Abstract
Dietary polyphenols present in fruits and vegetables have been reported to manifest beneficial health effects on humans. Polyphenol metabolites including their sulfated derivatives have been shown to be biologically active. Primarily due to the difficulty in preparing regiospecific sulfated polyphenols for detailed investigations, the exact functions of sulfated polyphenols, however, remain unclear. The current study aimed to develop a procedure for the regioselective production of sulfated polyphenols using E. coli cells expressing human cytosolic sulfotransferases (SULTs). Two regioisomers of sulfated genistein were produced by E. coli cells expressing human SULT1A3, SULT1C4, or SULT1E1, and purified using Diaion HP20 resin, followed by high pressure liquid chromatography (HPLC). Structural analysis using mass spectrometry (MS) and nuclear magnetic resonance (NMR) revealed that E. coli cells expressing SULT1A3 preferentially produced genistein 4’-sulfate, whereas E. coli cells expressing SULT1C4 preferentially produced genistein 7-sulfate. To improve the bioproductivity, the effects of several factors including the concentrations of glucose and SO42−, and growth temperature were investigated. The bioproduction procedure established in this study will be valuable for the production of regioselective sulfated polyphenols for use in future studies on their biological functions.
Keywords: Cytosolic sulfotransferase, regioselective sulfation, polyphenol metabolites, sulfated genistein, bioproduction
Introduction
Dietary polyphenols present in various fruits and vegetables have been reported to manifest beneficial health effects on humans (1, 2). These conclusions were mostly based on in vitro studies using aglycone or glycoside forms of flavonoids (3, 4). In human body, however, many phenolic compounds may be subjected to biotransformation via conjugation reactions such as sulfation and glucuronidation, (5, 6). Intriguingly, recent studies indicated that sulfated polyphenols may still be biologically active. For example, like unconjugated resveratrol, resveratrol 3-sulfate and resveratrol disulfate both were capable of inhibiting the E. coli LPS-induced release of TNF-α by macrophages (7). In another study, daidzein 7-sulfate was shown to affect transcriptional and anti-proliferative activities of estrogen receptor-β in cancer cells, whereas daidzein 4’-sulfate and daidzein 7, 4’-disulfate were inactive (8). While the biological activity of these polyphenol metabolites has been noted, the precise function of respective sulfated forms of flavonoids remains unclear due mostly to the difficulty in preparing regiospecific sulfated polyphenol compounds. Regioselective production of sulfated compounds by chemical synthesis is complicated and involves multiple reaction steps. On the other hand, enzymatic production, while allowing for regiospecifically producing sulfated polyphenols, is not suitable for their mass production. Recently, microbial bioconversion using genetically engineered bacteria, particularly E. coli, has been developed for the regioselective glucuronidation or methylation of polyphenol compounds (9, 10). Similar procedures, however, have not yet been developed for the production of sulfated polyphenols.
Sulfate conjugation is known to be involved in the biotransformation and elimination of xenobiotics such as drugs and some dietary compounds, and for the homeostasis of key endogenous compounds such as steroid/thyroid hormones, catecholamines, and bile acids (11-13). The cytosolic sulfotransferases (SULTs) catalyze the transfer of a sulfonate group from the sulfate donor, 3’-phosphoadenosine 5’-phosphosulfate (PAPS), to various compounds containing hydroxyl and/or amino group(s) (14). In higher organisms, PAPS is synthesized in the cytosol by a bifunctional PAPS synthetase (PAPSS) with both the ATP sulfurylase and APS kinase activities (15-17). In bacteria, fungi, algae, and plants, these enzymatic activities are, however, associated with distinct enzymes encoded by separate genes (18, 19). In humans, 13 SULTs that fall into four major gene families (SULT1, SULT2, SULT4 and SULT6) have been identified (20, 21). Of the 13 human SULTs, the seven SULT1 family members, SULT1A1, SULT1A2, SULT1A3, SULT1B1, SULT1C2, SULT1C4 and SULT1E1, have been shown to be capable of catalyzing the sulfation of dietary polyphenols such as isoflavones (e.g., genistein, daidzein) and flavonols (e.g., quercetin, kaempherol) (22). Interestingly, the sulfation of polyphenols occurred in a regioselective manner and that the regiospecificity varied depending on the responsible SULT enzymes. For example, SULT1A3 catalyzes the 4’- sulfation of genistein and daidzein, whereas SULT1E1 catalyzes the sulfation at 7- and 4’-position of those polyphenols (23). A variety of regioisomers of sulfated polyphenols therefore may be produced in human body.
In this communication, we report the establishment of a procedure for the regioselective production of sulfated polyphenols using E. coli cells expressing different human SULTs. Genistein was chosen as a model polyphenol in this study because genistein has been shown to be regioselectively sulfated by human SULTs (23). To our knowledge, this is the first study using genetically engineered bacteria for the regioselective production of sulfated polyphenols.
Materials and Methods
Materials
Genistein was purchased from Wako Pure Chemical Industries (Osaka, Japan). Trifluoroacetic acid (TFA) and sulfatase (from Helix pomatia) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Isopropyl β-D-thiogalactopyranoside (IPTG) was a product of Takara Bio (Shiga, Japan). BL21 Escherichia coli host strain was from Stratagene (La Jolla, CA, USA). pGEX-2TK prokaryotic GST fusion vector was from GE Healthcare Biosciences (Amersham Place, Little Chalfont, England). M9 medium and LB medium were prepared as reported previously (24). All other chemicals were of the highest grade commercially available.
Bioconversion of Genistein using human SULT-expressing E. coli cells
By employing the reverse transcription-polymerase chain reaction technique, we had previously cloned the cDNAs encoding human SULT1A3 (Gene ID: NM_177552), SULT1C4 (Gene ID: NM_006588), and SULT1E1 (Gene ID: NM_005420) (22). These three human SULT cDNAs were individually ligated into the pGEX-2TK prokaryotic expression vector. pGEX-2TK vector harboring SULT1A3, SULT1C4, or SULT1E1 cDNA was transformed into competent E. coli BL21 cells. Transformed cells were grown to OD600 nm=0.6~1.0 in 200 mL LB medium containing 100 μg/mL of ampicillin, and the expression of the recombinant SULT enzyme was induced using 0.25 mM IPTG. Expression of respective SULT was confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). SDS-PAGE was performed on 10% polyacrylamide gels based on Laemmli’s method (25). Following a 9-hour induction at 24°C, the cells were collected by centrifugation and resuspended in 100 mL of M9 medium supplemented with 0.5 mM genistein, 5% dimethyl sulfoxide (DMSO), 50 mM glucose, 2 mM MgSO4, 100 μg/mL ampicillin, and 0.25 mM IPTG, and the cell density was adjusted to OD600 nm = 2.5-3.0. This cell density was empirically chosen based on tolerance to potential toxicity of 5% DMSO needed to dissolve genistein to a 0.5 mM concentration and efficiency in the production of sulfated metabolite. The cell suspension thus prepared was incubated at 24°C or 37°C for 24 h. Afterwards, the cells were spun down and the supernatant was collected for use as described below.
Preparation of Sulfated Genistein
The cultured supernatant prepared as described above was fractionated using 20 mL Diaion HP20 resin (Nippon Rensui, Tokyo, Japan). After fractionation, the resin was washed with H2O, and genistein and its metabolites were eluted using 100 mL methanol. Ten mL H2O was added to the eluate, and the mixture was evaporated and the remaining residue was lyophilized. The freeze-dried powder was dissolved in 2 mL DMSO, and the solubilized genistein and its metabolites were separated by HPLC using a Shimadzu (Kyoto, Japan) Prominence HPLC system fitted with a photodiode array detector (258 nm). A 5 μm Capcell PAK C18 MGII column (4.6 × 250 mm; SHISEIDO, Tokyo, Japan) maintained at 40°C was used, and a gradient elution using 0.05 % TFA in H2O and methanol at a flow rate of 1 mL/min for 55 min was applied. The methanol concentrations used were as follows: 0% (0-5 min), 0-70% (5-40 min), 70-100% (40-45 min), 100% (45-50 min), 100-0% (50-55) min, and 0% (55-60min). The fractionated sample was evaporated. After evaporation, the sample was re-fractionated with 20 mL Diaion HP20 and eluted with methanol, and the eluate was again evaporated. The sample thus purified was dissolved in methanol and analyzed by mass spectrometer. For NMR analysis, the purified sample was dissolved in DMSO-d6 (Aldrich, Chicago, IL, USA).
Mass Spectrometry (MS) Analysis
Samples prepared as described above were analyzed using a Q-Exactive hybrid quadrupole-orbitrap mass spectrometer (Thermo Fisher scientific, Rockford, IL, USA) with a heated electrospray ionization source generated by direct infusion injection using a syringe pump. Data were acquired via Target-MS2 scan. Typical mass spectrometric analysis conditions used were as follows: polarity, negative ionization mode; spray voltage 2.0 kV; sheath gas flow rate, 10; auxiliary gas, 0; sweep gas, 0; heated capillary temperature, 270°C. The resolution was set at 140,000, and the AGC target was 2E5. The maximum injection time was 100 ms, and the normalized collision energy was 20%. The raw data files were analyzed using the Qual browser software in Xcalibur (Thermo fisher scientific, Rockford, IL., USA).
NMR Analysis
The sample dissolved in DMSO-d6 was analyzed using a Bruker Advance 400 instrument (400 MHz, 9.4 T) (Bruker, Karlsruhe, Germany). The chemical shifts (δ) for proton were given in parts per million (ppm) relative to tetramethylsilane (TMS) as an internal standard. For the 1H NMR experiment, 32 transients were acquired with a spectral width of 8000 Hz. All NMR data were processed using XWINNMR (Bruker, Karlsruhe, Germany). The chemical structures of two sulfated forms of genistein were determined based on the NMR data.
Quantitative Analysis of Sulfated Genistein
To quantitatively determine the portion of genistein that was present in sulfated form, the sample was treated with type H-1 sulfatase from Helix pomatia, and the sulfatase-treated sample was compared with untreated control with the same amount of unconjugated genistein. Treatment with sulfatase was performed according to the manufacturer's instructions with a slight modification. The reaction mixture, with a final volume of 100 μL, contained sulfatase (0.1 unit), fractionated genistein sulfate preparation, and 150 mM of Tris-HCl buffer (pH 8.0). Upon incubation for 4 hrs at 37°C, 50 µL of methanol was added and the mixture heated at 98°C for 3 minutes to stop the reaction. The heated sample was centrifuged at 1,5000 × g for 10 min, and the supernatant collected was analyzed by HPLC for unconjugated genistein, compared with a standard curve prepared using commercial genistein.
Results and Discussion
Production of Sulfated Genistein by E. coli Cells Expressing Human SULTs
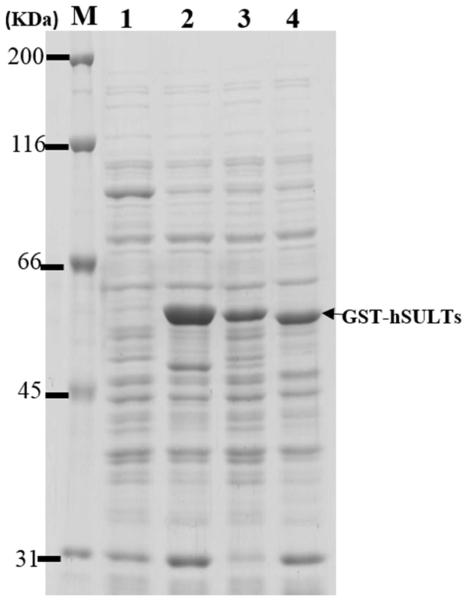
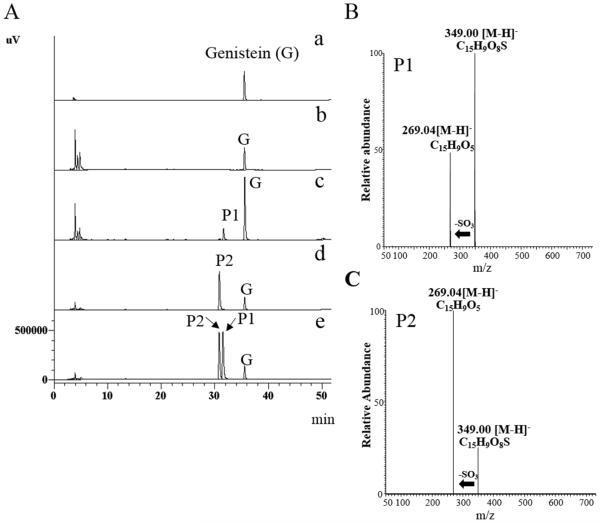
In this study, genistein was chosen as a model compound for investigating the production of regiospelectively sulfated product(s) using E. coli BL21 cells expressing human SULT1A3, SULT1C4, or SULT1E1. A previous study demonstrated that SULT1A3 catalyzed exclusively 4’-sulfation of genistein, whereas SULT1E1 catalyzed both 4’-sulfation and 7-sulfation (23). In the case of SULT1C4, while it has been shown to display strong sulfating activity toward genistein, its regioselectivity in mediating the sulfation of genistein remained unclear (22). It is noted that three other human SULT1 enzymes, SULT1A1, SULT1A2, and SULT1B1, have also been reported to display sulfating activity toward genistein (22). Our preliminary screening revealed, however, that SULT1A1, SULT1A2, and SULT1B1 displayed low regioselectivity toward genistein (date not shown). The remaining human SULT1 enzyme, SULT1C2, on the other hand, has been reported to be incapable of sulfating genistein (22). E. coli BL21 cells transformed with pGEX-2TK-SULT1A3, SULT1C4, or SULT1E1 cDNA were incubated in LB medium and the expression of GST-SULT fusion protein was induced by 0.25 mM IPTG and confirmed by SDS-PAGE (Fig. 1). These recombinant SULTs were expressed in the highly soluble GST fusion protein form in order to avoid inclusion body formation in E. coli cells. The bioconversion of genistein using E. coli expressing human SULT1A3, SULT1C4, or SULT1E1 was performed as described in “Materials and Methods”. Following a 24-hour incubation, the cell suspension was centrifuged, and the supernatant collected was subjected to HPLC analysis. As shown in Figure 2 (A), the aglycon form and two products (designated P1 and P2) derived from genistein were detected. Interestingly, the E. coli cells expressing GST-SULT1A3 preferentially produced P1, whereas E. coli cells expressing GST-SULT1C4 preferentially produced P2. For E. coli cells expressing GST-SULT1E1, approximately equal amounts of P1 and P2 were detected (Fig. 2A). Both P1 and P2 were hydrolyzed to became parent genistein by sulfatase (date not shown), indicating their identity being (sulfated) metabolites of genistein. It is interesting to point out that in the aforementioned bioconversion experiments, sulfated products of genistein were found to accumulate extracellularly, but not intracellularly. Previous studies have revealed that E. coli cells may express 37 multidrug resistance transporters, belonging to resistance-nodulation-division (RND), major facilitator superfamily (MSF), ATP binding cassettle (ABC), and small multidrug resistance (SMR) families, that function to eliminate various xenobiotics and drugs, as well as organic anion compounds (26-29). It is possible that sulfated products of genistein, which are also organic anion compounds, might have been eliminated extracellularly by these transporters. Moreover, once out of the cells, sulfated genistein may be unable to cross the cell membrane of E. coli cells due to the negative charged sulfate group. As a result, sulfated products generated by SULT-expressing E. coli cells all accumulated in the extracellular fraction.
Figure 1. Expression of GST-SULT in E. coli BL21 cells.
E. coli BL21 cells were transformed with pGEX-2TK or pGEX-2TK harboring cDNA encoding human SULT, grown to 0.4 OD600 nm, and induced with IPTG for the expression of GST-SULT (see Materials and Methods). Recombinant GST-SULT expression was verified by SDS-PAGE. Lane M, molecular maker; Lane 1, Mock E. coli BL21 cells; Lane 2, E. coli BL21 cells expressing GST-SULT1A3; Lane 3, E. coli BL21 cells expressing GST-SULT1C4; Lane 4, E. coli BL21 cells expressing GST-SULT1E1.
Figure 2. HPLC analyses of spent E. coli M9 media and MS/MS spectra of purified sulfated products (P1 and P2).
(A) Medium samples were analyzed by HPLC equipped a photodiode array detector (258 nm) and a C18 reverse-phase column. The bioconversion of genistein was performed for 24 h at 24°C using E. coli cells expressing GST-SULT as described in Materials and methods. (a) Genistein; (b) Spent medium (M9) of E. coli BL21 cells transformed with empty pGEX-2TK vector (Mock); (c) Spent medium of E. coli BL21 cells expressing GST-SULT1A3; (d) Spent medium of E. coli BL21 cells expressing GST-SULT1C4; (e) Spent medium of E. coli BL21 cells expressing GST-SULT1E1. Genistein (f) and its metabolic products, P1 and P2, were indicated in each panel. (B) MS/MS spectrum of P1; (C) MS/MS spectrum of P2. MS/MS data were obtained from the 349.00 m/z ion as the precursor for high energy collisional dissociation.
Structural Determination of Sulfated Genistein Produced by E. coli Cells Expressing Human SULT
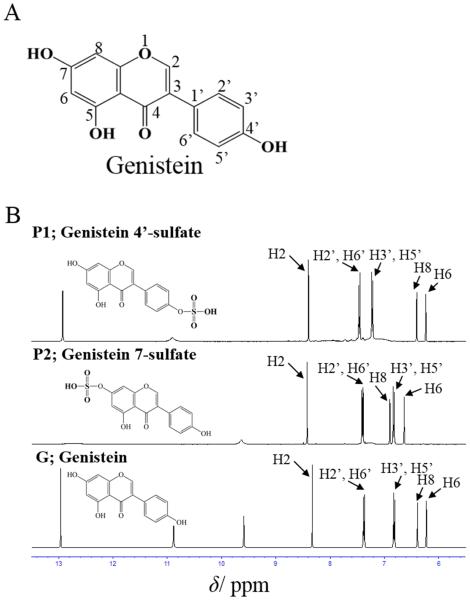
To clarify their chemical structure, P1 and P2, purified as described in “Materials and Methods”, were subjected to the MS and NMR analyses. Negative ion mode full-scan mass spectrometry showed that the highest relative abundance for both P1 and P2 was at m/z 349.00, which corresponded to the theoretical monoisotopic mass for the [M-H]− ion of mono-sulfated genistein (genistein 7-sulfate or genistein 4’-sulfate). As shown in Fig 2 (B, C), further selected ion monitoring of each parent ion and the subsequent high-energy collision dissociation fragmentation of the parent ions produced authentic genistein lacking one SO3− group (m/z 79.96) at m/z 269.04. These results indicated that P1 and P2 were indeed mono-sulfated genistein. To further determine the chemical structure of P1 and P2, 1H NMR analysis was performed (Fig. 3A, 3B). The 1H NMR spectra (400MHz, DMSO-d6) of genistein and its sulfated products, P1 and P2, obtained were: genistein: δ 8.33s (H2), 7.37d (J = 8.6 Hz, H2’ and H6’), 6.81d (J = 8.7 Hz, H3’ and H5’), 6.38d (J = 2.1 Hz, H8), 6.23d (J = 2.1 Hz, H6); P1 (genistein 4’-sulfate): δ 8.4s (H2), 7.46d (J = 8.6 Hz, H2’ and H6’), 7.22d (J = 8.6 Hz, H3’ and H5’), 6.4d (J = 2.0 Hz, H8), 6.24d (J = 2.0 Hz, H6); P2 (genistein 7-sulfate): δ 8.42s (H2), 7.40d (J = 8.6 Hz, H2’ and H6’), 6.9d (J = 2.1 Hz, H8), 6.82d (J = 8.6 Hz, H3’ and H5’), 6.63d (J = 8.6 Hz, H6). Compared with that of genistein, the 1H NMR spectrum of P1 showed a lower field shift (Δδ −0.41) at H3’ and H5’, indicating that the hydroxyl group at 4’ position of genistein was substituted by a sulfate group. On the other hand, the 1H NMR spectrum of P2 showed a lower shift (H6:Δδ −0.4, Η8: Δδ −0.5) at H6 and H8, indicating that the hydroxyl group at 7 position of genistein was substituted by a sulfate group. These 1H NMR spectra were in agreement with those of genistein 4’-sulfate and genistein 7-sulfate prepared by organic synthesis (23). Based on these results, it can be concluded that P1 and P2 were, respectively, genistein 4’-sulfate and genistein 7-sulfate. E. coli cells expressing GST-SULT1A3 therefore preferentially produced genistein 4’-sulfate, whereas E. coli cells expressing GST-SULT1C4 preferentially produced genistein 7-sulfate.
Figure 3. Comparison of 1H NMR spectra of genistein and its sulfated products (P1; genistein 4’-sulfate, P2; geniste 7-sulfate).
(A) Chemical structure of genistein. (B) 1H NMR spectra of P1, P2, and genistein. NMR was performed as describe in Materials and Methods. The signals were assigned based on a previous report (23). Arrows indicate assigned positions of protons.
Effect of Glucose, Inorganic Sulfate, and Temperature on Microbial Production of Sulfated Genistein
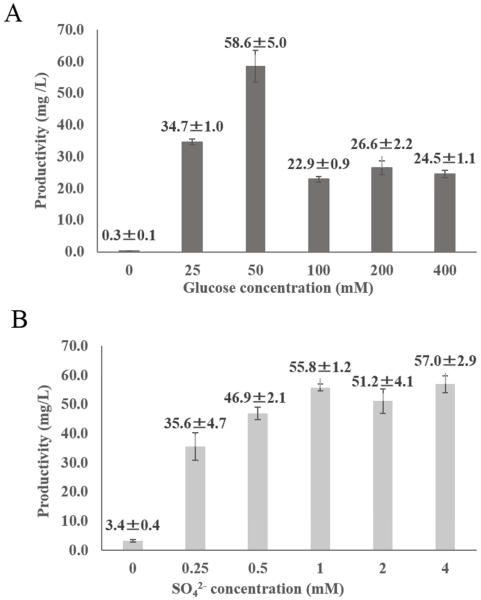
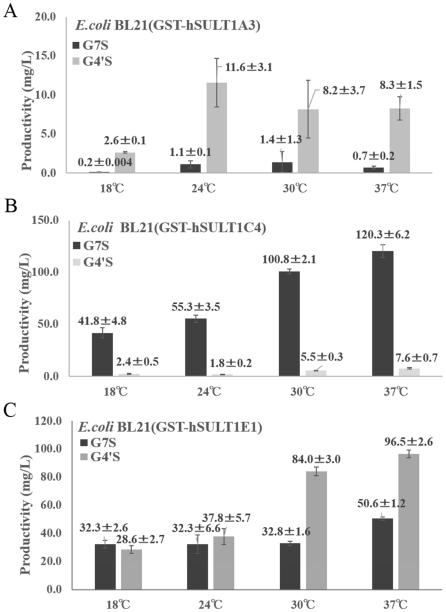
An important issue is whether the culture condition such as the concentration of glucose or inorganic sulfate (SO42−) and temperature may affect the production of sulfated genistein by E. coli cells expressing human SULT. It is known that ATP and SO42− are essential precursors for the synthesis of the sulfonate donor, PAPS (5-7). We therefore decided to investigate the production of sulfated genistein by GST-SULT1C4-expressing E. coli cells incubated in media containing different concentrations of glucose and MgSO4. As shown in Fig. 4 (A, B), in the medium without glucose or MgSO4 the production of genistein sulfate was markedly low. In medium supplemented with glucose, however, the production of genistein 7-sulfate was substantially enhanced, with the optimum concentration of glucose being 50 mM. At elevated concentrations of glucose (100 - 400 mM), the production of genistein 7-sulfate was suppressed. In regard to the effects of inorganic sulfate, the production of genistein 7-sulfate was quite efficient at MgSO4 concentrations above 1 mM. The effects of glucose or MgSO4 on the production of sulfated genistein by GST-SULT1A3- or GST-SULT1E-expressing E. coli cells was examined and similar results were obtained (date not shown). These results, therefore, indicated that both glucose and SO42− are essential factors for the production of sulfated genistein by SULT-expressing E. coli cells. Another factor that may affect the production of sulfated genistein is the temperature at which the E. coli culture is maintained. To find out whether this is the case, the production of sulfated genistein by GST-SULT-expressing E. coli cells at different incubation temperatures (18°C, 24°C, 30°C or 37°C) was examined. As shown in Fig. 5 (A-C), all GST-SULT-expressing E. coli cell cultures showed low productivity on production of each regioselective sulfated genistein at 18 °C. Also, there was not much difference in the production of genistein 4’-sulfate by GST-SULT1A3-expressing E. coli cells incubated at different temperatures ranging 24°C to 37°C (Fig. 5A). In contrast, there was a significant temperature effect on the production of genistein 7-sulfate by GST-SULT1C4-expressing E. coli cells. The production of genistein 7-sulfate by GST-SULT1C4-expressing E. coli cells increased in a temperature-dependent manner in the temperature range from 18°C to 37°C (Fig. 5B). These results indicated that incubation temperature was also an important factor that may affect the production of sulfated compound by GST-SULT-expressing E. coli cells. Taken together, the above-mentioned results indicated that the M9 medium containing 50 mM glucose and 1 mM (or higher concentration of) MgSO4 appeared to be the optimal for the regioselective production of genistein sulfate using this bioconversion method. In addition, using E. coli cells expressing GST-SULT1C4 at 37°C provided the highest efficiency in the regioselective production of genistein 7-sulfate. Similarly, the production of genistein 4’-sulfate was most efficient when GST-SULT1E1-expressing E. coli cells were incubated at 37°C (Fig. 5C). GST-SULT1E1-expressing E. coli cells however, exhibited low regioselctivity in the production of genistein sulfate. Therefore it may be better to use GST-SULT1A3-expressing E. coli cells for the selective production of genistein 4’-sulfate.
Figure 4. Effects of glucose and SO42− on the production of sulfated genistein by E. coli BL21 cells expressing GST-SULT1C4.
(A) Effect of the concentration of glucose (0, 25, 50, 100, 200 or 400 mM) on the production of genistein 7-sulfate by E. coli BL21 cells expressing GST-SULT1C4. The bioconversion of genistein was performed for 24 h at 24°C in the presence of 500 μM genistein, different concentrations of glucose, and 2 mM MgSO4 as described in Materials and methods. (B) Effect of the concentration of SO42− (0, 0.25, 0.5, 1, 2, or 4 mM) on the production of genistein 7-sulfate by E. coli BL21 cells expressing GST-SULT1C4. The bioconversion of genistein was performed for 24 h at 24°C in the presence of 500 μM genistein, 50 mM glucose, and different concentrations of MgSO4 as described in Materials and methods.
Figure 5. Effect of temperature on microbial production of sulfated genistein.
Effect of temperature (18°C, 24°C, 30°C or 37°C) on the production of sulfated genistein by E. coli BL21 cells expressing GST-SULT. (A) Spent medium of E. coli BL21 cells expressing GST-SULT1A3; (B) Spent medium of E. coli BL21 cells expressing GST-SULT1C4; (C) Spent medium of E. coli BL21 cells expressing GST-SULT1E1. Quantitative analysis of sulfated genistein was performed as described in Materials and Methods. Data shown represent means ± SD derived from three determinations.
To summarize, we developed in the current study a bioconversion procedure for the regioselective production of sulfated genistein using E. coli expressing different human SULTs. Glucose, SO42−, and incubation temperature were shown to be key factors that affected the efficiency of the production. The procedure developed may also be used for the regioselective production of other polyphenols (Shimohira et al., unpublished data). In recent years, it has been an interesting issue regarding the physiological function of phase II metabolites of polyphenols, as well as other dietary xenobiotics (23, 30). The regioselective production procedure established in this study will allow for large scale production of sulfated derivatives of polyphenols and other compounds for use in the functional studies in order to reveal in detail their physiological implications.
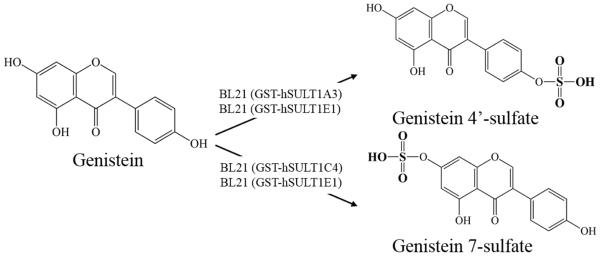
Figure 6. Regioselective production of sulfated genistein by E. coli BL21 cells expressing human GST-SULT.
The figure shows that E. coli BL21 cells expressing GST-SULT1A3 produces preferentially genistein 4’-sulfate; whereas E. coli BL21 cells expressing GST-SULT1C4 produces preferentially genistein 7-sulfate. E. coli BL21 cells expressing hSULT1E1, on the other hand, displayed low regioselectivity on sulfation of genistein. The bioproduction method developed in this study is thus useful for selectively producing regiospecific sulfated polyphenols.
Acknowledgment
This work was supported by JSPS KAKENHI Grant Numbers 25850074 (K.K.), 26450130 (M.S.), 15H04502 (Y.S.), and a grant from National Institutes of Health (Grant No. R03HD071146; to M.C.L).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Kris-Etherton PM, Hecker KD, Bonanome A, Coval SM, Binkoski AE, Hilpert KF, Griel AE, Etherton TD. Bioactive compounds in foods: their role in the prevention of cardiovascular disease and cancer. Am. J. Med. 2002;113:71S–88S. doi: 10.1016/s0002-9343(01)00995-0. [DOI] [PubMed] [Google Scholar]
- 2.Ullah MF, Khan MW. Food as medicine: potential therapeutic tendencies of plant derived polyphenolic compounds. Asian. Pac. J. Cancer. Prev. 2008;9:187–195. [PubMed] [Google Scholar]
- 3.García-Lafuente A, Guillamón E, Villares A, Rostagno MA, Martínez JA. Flavonoids as anti-inflammatory agents: implications in cancer and cardiovascular disease. Inflamm. Res. 2009;58:537–552. doi: 10.1007/s00011-009-0037-3. [DOI] [PubMed] [Google Scholar]
- 4.Crozier A, Jaganath IB, Clifford MN. Dietary phenolics: chemistry, bioavailability and effects on health. Nat. Prod. Rep. 2009;26:1001–1043. doi: 10.1039/b802662a. [DOI] [PubMed] [Google Scholar]
- 5.Manach C, Scalbert A, Morand C, Rémésy C, Jiménez L. Polyphenols: food sources and bioavailability. Am. J. Clin. Nutr. 2004;79:727–747. doi: 10.1093/ajcn/79.5.727. [DOI] [PubMed] [Google Scholar]
- 6.Yang CS, Sang S, Lambert JD, Lee MJ. Bioavailability issues in studying the health effects of plant polyphenolic compounds. Mol. Nutr. Food. Res. 2008;52:S139–S151. doi: 10.1002/mnfr.200700234. [DOI] [PubMed] [Google Scholar]
- 7.Walker J, Schueller K, Schaefer LM, Pignitter M, Esefelder L, Somoza V. Resveratrol and its metabolites inhibit pro-inflammatory effects of lipopolysaccharides in U-937 macrophages in plasma-representative concentrations. Food Funct. 2014;5:74–84. doi: 10.1039/c3fo60236b. [DOI] [PubMed] [Google Scholar]
- 8.Totta P, Acconcia F, Virgili F, Cassidy A, Weinberg PD, Rimbach G, Marino M. Daidzein-sulfate metabolites affect transcriptional and antiproliferative activities of estrogen receptor-beta in cultured human cancer cells. J. Nutr. 2005;135:2687–2693. doi: 10.1093/jn/135.11.2687. [DOI] [PubMed] [Google Scholar]
- 9.Kim SY, Lee HR, Park KS, Kim BG, Ahn JH. Metabolic engineering of Escherichia coli for the biosynthesis of flavonoid-O-glucuronides and flavonoid-O-galactoside. Appl. Microbiol. Biotechnol. 2015;99:2233–2242. doi: 10.1007/s00253-014-6282-6. [DOI] [PubMed] [Google Scholar]
- 10.Kim MJ, Kim BG, Ahn JH. Biosynthesis of bioactive O-methylated flavonoids in Escherichia coli. Appl. Microbiol. Biot. 2013;97:7195–7720. doi: 10.1007/s00253-013-5020-9. [DOI] [PubMed] [Google Scholar]
- 11.Falany CN. Enzymology of human cytosolic sulfotransferases. FASEB J. 1997;11:206–216. doi: 10.1096/fasebj.11.4.9068609. [DOI] [PubMed] [Google Scholar]
- 12.Hemmerich S, Verdugo D, Rath VL. Strategies for drug discovery by targeting sulfation pathways. Drug. Discov. Today. 2004;9:967–975. doi: 10.1016/S1359-6446(04)03261-1. [DOI] [PubMed] [Google Scholar]
- 13.Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, Martin JL, McManus ME. Human sulfotransferases and their role in chemical metabolism. Toxicol. Sci. 2006;90:5–22. doi: 10.1093/toxsci/kfj061. [DOI] [PubMed] [Google Scholar]
- 14.Lipmann F. Biological sulfate activation and transfer. Science. 1958;128:575–580. doi: 10.1126/science.128.3324.575. [DOI] [PubMed] [Google Scholar]
- 15.Robbins PW, Lipmann F. Separation of the two enzymatic phases in active sulfate synthesis. J. Biol. Chem. 1958;233:681–685. [PubMed] [Google Scholar]
- 16.Robbins PW, Lipmann F. Enzymatic synthesis of adenosine-5'-phosphosulfate. J. Biol. Chem. 1958;233:686–690. [PubMed] [Google Scholar]
- 17.Li H, Deyrup A, Mensch JR, Jr., Domowicz M, Konstantinidis AK, Schwartz NB. The isolation and characterization of cDNA encoding the mouse bifunctional ATP sulfurylase-adenosine 5'-phosphosulfate kinase. J. Biol. Chem. 1995;270:29453–29459. doi: 10.1074/jbc.270.49.29453. [DOI] [PubMed] [Google Scholar]
- 18.Leyh TS. The physical biochemistry and molecular genetics of sulfate activation. Crit. Rev. Biochem. Mol. Biol. 1993;28:515–542. doi: 10.3109/10409239309085137. [DOI] [PubMed] [Google Scholar]
- 19.Thomas D, Surdin-Kerjan Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1997;61:503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, Coughtrie MWH. A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics. 2004;14:199–211. doi: 10.1097/00008571-200403000-00009. [DOI] [PubMed] [Google Scholar]
- 21.Freimuth RR, Wiepert M, Chute CG, Wieben ED, Weinshilboum RM. Human cytosolic sulfotransferase database mining: identification of seven novel genes and pseudogenes. Pharmacogenomics. J. 2004;4:54–65. doi: 10.1038/sj.tpj.6500223. [DOI] [PubMed] [Google Scholar]
- 22.Pai TG, Suiko M, Sakakibara Y, Liu MC. Sulfation of flavonoids and other phenolic dietary compounds by the human cytosolic sulfotransferases. Biochem. Biophys. Res. Commun. 2001;5:1175–1179. doi: 10.1006/bbrc.2001.5316. [DOI] [PubMed] [Google Scholar]
- 23.Nakano H, Ogura K, Takahashi E, Harada T, Nishiyama T, Muro K, Hiratsuka A, Kadota S, Watabe T. Regioselective monosulfation and disulfation of the phytoestrogens daidzein and genistein by human liver sulfotransferases. Drug Metab Pharmacokinet. 2004;19:216–226. doi: 10.2133/dmpk.19.216. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 1989. [Google Scholar]
- 25.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Rouch DA, Cram DS, DiBerardino D, Littlejohn TG, Skurray RA. Efflux-mediated antiseptic resistance gene qacA from Staphylococcus aureus: common ancestry with tetracycline- and sugar-transport proteins. Mol. Microbiol. 1990;4:2051–2062. doi: 10.1111/j.1365-2958.1990.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 27.Wilmut I, Beaujean N, Sousa PA, Dinnyes A, King TJ, Paterson LA, Wells DN, Young LE. Somatic cell nuclear transfer. Nature. 2002;419:583–586. doi: 10.1038/nature01079. [DOI] [PubMed] [Google Scholar]
- 28.McMurry L, Petrucci RE, Jr., Levy SB. Active efflux of tetracycline encoded by four genetically different tetracycline resistance determinants in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1980;77:3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamazaki S, Sakakibara H, Takemura H, Yasuda M, Shimoi K. Quercetin-3-O-glucronide inhibits noradrenaline binding to α2-adrenergic receptor, thus suppressing DNA damage induced by treatment with 4-hydroxyestradiol and noradrenaline in MCF-10A cells. J. Steroid. Biochem. Mol. Biol. 2014;143:122–129. doi: 10.1016/j.jsbmb.2014.02.014. [DOI] [PubMed] [Google Scholar]