the metal with the RNA, which is so strong that it forms metal-RNA structures that do not dissociate (or denature) completely, even in 8 M urea.
Interestingly, there were eight additional cations that were inhibitory to the activity of the ribozyme without being disruptive to gel migration (Figure 7B). Two possible reasons for this inhibition are that the metal stabilizes an RNA structure within the allosteric domain that precludes the formation of the active state of the ribozyme, or that the metal binds at or near the active site and more directly inhibits catalysis. The nature of this inhibitory effect was not further investigated in this study.
The only metal in addition to the five described above that activates the ribozyme is Fe2+. These findings highlight the fact that the ribozyme does not make use of a universal divalent binding pocket to bring about allosteric activation. The cation pocket formed by class II ribozymes must therefore be sensitive to one or more characteristics of the six divalent metal ions that induce allosteric activation, or can reject other cations because they each have characteristics that are easily distinguished by RNA.
Upon sorting cationic metals according to several physical properties (Figure 8), some general trends that might be important for triggering ribozyme function become more apparent. For example, the ionic radii of the cations that induce allosteric ribozyme function tend to be smaller than those that do not activate the ribozyme. However, the fact that Cd2+ and Ca2+ have near identical ionic radii but are opposite in their ability to induce ribozyme function, indicates that this physical property cannot be the sole reason for metal ion discrimination. In addition, the range of ionic radii that are exhibited by cations that activate the ribozymes is quite broad, which suggests that the RNA most probably is not discriminating, based on the sub-angstrom differences in cation size. This hypothesis is further supported by the fact that Mg2+ has an ionic radius that is in the middle of the range covered by activating cations, yet the allosteric site of the RNA can discriminate against this metal (as is also the case with several other non-activating cations).
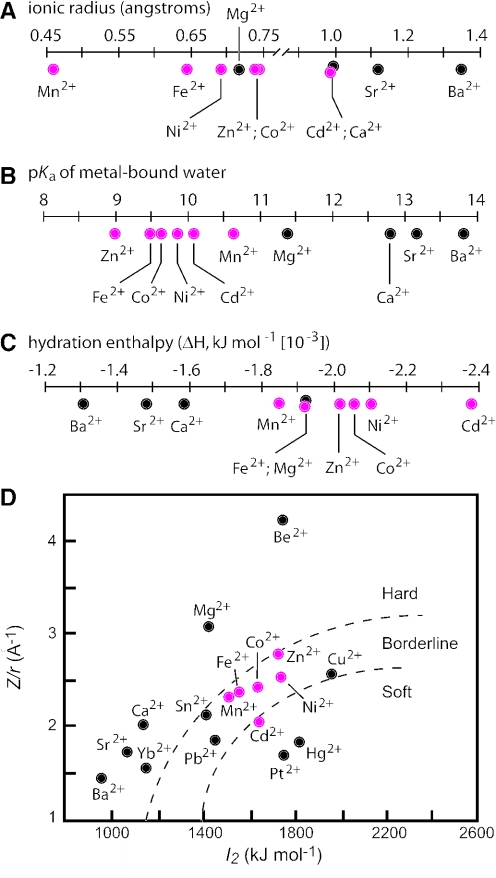
Figure 8.
Some physicochemical characteristics of the metal ions used in this study. Metal ions are classified according to (A) ionic radius (B) pKa of metal-bound water (C) hydration enthalpy and (D) ‘hard’ versus ‘soft’ character. Red and black circles reflect cations that are active or inactive, respectively, as allosteric effectors for the class II ribozyme. Plots of physical parameters were generated from parameters published elsewhere (35).
Similarly, the activating and non-activating metal ions tend to cluster when plotting their hydration enthalpy values. However, Mg2+ falls into the same ranges as the activating metals, which again indicates that this physical property likewise cannot be the exclusive explanation for the observed discrimination. It is quite intriguing that there is a more clear correlation between the activity of the cation and its pKa for a water ligand. Specifically, the ribozyme-active metals have pKa values for metal-bound water ligands of between 9 and 10.6, whereas Mg2+ and other non-activating cations plotted have pKa values of 11.4 or higher. However, it is unlikely that the protonation state of metal-bound water molecules is the critical determinant for function, as the activating metals typically exhibit near identical apparent KD values and activate the ribozyme to the same maximum rate constant. This is unlikely to occur since the different metals would carry water ligands that were protonated to different extents under the assay conditions used, which would be expected to cause differences in binding affinities to RNA.
The preferred coordination number (CN), or the number of ligands that interact with the metal, could also be a factor that is used by the RNAs to distinguish between metal types. Mg2+ generally prefers six ligands, but it will coordinate 4, 5 or 7 ligands with less frequency (31). However, there are no obvious distinctions between Mg2+ and the divalent cations that trigger allosteric function. For example, Mn2+ is very similar in CN character compared with Mg2+, while Zn2+ distributes more readily among CN of 4, 5 or 6. It should be noted cobalt hexamine fails to induce activity with either class II or class IV ribozymes (data not shown), which is consistent with the inner sphere coordination of the metal ion.
The characteristics described previously are also related to the ‘hard’ or ‘soft’ character of metals. In describing metals, hard generally indicates an electron cloud that is difficult to deform and has low polarization potential, while soft generally indicates an easily deformable electron cloud that permits high polarization and favors bonding interactions of a more covalent character. Figure 8D plots Z/r versus I2, the second ionization constant, and is one way of representing hard versus soft nature (32). It is striking from this figure that the Class II switch has a binding pocket that recognizes a cluster of ‘borderline’ metals (and Cd2+ which is considered a soft metal). This characteristic is perhaps best for explaining the high level of discrimination against Mg2+, while allowing a cluster of borderline metals to act as activators. It may be that relatively small binding motifs can be created, which recognize metals based on their hard-soft character.
The commonality of divalent metal binding pockets in RNA
The particular type of metal-binding pockets that presumably are present in each of the classes of allosteric RNAs isolated in this study appear to be far more common in sequence space than are metal-binding pockets that have greater specificity for individual metal ions. First, we find that all classes of allosteric RNAs identified in this study exhibit very similar cation recognition patterns. In addition, we used further rounds of mutation and in vitro selection in an attempt to create ribozymes possessing increased specificities and affinities for individual divalent metals, as has similarly been done with metal-dependent catalytic DNA (33,34). Specifically, a degenerate population of ribozymes was created again by introducing mutations into the binding domain of the class II ribozyme. This mutagenized pool was then subjected to two different selection strategies. The first reselection was conducted in an attempt to enrich for ribozymes that are activated by Ni2+, but not with any of the other divalent metals. Only Ni2+ was used in for positive selection, and a combination of Cd2+, Co2+, Mn2+ and Zn2+ were used in each negative selection to remove from the population variants that retain significant dependency on these cations. The second reselection was performed to enrich for ribozymes with improved affinity by lowering the metal concentration in the selection step.
After many rounds of selection in both cases, the population rebounded only to the level expected for the performance of the parent molecule under the reaction conditions used (data not shown). This suggests that there were not abundant solutions in the sequence space near the class II ribozyme for significantly improved specificity or affinity. It may be that the collection of metals that activate the class II ribozyme have properties similar enough such that small metal-binding pockets made of RNA will not be capable of divalent-specific discrimination, and that the only possible way to discriminate between them would be to have a far more complex binding pocket formed by a larger RNA. Likewise, tighter binding may also be possible only with a more complex fold or with a sequence that is simply too distant in sequence space from the parent molecule to have been found in our reselection effort.
CONCLUSIONS
Our findings demonstrate that it is possible to isolate RNA molecules with activities that are regulated selectively by a set of metal ions. The ribozymes studied here are able to bind Cd2+, Co2+, Mn2+, Ni2+, Zn2+ and Fe2+, while discriminating against Mg2+ and numerous other divalent cations. The five classes of ribozymes described herein do not exhibit an ability to discriminate among this collection of effectors, suggesting that the metal-binding allosteric site recognizes equally well, one or more common features of these metal ions, while excluding cations that lack these features. These ribozymes have binding domains that prefer metals that are borderline in hard–soft characterization.
The results from our more extensive cation survey also reveals that a number of divalent and trivalent metal ions are presumably ignored by the allosteric site of class II ribozymes, although they do not inhibit allosteric activity, when the reaction is supplemented with a cation that is known to induce ribozyme activity. In contrast, 13 divalent and trivalent cations are inhibitory, of which five of these cause a general disruption of RNA structure. It is most likely that this later set of cations (Be2+, Fe3+, Al3+, Ru2+ and Dy2+) would cause similar disruptions in other RNAs, which could define a mechanism for toxicity of these cations at high concentrations in vivo.
The selection described herein highlights a practical advantage of the method of allosteric selection. Immobilization of cations for the purpose of generating aptamers by in vitro selections using affinity-based chromatographic separation would limit the number of coordination sites that could be exploited by the RNA. By presenting the ligand free in solution, the RNA is granted full access to the small cation target. Similar allosteric selections with starting pools of RNA variant based on larger random-sequence domains and precisely tailored reaction conditions could yet lead to the isolation of highly-specific cation sensors, perhaps even for those metal ions that proved in this study to be inhibitory or disruptive to RNA structure and function.
Acknowledgments
We thank Ann Valentine for helpful comments on the manuscript. Funding for this study and to pay the Open Access publication charges for this article was provided by NIH and NSF.
REFERENCES
- 1.Patel D.J., Suri A.K. Structure, recognition and discrimination in RNA aptamer complexes with cofactors, amino acids, drugs and aminoglycoside antibiotics. J. Biotechnol. 2000;74:39–60. doi: 10.1016/s1389-0352(99)00003-3. [DOI] [PubMed] [Google Scholar]
- 2.Brody E.N., Gold L. Aptamers as therapeutic and diagnostic agents. J. Biotechnol. 2000;74:5–13. doi: 10.1016/s1389-0352(99)00004-5. [DOI] [PubMed] [Google Scholar]
- 3.Koizumi M., Soukup G.A., Kerr J.N., Breaker R.R. Allosteric selection of ribozymes that respond to the second messengers cGMP and cAMP. Nature Struct. Biol. 1999;6:1062–1071. doi: 10.1038/14947. [DOI] [PubMed] [Google Scholar]
- 4.Sassanfar M., Szostak J.W. An RNA motif that binds ATP. Nature. 1993;364:550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 5.Lorsch J.R., Szostak J.W. In vitro selection of RNA aptamers specific for cyanocobalamin. Biochemistry. 1994;33:973–982. doi: 10.1021/bi00170a016. [DOI] [PubMed] [Google Scholar]
- 6.Winkler W., Nahvi A., Breaker R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 7.Winkler W.C., Cohen-Chalamish S., Breaker R.R. An mRNA structure that controls gene expression by binding FMN. Proc. Natl Acad. Sci. USA. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nahvi A., Sudarsan N., Ebert M.S., Zou X., Brown K.L., Breaker R.R. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002;9:1043. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 9.Epshtein V., Mironov A.S., Nudler E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl Acad. Sci. USA. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mironov A.S., Gusarov I., Rafikov R., Lopez L.E., Shatalin K., Kreneva R.A., Perumov D.A., Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- 11.Winkler W.C., Nahvi A., Roth A., Collins J.A., Breaker R.R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 12.Schneider D., Tuerk C., Gold L. Selection of high affinity RNA ligands to the bacteriophage R17 coat protein. J. Mol. Biol. 1992;228:862–869. doi: 10.1016/0022-2836(92)90870-p. [DOI] [PubMed] [Google Scholar]
- 13.Klug S.J., Famulok M. All you wanted to know about SELEX. Mol. Biol. Rep. 1994;20:97–107. doi: 10.1007/BF00996358. [DOI] [PubMed] [Google Scholar]
- 14.Robertson M.P., Ellington A.D. In vitro selection of an allosteric ribozyme that transduces analytes to amplicons. Nat. Biotechnol. 1999;17:62–66. doi: 10.1038/5236. [DOI] [PubMed] [Google Scholar]
- 15.Soukup G.A., Emilsson G.A., Breaker R.R. Altering molecular recognition of RNA aptamers by allosteric selection. J. Mol. Biol. 2000;298:623–632. doi: 10.1006/jmbi.2000.3704. [DOI] [PubMed] [Google Scholar]
- 16.Breaker R.R. Engineered allosteric ribozymes as biosensor components. Curr. Opin. Biotechnol. 2002;13:31–39. doi: 10.1016/s0958-1669(02)00281-1. [DOI] [PubMed] [Google Scholar]
- 17.Emilsson G.M., Nakamura S., Roth A., Breaker R.R. Ribozyme speed limits. RNA. 2003;9:907–918. doi: 10.1261/rna.5680603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sreedhara A., Cowan J.A. Structural and catalytic roles for divalent magnesium in nucleic acid biochemistry. BioMetals. 2002;15:211–223. doi: 10.1023/a:1016070614042. [DOI] [PubMed] [Google Scholar]
- 19.Ciesiolka J., Gorski J., Yarus M. Selection of an RNA domain that binds Zn2+ RNA. 1995;1:538–550. [PMC free article] [PubMed] [Google Scholar]
- 20.Soukup G.A., Breaker R.R. Engineering precision RNA molecular switches. Proc. Natl Acad. Sci. USA. 1999;96:3584–3589. doi: 10.1073/pnas.96.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang J., Breaker R.R. Rational design of allosteric ribozymes. Chem. Biol. 1997;4:453–459. doi: 10.1016/s1074-5521(97)90197-6. [DOI] [PubMed] [Google Scholar]
- 22.Soukup G.A., Breaker R.R. Design of allosteric hammerhead ribozymes activated by ligand-induced structure stabilization. Structure. 1999;7:783–791. doi: 10.1016/s0969-2126(99)80102-6. [DOI] [PubMed] [Google Scholar]
- 23.Hertel K.J., Pardi A., Uhlenbeck O.C., Koizumi M., Ohtsuka E., Uesugi S., Cedergren R., Eckstein F., Gerlach W.L., Hodgson R. Numbering system for the hammerhead. Nucleic Acids Res. 1992;20:3252. doi: 10.1093/nar/20.12.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Rear J.L., Wang S., Feig A.L., Beigelman L., Uhlenbeck O.C., Herschlag D. Comparison of the hammerhead cleavage reactions stimulated by monovalent and divalent cations. RNA. 2001;7:537–545. doi: 10.1017/s1355838201002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blount K.F., Uhlenbeck O.C. The hammerhead ribozyme. Biochem. Soc. Trans. 2002;30:1119–1122. doi: 10.1042/bst0301119. [DOI] [PubMed] [Google Scholar]
- 26.Stage-Zimmermann T.K., Uhlenbeck O.C. Hammerhead ribozyme kinetics. RNA. 1998;4:875–889. doi: 10.1017/s1355838298980876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feig A.L., Panek M., Horrocks W.D., Jr, Uhlenbeck O.C. Probing the binding of Tb(III) and Eu(III) to the hammerhead ribozyme using luminescence spectroscopy. Chem. Biol. 1999;6:801–810. doi: 10.1016/s1074-5521(99)80127-6. [DOI] [PubMed] [Google Scholar]
- 28.Mandal M., Lee M., Barrick J.E., Weinberg Z., Emilsson G.A., Ruzzo W.L., Breaker R.R. A glycine-dependent riboswitch that uses cooperative binding to control gene expression in bacteria. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 29.Breaker R.R., Joyce G.F. Inventing and improving ribozyme function: rational design versus iterative selection methods. Trends Biotechnol. 1994;12:268–275. doi: 10.1016/0167-7799(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 30.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glusker J.P., Katz A.K., Bock C.W. Metal ions in biological systems. Rigaku J. 1999;16:8–16. [Google Scholar]
- 32.Silva J.J.R.F.D. The Biological Chemistry of the Elements: The Inorganic Chemistry of Life. New York: Oxford University Press; 1991. [Google Scholar]
- 33.Bruesehoff P.J., Li J., Augustine A.J., III, Lu Y. Improving metal ion specificity during in vitro selection of catalytic DNA. Comb. Chem. High Throughput Screen. 2002;5:327–335. doi: 10.2174/1386207023330264. [DOI] [PubMed] [Google Scholar]
- 34.Wang W., Billen L.P., Li Y. Sequence diversity, metal specificity, and catalytic proficiency of metal-dependent phosphorylating DNA enzymes. Chem. Biol. 2002;9:507–517. doi: 10.1016/s1074-5521(02)00127-8. [DOI] [PubMed] [Google Scholar]
- 35.Richens D.T. The Chemistry of Aqua Ions: Synthesis, Structure, and Reactivity: A Tour Through the Periodic Table of Elements. New York: J. Wiley; 1997. [Google Scholar]