Abstract
The nucleosome formation potential of introns, intergenic spacers and exons of human genes is shown here to negatively correlate with among-tissues breadth of gene expression. The nucleosome formation potential is also found to negatively correlate with the GC content of genomic sequences; the slope of regression line is steeper in exons compared with noncoding DNA (introns and intergenic spacers). The correlation with GC content is independent of sequence length; in turn, the nucleosome formation potential of introns and intergenic spacers positively (albeit weakly) correlates with sequence length independently of GC content. These findings help explain the functional significance of the isochores (regions differing in GC content) in the human genome as a result of optimization of genomic structure for epigenetic complexity and support the notion that noncoding DNA is important for orderly chromatin condensation and chromatin-mediated suppression of tissue-specific genes.
Genomes of warm-blooded vertebrates consist of the isochores, relatively homogenous regions differing in GC content [reviewed in (1–4)]. There are various hypotheses explaining the appearance of isochores. The main controversy is mutation bias (in a broad sense, including repair bias and biased gene conversion) versus selection (5–13). One selectionist explanation suggests that selection was for the increase of thermostability of DNA helix (caused by GC-enrichment) in certain genomic regions as a result of adaptation to elevated temperature in warm-blooded vertebrates (2,14). Another hypothesis implies that there was an optimization of physical properties of DNA molecule for active transcription in the GC-rich regions and for gene suppression in the GC-poor regions in the more complexly organized vertebrates (‘epigenetic optimization’) (15–17).
The ‘thermostability’ hypothesis was criticized because no correlation was found between ambient temperature and GC content in bacteria and poikilothermal vertebrates (18–21), and because thermostability of genomic sequences of warm-blooded vertebrates increases with the elevation of GC content slower than in random sequences, which indicates that the increase of thermostability is not a leading force for GC-enrichment (15,16). At the same time, bendability of genomic sequences of warm-blooded vertebrates increases faster with the elevation of GC content and curvature drops faster than in random sequences (15,16). The former property is believed to associate with open chromatin while the latter, with condensed chromatin (22–26), which is in agreement with the ‘epigenetic optimization’ concept. Also, the thermostability of corresponding RNA/DNA and RNA/RNA duplexes increases faster with the elevation of GC content (in contrast to thermostability of DNA/DNA duplex), which also suggests a possible involvement of transcription and/or antisense RNA-mediated regulation into the causes of GC-enrichment (27). The GC-rich sequences are predominantly found in the central, open chromatin of the interphase nuclei of warm-blooded vertebrates, whereas the GC-poor sequences, in the peripheral, more compact chromatin (28). The ‘epigenetic optimization’ model also gained certain support in the fact that housekeeping genes tend to be located in the GC-rich isochores, whereas tissue-specific genes, in the GC-poor isochores (17,29,30).
Recently, it was found that the highly and broadly expressed (i.e. expressed in many tissues) genes are shorter, both in their intronic and coding sequences than genes expressed in tissue-specific fashion (31–34). Because transcription and translation are energetically costly, this shortness was interpreted as a result of selection for economy (31–33). However, it was argued that it is not that housekeeping genes become shorter but that tissue-specific genes are getting longer (34). The tissue-specific proteins have more complex architectures that explain the increase in their length (34). Not only introns, but also the intergenic spacers around the highly expressed and housekeeping genes are similarly (or even more regularly) shorter compared with tissue- and development-specific genes (33,34). As a result, the ‘gene nest’ proportion (ratio of intra- plus intergenic noncoding to coding DNA lengths) negatively correlates with the breadth and level of gene expression (34). Therefore, the greater amount of intra- and intergenic noncoding DNA, in which tissue- and development-specific genes are embedded, was supposed to be involved in chromatin-mediated suppression of these genes (which suggests another dimension to the ‘epigenetic optimization’ concept). At the same time, if GC content of intronic and intergenic sequences was controlled using multifactor statistical analyses, the correlation between the ‘gene nest’ proportion and expression breadth disappeared (34). This fact indicates that GC content is an important property of the ‘gene nest’. Here, I study the relationships between expression breadth, GC content and nucleosome formation potential of exons and noncoding sequences within and around a given gene in the human genome.
MATERIALS AND METHODS
The data on the expression levels of human genes were taken from the Gene Expression Atlas (35). They present the results of oligonucleotide microarray experiments. The uniform platform for all tissues was Affymetrix U95A. Only probes that presented the characterized genes (i.e. those with links to the RefSeq database) were used, with signals from probes on the chip corresponding to the same gene being averaged (with total of 7708 genes). Only data for normal tissues were used, samples and replicates representing the same tissue were averaged (with total of 32 tissues). As was recommended, a gene was regarded as expressed if its signal level exceeded a conservative threshold of 200 arbitrary units (35). It should also be noted that there is a strong correlation (Spearman r = 0.89, P < 10−8) between the expression breadth (number of tissues where a given gene is expressed) and the expression level averaged over all tissues studied (34). Genomic sequences were extracted from the RefSeq database (36). The intronic sequences were found for 6874 genes, and the intergenic sequences (both upstream and downstream ones), for 5104 genes. In the case of alternative splicing variants, the longest coding sequence was taken. The average length and GC content of upstream and downstream intergenic sequences were taken as the corresponding values of the intergenic spacer for a given gene. The nucleosome formation potential of nucleotide sequences was determined using the method by Levitsky and co-workers (37–39), constructed and verified on the basis of large experimental datasets of nucleosome positioning sequences (40–42). The nucleosome formation potential was averaged over each sequence length; for introns, exons and intergenic spacers it was also weight-averaged for each gene.
RESULTS AND DISCUSSION
The nucleosome formation potential shows a negative correlation with GC content (Figure 1), which is in accordance with the expectations from the physical properties of DNA helix associated with GC content (16). It was previously reported that GC content of both intronic and intergenic sequences should be included as covariate into multifactor statistical analyses for the correlation between the ‘gene nest’ proportion and expression breadth to disappear completely (34). This fact was interpreted in the sense that either both intronic and intergenic GC content taken together better reflects the isochore affiliation of a given gene (i.e. the regional effect) or that the local variation of GC content among intronic and intergenic sequences is also an important property of the ‘gene nest’. The negative correlation between nucleosome formation potential and GC content (Figure 1) suggests that the combined impact of both intergenic and intronic GC content on the expression breadth is a local effect related to nucleosome formation.
Figure 1.
Regression of nucleosome formation potential on GC-content in the human genome. (A) Exons (intercept = 4.6 ± 0.1, slope = −0.08 ± 0.00, r = −0.75, P < 10−8). (B) Introns (intercept = 2.3 ± 0.0, slope = −0.02 ± 0.00, r = −0.78, P < 10−8). (C) Intergenic spacers (intercept = 2.4 ± 0.0, slope = −0.03 ± 0.00, r = −0.73, P < 10−8). Dashed lines, confidence limits; dotted lines, prediction limits, both for P = 0.95; and a.u., arbitrary units.
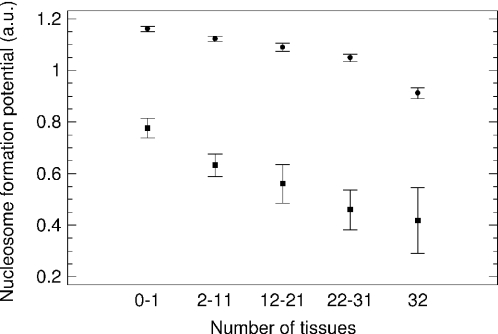
Introns of broadly expressed genes show a lower nucleosome formation potential compared with introns of tissue-specific genes (Figure 2). Intergenic spacers show a similar picture (not shown in Figure 2 because they mostly overlap with introns). Exons show a similar trend with introns' (and intergenic spacers') reduction of nucleosome formation potential with increasing gene expression breadth. However, their mean potential is lower than potential of introns and intergenic spacers for all gene expression groups (Figure 2). In the intergenic spacers, the nucleosome formation potential drops similarly to introns with the increase of GC content, whereas in exons it drops significantly faster (confer slopes in Figure 1). It is noteworthy that in the GC-poor sequences, exons have a similar or even a higher nucleosome formation potential compared with noncoding DNA, but the picture is reversing with the elevation of GC content (Figure 1). In genes located in the heavy isochores [i.e. with GC content above 50% (1)], exons have a higher GC content than introns by ∼5% (absolute percentage) (16). Therefore, judging by the data shown in Figure 1, introns seem to be the main source of nucleosome formation potential in the genes of heavy isochores. A greater scatter of exon points compared with noncoding sequences (Figure 1) can be explained by the shorter length of exons (and thus, higher statistical noise) and/or by exons' informational load that may interfere with requirements of nucleosome formation.
Figure 2.
The nucleosome formation potential in human exons (squares) and introns (circles) of genes expressed in a different number of tissues (ANOVA and Kruskal–Wallis, in both cases P < 10−8). The data for intergenic spacers are not shown because they mostly overlap with introns; a.u., arbitrary units.
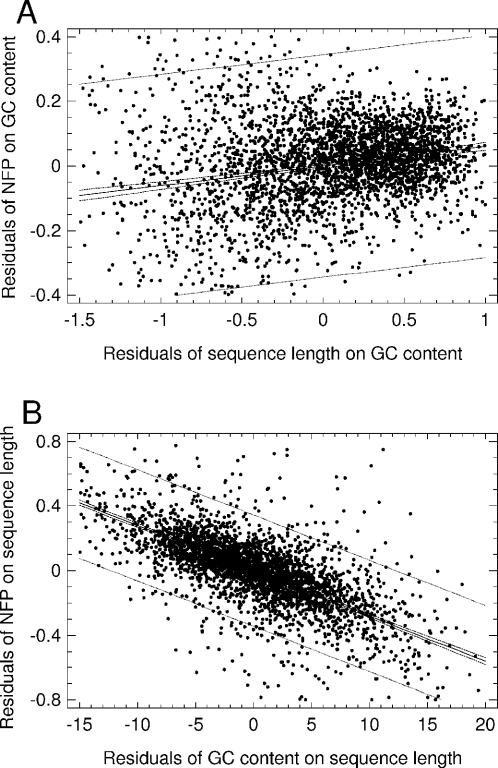
The nucleosome formation potential correlates positively with intronic and intergenic sequence lengths (for log-transformed lengths: for introns, r = 0.39, P < 10−8; for intergenic spacers, r = 0.43, P < 10−8). A part of this correlation is independent of GC content (Figure 3A). (For exons, the partial correlation between nucleosome formation potential and sequence length independently of GC content was not significant.) In turn, the negative correlation between nucleosome formation potential and GC content is partially independent of sequence length (Figure 3B). These facts indicate that both the GC content and the length of introns and intergenic spacers are related to chromatin condensation in the human genome.
Figure 3.
Multiple regression analysis of nucleosome formation potential in human intergenic spacers. (A) Regression on (log-transformed) sequence length at fixed GC-content (partial r = 0.23, P < 10−6; for introns, the picture was similar, partial r = 0.14, P < 10−6). (B) Regression on GC-content at fixed sequence length (r = −0.68, P < 10−8; for introns, partial r = −0.74, P < 10−8). Dashed lines, confidence limits; dotted lines, prediction limits, both for P = 0.95; and NFP, nucleosome formation potential.
It was previously reported that nucleosome formation potential is higher in introns compared with exons; however, no statistical analysis was made, and the authors' histograms of nucleosome formation potential for introns and exons mostly overlapped [Figure 1a in (38)]. This overlap is probably due to a large number of GC-poor introns and exons in the authors' dataset. It was also found (on a limited dataset of ∼200 genes) that nucleosome formation potential is higher in the promoter regions of tissue-specific genes compared with the promoters of housekeeping genes (37). The data presented here show that the same is true not only for the promoters but also for the bulk of noncoding DNA, which suggests its involvement in chromatin-mediated suppression of tissue-specific genes. It is now well recognized that epigenetic mechanisms, operating on the large domains rather than on individual promoters, are used in maintaining and stably transmitting chromatin states through the cell cycle (43,44). A high local concentration of nucleosomes is necessary for the higher-order chromatin condensation, which is a distinct level of transcriptional regulation (44,45). The present data also help explain why GC content rises near gene transcription start site [Figure 1A in (46)]. The elevation of GC content is associated with the reduction of nucleosome formation potential that should facilitate the binding of transcription factors (after the higher levels of chromatin condensation are unfolded).
It was long argued that noncoding DNA might be necessary for correct chromatin structure because exons are under strong selection pressure for informational content (38,47–49). It was even shown in several cases that after experimentally removing the introns, genes lose the ability to form nucleosomes (50,51). The present data suggest that both the length and the GC content of noncoding DNA (and thus the isochoric structure) in the human genome can be relevant to chromatin-mediated suppression of tissue-specific genes.
It should be noted that the existence of the isochores in the genomes of warm-blooded vertebrates, which was initially discovered using the CsCl density gradient ultracentrifugation, was questioned after the completion of the human genome (52). However, the statistical test for isochoric structure used in (52) was criticized, and the later works confirmed the existence of isochores (53–55). While too complicated tests may be prone to unexpected errors and interpretation problems, even a simple analysis of intron–exon contrasts demonstrated that notwithstanding a high global compositional heterogeneity, genomes of warm-blooded vertebrates show an unusual local homogeneity: the mean absolute intron–exon difference in GC content is more than 2-fold lower in them than in the genomes of other organisms [Figure 1A in (56)]. The greater length of mammal introns compared with introns of other studied organisms makes this small intron-exon contrast even more unusual. These data support the existence of isochores in the mammal genomes.
The noncoding DNA was also supposed to play a ‘buffering’ role, damping the effect of solvent fluctuations on the nuclear machinery (57). Recently, it was shown in comparison of two closely related amphibian species differing in genome size that chromatin condensation was steadier and its reaction to changes in solvent composition (caused by elevated extracellular salinity) was more inertial in the species with the larger genome, which is in agreement with the ‘buffering’ function of noncoding DNA (58). The ability of DNA to act as a ‘buffer’ to control the concentration (more exactly, the activity) of DNA-binding proteins was even used for the development of experimental methods for the investigation of histone–DNA interactions [reviewed in (59)]. It is also possible that there can be local buffering in regard to a given locus. A large amount of noncoding DNA within and around a tissue-specific gene may furnish high local histone concentration and thus secure suppression of transcriptional noise. [Transcriptional background noise is believed to be a great threat to cellular function as the number of genes increases during evolution (60,61).] The role of DNA in providing high local concentration of sequence-specific DNA-tropic proteins is discussed in (62). The same rationale can be valid for less specific histone–DNA interactions.
Acknowledgments
This work was supported by the Russian Foundation for Basic Research (RFBR) and by the Programme of the Presidium of the Russian Academy of Sciences ‘Molecular and Cellular Biology’ (MCB RAS). The Open Access publication charges for this article were waived by Oxford University Press.
REFERENCES
- 1.Bernardi G. Isochores and the evolutionary genomics of vertebrates. Gene. 2000;241:3–17. doi: 10.1016/s0378-1119(99)00485-0. [DOI] [PubMed] [Google Scholar]
- 2.Bernardi G. The compositional evolution of vertebrate genomes. Gene. 2000;259:31–43. doi: 10.1016/s0378-1119(00)00441-8. [DOI] [PubMed] [Google Scholar]
- 3.Bernardi G. Structural and Evolutionary Genomics. Natural Selection in Genome Evolution. Amsterdam: Elsevier; 2004. [Google Scholar]
- 4.Eyre-Walker A., Hurst L.D. The evolution of isochores. Nature Rev. Genet. 2001;2:549–555. doi: 10.1038/35080577. [DOI] [PubMed] [Google Scholar]
- 5.Eyre-Walker A. Evidence of selection on silent site base composition in mammals: potential implications for the evolution of isochores and junk DNA. Genetics. 1999;152:675–683. doi: 10.1093/genetics/152.2.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fryxell K.J., Zuckerkandl E. Cytosine deamination plays xa primary role in the evolution of mammalian isochores. Mol. Biol. Evol. 2000;17:1371–1383. doi: 10.1093/oxfordjournals.molbev.a026420. [DOI] [PubMed] [Google Scholar]
- 7.Galtier N., Piganeau G., Mouchiroud D., Duret L. GC content evolution in mammalian genomes, the biased gene conversion hypothesis. Genetics. 2001;159:907–911. doi: 10.1093/genetics/159.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith N.G., Eyre-Walker A. Synonymous codon bias is not caused by mutation bias in human. Mol. Biol. Evol. 2001;18:982–986. doi: 10.1093/oxfordjournals.molbev.a003899. [DOI] [PubMed] [Google Scholar]
- 9.Duret L., Semon M., Piganeau G., Mouchiroud D., Galtier N. Vanishing GC-rich isochores in mammalian genomes. Genetics. 2002;162:1837–1847. doi: 10.1093/genetics/162.4.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lercher M.J., Smith N.G., Eyre-Walker A., Hurst L.D. The evolution of isochores: evidence from SNP frequency distributions. Genetics. 2002;162:1805–1810. doi: 10.1093/genetics/162.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arndt P.F., Petrov D.A., Hwa T. Distinct changes of genomic biases in nucleotide substitution at the time of Mammalian radiation. Mol. Biol. Evol. 2003;20:1887–1896. doi: 10.1093/molbev/msg204. [DOI] [PubMed] [Google Scholar]
- 12.Galtier N. Gene conversion drives GC content evolution in mammalian histones. Trends Genet. 2003;19:65–68. doi: 10.1016/s0168-9525(02)00002-1. [DOI] [PubMed] [Google Scholar]
- 13.Meunier J., Duret L. Recombination drives the evolution of GC-content in the human genome. Mol. Biol. Evol. 2004;21:984–990. doi: 10.1093/molbev/msh070. [DOI] [PubMed] [Google Scholar]
- 14.Bernardi G., Bernardi G. Compositional constraints and genome evolution. J. Mol. Evol. 1986;24:1–11. doi: 10.1007/BF02099946. [DOI] [PubMed] [Google Scholar]
- 15.Vinogradov A.E. Bendable genes of warm-blooded vertebrates. Mol. Biol. Evol. 2001;18:2195–2200. doi: 10.1093/oxfordjournals.molbev.a003766. [DOI] [PubMed] [Google Scholar]
- 16.Vinogradov A.E. DNA helix: the importance of being GC-rich. Nucleic Acids Res. 2003;31:1838–1844. doi: 10.1093/nar/gkg296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vinogradov A.E. Isochores and tissue-specificity. Nucleic Acids Res. 2003;31:5212–5220. doi: 10.1093/nar/gkg699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galtier N., Lobry J.R. Relationships between genomic G+C content, RNA secondary structures, and optimal growth temperature in prokaryotes. J. Mol. Evol. 1997;44:632–636. doi: 10.1007/pl00006186. [DOI] [PubMed] [Google Scholar]
- 19.Hurst L.D., Merchant A.R. High guanine–cytosine content is not an adaptation to high temperature, a comparative analysis amongst prokaryotes. Proc. R. Soc. Lond., B., Biol. Sci. 2001;268:493–497. doi: 10.1098/rspb.2000.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belle E.M., Smith N., Eyre-Walker A. Analysis of the phylogenetic distribution of isochores in vertebrates and a test of the thermal stability hypothesis. J. Mol. Evol. 2002;55:356–363. doi: 10.1007/s00239-002-2333-1. [DOI] [PubMed] [Google Scholar]
- 21.Ream R.A., Johns G.C., Somero G.N. Base compositions of genes encoding alpha-actin and lactate dehydrogenase-A from differently adapted vertebrates show no temperature-adaptive variation in G + C content. Mol. Biol. Evol. 2003;20:105–110. doi: 10.1093/molbev/msg008. [DOI] [PubMed] [Google Scholar]
- 22.Radic M.Z., Lundgren K., Hamkalo B.A. Curvature of mouse satellite DNA and condensation of heterochromatin. Cell. 1987;50:1101–1108. doi: 10.1016/0092-8674(87)90176-0. [DOI] [PubMed] [Google Scholar]
- 23.Blomquist P., Belikov S., Wrange O. Increased nuclear factor 1 binding to its nucleosomal site mediated by sequence-dependent DNA structure. Nucleic Acids Res. 1999;27:517–525. doi: 10.1093/nar/27.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anselmi C., Bocchinfuso G., De Santis P., Savino M., Scipioni A. Dual role of DNA intrinsic curvature and flexibility in determining nucleosome stability. J. Mol. Biol. 1999;286:1293–1301. doi: 10.1006/jmbi.1998.2575. [DOI] [PubMed] [Google Scholar]
- 25.Anselmi C., Bocchinfuso G., De Santis P., Savino M., Scipioni A. A theoretical model for the prediction of sequence-dependent nucleosome thermodynamic stability. Biophys. J. 2000;79:601–613. doi: 10.1016/S0006-3495(00)76319-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiyama R., Trifonov E.N. What positions nucleosomes?—A model. FEBS Lett. 2002;523:7–11. doi: 10.1016/s0014-5793(02)02937-x. [DOI] [PubMed] [Google Scholar]
- 27.Vinogradov A.E. Silent DNA: speaking RNA language? Bioinformatics. 2003;9:2167–2170. doi: 10.1093/bioinformatics/btg320. [DOI] [PubMed] [Google Scholar]
- 28.Saccone S., Federico C., Bernardi G. Localization of the gene-richest and the gene-poorest isochores in the interphase nuclei of mammals and birds. Gene. 2002;300:169–178. doi: 10.1016/s0378-1119(02)01038-7. [DOI] [PubMed] [Google Scholar]
- 29.Mouchiroud D., Fichant G., Bernardi G. Compositional compartmentalization and gene composition in the genome of vertebrates. J. Mol. Evol. 1987;26:198–204. doi: 10.1007/BF02099852. [DOI] [PubMed] [Google Scholar]
- 30.Pesole G., Bernardi G., Saccone C. Isochore specificity of AUG initiator context of human genes. FEBS Lett. 1999;464:60–62. doi: 10.1016/s0014-5793(99)01675-0. [DOI] [PubMed] [Google Scholar]
- 31.Castillo-Davis C.I., Mekhedov S.L., Hartl D.L., Koonin E.V., Kondrashov F.A. Selection for short introns in highly expressed genes. Nature Genet. 2002;31:415–418. doi: 10.1038/ng940. [DOI] [PubMed] [Google Scholar]
- 32.Eisenberg E., Levanon E.Y. Human housekeeping genes are compact. Trends Genet. 2003;19:362–365. doi: 10.1016/S0168-9525(03)00140-9. [DOI] [PubMed] [Google Scholar]
- 33.Urrutia A.O., Hurst L.D. The signature of selection mediated by expression on human genes. Genome Res. 2003;13:2260–2264. doi: 10.1101/gr.641103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vinogradov A.E. Compactness of human housekeeping genes: selection for economy or genomic design. Trends Genet. 2004;20:248–253. doi: 10.1016/j.tig.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Su A.I., Cooke M.P., Ching K.A., Hakak Y., Walker J.R., Wiltshire T., Orth A.P., Vega R.G., Sapinoso L.M., Moqrich A., et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl Acad. Sci. USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence project: update and current status. Nucleic Acids Res. 2003;31:34–37. doi: 10.1093/nar/gkg111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levitsky V.G., Podkolodnaya O.A., Kolchanov N.A., Podkolodny N.L. Nucleosome formation potential of eukaryotic DNA: calculation and promoters analysis. Bioinformatics. 2001;17:998–1010. doi: 10.1093/bioinformatics/17.11.998. [DOI] [PubMed] [Google Scholar]
- 38.Levitsky V.G., Podkolodnaya O.A., Kolchanov N.A., Podkolodny N.L. Nucleosome formation potential of exons, introns, and Alu repeats. Bioinformatics. 2001;17:1062–1064. doi: 10.1093/bioinformatics/17.11.1062. [DOI] [PubMed] [Google Scholar]
- 39.Levitsky V.G. RECON: a program for prediction of nucleosome formation potential. Nucleic Acids Res. 2004;32:W346–W349. doi: 10.1093/nar/gkh482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ioshikhes I., Trifonov E.N. Nucleosomal DNA sequence database. Nucleic Acids Res. 1993;21:4857–4859. doi: 10.1093/nar/21.21.4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Widlund H.R., Cao H., Simonsson S., Magnusson E., Simonsson T., Nielsen P.E., Kahn J.D., Crothers D.M., Kubista M. Identification and characterization of genomic nucleosome-positioning sequences. J. Mol. Biol. 1997;267:807–817. doi: 10.1006/jmbi.1997.0916. [DOI] [PubMed] [Google Scholar]
- 42.Cao H., Widlund H.R., Simonsson T., Kubista M. TGGA repeats impair nucleosome formation. J. Mol. Biol. 1998;281:253–260. doi: 10.1006/jmbi.1998.1925. [DOI] [PubMed] [Google Scholar]
- 43.Farkas G., Leibovitch B.A., Elgin S.C.R. Chromatin organization and transcriptional control of gene expression in Drosophila. Gene. 2000;253:117–136. doi: 10.1016/s0378-1119(00)00240-7. [DOI] [PubMed] [Google Scholar]
- 44.Horn P.J., Peterson C.L. Molecular biology. Chromatin higher order folding—wrapping up transcription. Science. 2002;297:1824–1827. doi: 10.1126/science.1074200. [DOI] [PubMed] [Google Scholar]
- 45.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 46.Aerts S., Thijs G., Coessens B., Staes M., Moreau Y., De Moor B. Toucan: deciphering the cis-regulatory logic of coregulated genes. Nucleic Acids Res. 2003;31:1753–1764. doi: 10.1093/nar/gkg268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zuckerkandl E. A general function of noncoding polynucleotide sequences. Mass binding of transconformational proteins. Mol. Biol. Rep. 1981;7:149–158. doi: 10.1007/BF00778746. [DOI] [PubMed] [Google Scholar]
- 48.Zuckerkandl E. Junk DNA and sectorial gene repression. Gene. 1997;205:323–343. doi: 10.1016/s0378-1119(97)00543-x. [DOI] [PubMed] [Google Scholar]
- 49.Trifonov E.N. Spatial separation of overlapping messages. Comput. Chem. 1993;117:27–31. [Google Scholar]
- 50.Lauderdale J.D., Stein A. Introns of the chicken ovalbumin gene promote nucleosome alignment in vitro. Nucleic Acids Res. 1992;20:6589–6596. doi: 10.1093/nar/20.24.6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu K., Sandgren E.P., Palmiter R.D., Stein A. Rat growth hormone gene introns stimulate nucleosome alignment in vitro and in transgenic mice. Proc. Natl Acad. Sci. USA. 1995;92:7724–7728. doi: 10.1073/pnas.92.17.7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 53.Bernardi G. Misunderstandings about isochores. Part 1. Gene. 2001;276:3–13. doi: 10.1016/s0378-1119(01)00644-8. [DOI] [PubMed] [Google Scholar]
- 54.Li W., Bernaola-Galvan P., Carpena P., Oliver J.L. Isochores merit the prefix ‘iso’. Comput. Biol. Chem. 2003;27:5–10. doi: 10.1016/s1476-9271(02)00090-7. [DOI] [PubMed] [Google Scholar]
- 55.Wen S.Y., Zhang C.T. Identification of isochore boundaries in the human genome using the technique of wavelet multiresolution analysis. Biochem. Biophys. Res. Commun. 2003;311:215–222. doi: 10.1016/j.bbrc.2003.09.198. [DOI] [PubMed] [Google Scholar]
- 56.Vinogradov A.E. Within-intron correlation with base composition of adjacent exons in different genomes. Gene. 2001;276:143–151. doi: 10.1016/s0378-1119(01)00638-2. [DOI] [PubMed] [Google Scholar]
- 57.Vinogradov A.E. Buffering: a possible passive-homeostasis role for redundant DNA. J. Theor. Biol. 1998;193:197–199. doi: 10.1006/jtbi.1997.0629. [DOI] [PubMed] [Google Scholar]
- 58.Vinogradov A.E. Genome size and chromatin condensation in vertebrates. Chromosoma. 2005 doi: 10.1007/s00412-004-0323-3. in press. [DOI] [PubMed] [Google Scholar]
- 59.Thastrom A., Lowary P.T., Widom J. Measurement of histone–DNA interaction free energy in nucleosomes. Methods. 2004;33:33–44. doi: 10.1016/j.ymeth.2003.10.018. [DOI] [PubMed] [Google Scholar]
- 60.Bird A.P. Gene number, noise reduction and biological complexity. Trends Genet. 1995;11:94–100. doi: 10.1016/S0168-9525(00)89009-5. [DOI] [PubMed] [Google Scholar]
- 61.Hurst L.D. Evolutionary genetics. The silence of the genes. Curr. Biol. 1995;5:459–461. doi: 10.1016/s0960-9822(95)00090-x. [DOI] [PubMed] [Google Scholar]
- 62.Droge P., Muller-Hill B. High local protein concentrations at promoters: strategies in prokaryotic and eukaryotic cells. Bioessays. 2001;23:179–183. doi: 10.1002/1521-1878(200102)23:2<179::AID-BIES1025>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]