Abstract
RNA metabolism is a major contributor to the pathogenesis of clinical disorders associated with premutation size alleles of the fragile X mental retardation (FMR1) gene. Herein, we determined the structural properties of numerous FMR1 transcripts harboring different numbers of both CGG repeats and AGG interruptions. The stability of hairpins formed by uninterrupted repeat-containing transcripts increased with the lengthening of the repeat tract. Even a single AGG interruption in the repeated sequence dramatically changed the folding of the 5′UTR fragments, typically resulting in branched hairpin structures. Transcripts containing different lengths of CGG repeats, but sharing a common AGG pattern, adopted similar types of secondary structures. We postulate that interruption-dependent structure variants of the FMR1 mRNA contribute to the phenotype diversity, observed in premutation carriers.
INTRODUCTION
Fragile X syndrome is the most frequent form of inherited mental retardation (1). The classical form of this disease is caused by DNA expansions of the CGG repeats, located in the promoter region of the FMR1 gene, associated with hypermethylation of the repeating tract along with the upstream CpG island (2–5). Transcription silencing and the loss of the FMR1 protein (FMRP) are the consequences of this mutation. Unaffected individuals generally have 5–55 CGG repeats, whereas fragile X patients possess >200 repeats (6). The premutation alleles found in the FMR1 gene contain unmethylated expansions of 50–200 CGG repeats (6,7). For the past several years, individuals harboring premutation alleles were referred to as asymptomatic. Recently, some of the premutation carriers have been demonstrated to exhibit one or more distinct clinical disorders: mild cognitive and behavioral deficits on the fragile-X spectrum, tremor ataxia syndrome (FXTAS) and premature ovarian failure (POF) (8).
The premutation alleles have a tendency to increase in repeat tract length upon maternal transmission and the risk of developing a full mutation, strongly depends on the size of CGG tracts in the premutation alleles (9,10). However, the CGG length polymorphism is not the only sequence variation observed in the repeat region of the FMR1 gene. A majority of the normal alleles of this gene harbor AGG interruptions disrupting the homogeneity of the CGG repeat stretch in periodic intervals (usually every 8–12 CGG repeats) (11–13). The most frequent are the alleles harboring two AGG interruptions, but cases of three and four interruptions were also observed. Loss of the heterogeneity of the CGG tract (AGG interruptions) leading to the lengthening of the uninterrupted trinucleotide repeat region is considered to be a crucial step towards the instability of the CGG repeats, resulting in the subsequent expansion of these sequences (12,13). Thus, at the DNA level, the AGG interruptions play an important stabilizing role for the CGG tracts.
In the case of affected individuals with permutations, as well as patients with unmethylated (or partially methylated) full expansions, the relationship between mutation and the phenotype is much more complex than the simple transcription silencing mechanism described above. Several studies showed that the FMR1 mRNA levels in the premutation carriers are significantly elevated (14–16). On the contrary, the amount of the FMRP was reduced, suggesting an impairment of the translation process and a compensatory induction of the FMR1 mRNA synthesis (16–19). It has also been postulated that the elevated amount of mRNA is important for the formation of the neuronal inclusions observed in a few symptomatic premutation carriers, as well as in mouse and Drosophila melanogaster CGG premutation models (8,20–22). The expanded CGG tracts in the 5′UTR FMR1 mRNA impede translation of FMRP in a length-dependent manner (16,18). The translation impairment is due to the stalling of the ribosome scanning process, hence the FMR1 mRNA was found to be associated with light polysomes or with inactive ribonucleoparticles (17,18).
The important role of RNA structure adopted by the GC-rich 5′UTR of the FMR1 mRNA in the regulation of the FMR1 gene expression was recurrently postulated (16,18,23–27). On the other hand, no detailed structural studies were reported to substantiate the involvement of the CGG RNA structure in the pathogenesis of the fragile X syndrome and other FMR1-related diseases. Herein, we show that CGG repeats present in the natural sequence context of the human 5′UTR FMR1 mRNA form stable secondary structures, which have a potential to influence the mRNA function. We demonstrate that single nucleotide substitutions (AGG interruptions) in the CGG tracts play an important role in the formation of the FMR1 mRNA structure, and both the length of the CGG tracts as well as their interruptions may have significant clinical implications for fragile X and other FMR1-related syndromes.
MATERIALS AND METHODS
DNA templates for in vitro transcription
DNA templates for in vitro transcription were obtained by PCR amplification. Transcripts fx1, fx2, fx3, fx4 and fx8 (Table 1) represent alleles of the FMR1 gene commonly found in the general population. DNA samples from healthy individuals, selected from our laboratory genomic DNA collection, were screened by PCR with primers FAX1 5′ CGCTCAGCTCCGTTTCGGTTTCACTTCCGGT and FAX2 5′ AGCCCCGCACTTCCACCACCAGCTCCTCCA. The lengths of the radiolabeled PCR products were compared to the M13 sequencing ladder to determine the number of the CGG repeats present. The interruption status of amplified alleles was established by DNA sequencing. The selected alleles were reamplified with reverse primer FX1 5′ GGGGGCGTGCGGCAGCGCGG and the modified forward primer extended by the T7 RNA polymerase promoter FX2T7gg 5′ TAATACGACTCACTATAGGCGTGCGGCAGCG. All PCR reactions were performed in the same conditions: total volume of 20 μl, 1 μM of each primer, 200 μM of dATP, dCTP and dTTP, 100 μM of dGTP supplemented with 100 μM of N7deazadGTP (Boehringer Mannheim), 10% (v/v) dimethyl sulfoxide (DMSO) (Serva) and 0.5 U of AmpliTaq polymerase (Perkin Elmer) in a buffer containing 10 mM Tris (pH 8.3), 50 mM KCl and 1.5 mM MgCl2. PCR thermal cycling conditions were: 95°C for 3 min followed by 30 cycles of 95°C for 30 s, 72°C for 90 s with the final extension at 72°C for 7 min.
Table 1.
Characteristics of in vitro transcripts used in this study
| Transcript | Number of repeats | Interruption status | Length of the transcript (nt)a |
|---|---|---|---|
| fx1 | 19 | (CGG)19 | 95 |
| fx2 | 23 | (CGG)23 | 107 |
| fx3 | 28 | (CGG)28 | 122 |
| fx4 | 23 | (CGG)9AGG(CGG)13 | 107 |
| fx5 | 28 | (CGG)9AGG(CGG)18 | 122 |
| fx6 | 28 | (CGG)18AGG(CGG)9 | 122 |
| fx7 | 28 | (CGG)9AGG(CGG)8AGG(CGG)9 | 122 |
| fx8 | 30 | (CGG)10AGG(CGG)9AGG(CGG)9 | 128 |
| fx9 | 47 | (CGG)9AGG(CGG)8AGG(CGG)28 | 179 |
aTotal length of the analyzed transcript, including 14 nt at the 5′ end and 24 nt at the 3′ end of the CGG repeats.
DNA templates for transcripts fx5, fx6 and fx7 (Table 1) were obtained using the PCR mutagenesis protocol with oligonucleotides harboring AGG interruptions: FX5oligo 5′ GGCGTGCGGCAGCG(CGG)9AGG(CGG)11, FX6oligo 5′ GGCGTGCGGCAGCG(CGG)18AGG(CGG)4 and FX7oligo 5′ GGCGTGCGGCAGCG(CGG)9AGG(CGG)8AGG(CGG)4. These oligonucleotides, radioactively labeled at the 5′ ends, were hybridized to the purified PCR product containing 28 CGG repeats with no AGG interruptions (template for the fx3 transcript) followed by the primer elongation step at 72°C for 30 min using 0.5 U of AmpliTaq DNA polymerase and 200 μM dNTPs. The elongation products were purified through a 6% denaturing polyacrylamide gel and PCR reamplified with the FX2T7gg and FX1 primers.
In vitro transcription and RNA labeling
In vitro transcription reactions and 5′ end labeling of RNA molecules were performed as described earlier (28). To label the 3′ end, the RNA fragments were incubated with 20 U of T4 RNA ligase (Epicentre Technologies) and 30 μCi of [32P]pCp (Amersham Corp.), at 4°C for 16–18 h. The labeled RNAs were purified by electrophoresis on denaturing 10% polyacrylamide gels, localized on the gels by autoradiography and recovered as described earlier (28).
Analysis of RNA conformation using native PAGE
In order to check for structure homogeneity of the RNA molecules, all labeled transcripts were analyzed on 5–10% polyacrylamide gels (400/300/0.8 mm, acrylamide/bisakrylamide–29/1) in non-denaturing conditions. The electrophoresis was conducted at a constant power of 10 or 20 W for 4–6 h in 0.5 × TB buffer (45 mM Tris–borate) at 20°C (identical to the temperature of chemical and enzymatic structure probing reactions). Prior to gel electrophoresis, the 32P-labeled transcripts were subjected to a denaturation/renaturation procedure in a solution containing 10 mM Tris–HCl (pH 7.2), 40 mM NaCl and 10 mM MgCl2, by heating the sample at 90°C for 1 min. and slowly cooling it to 20°C (∼1°C/min), and mixed with an equal volume of 7% sucrose with dyes. Electrophoresis performed in the presence of 1–10 mM Mg2+ and constant buffer circulation did not reveal any significant differences in the formation of stable conformers. The specific conditions for the temperature and pH dependence experiments are described in the legend to the Supplementary Figure 2.
Two different electrophoretic migration standards were used: ds69 and ds107. The ds69 represents dsRNA molecule, 69 bp long, obtained by hybridization of two complementary RNA oligomers: 5′ GGG(CUG)21CCC and 5′ GGG(CAG)21 CCC. The second marker, ds107, was obtained by annealing of the fx4 transcript with its complementary molecule containing 23 CCG repeats.
A number of transcripts analyzed in this study migrated on the native gels as two distinct conformers. In all cases, the contribution of the less prevalent conformer was too high to be neglected in the structure studies. Two assays were used to obtain conformer-specific structural data. First, the preparative amount of intact conformers was separated on a native 8% polyacrylamide gel, exposed to the X ray film and then separately excised and eluted from the gel with 20 mM Tris–HCl, pH 7.2. The conformer-specific structure probing was performed without the initial denaturation/renaturation step as described below. Alternatively, structure probing reactions were performed on the mixture of coexisting stable conformers and partially nicked RNA molecules that were resolved on native polyacrylamide gels. Nicked transcripts (due to the nuclease or lead ion hydrolysis), which migrate on native gels at the same rate as intact conformers, were eluted from the gel (with 0.3 M potassium acetate, pH 5.1, 1 mM EDTA and 0.1% SDS), precipitated and analyzed on denaturing polyacrylamide gels. Although, both methods led to identical results, the first, more straightforward approach was used more frequently. In order to rule out the possibility of sequence heterogeneity between the stable coexisting conformers, RNA sequencing analysis of each conformer was conducted using RNA Sequencing Kit (Pharmacia Biotech Inc.) according to the manufacturer's recommendations.
Nuclease digestions, lead cleavages and analysis of reaction products
Prior to the structure probing reactions, the labeled transcripts were subjected to the denaturation/renaturation procedure as described above and supplemented with the unlabeled RNA carrier to the final concentration of 8 μM. Three different types of unlabeled RNAs were successfully used as carrier [homologous CGG transcripts, crude CGG in vitro transcription products and yeast tRNA (Boehringer Mannheim)] with no influence on the results of structure probing analyses. In addition, the electrophoretic migration of the CGG transcripts in non-denaturing conditions was unaffected by different types of carrier RNAs. Limited digestions with lead ions and nucleases S1, T1, V1, P1 were carried out in 10 mM Tris–HCl (pH 7.2), 40 mM NaCl and 1 or 10 mM MgCl2. No significant differences in the results of experiments conducted at varying MgCl2 concentrations were observed. In the case of S1 reactions, ZnCl2 was also present at the 1 mM concentration. The U2 ribonuclease reactions were performed in a buffer containing 10 mM sodium acetate, pH 5.5. Unless otherwise indicated, the reactions were carried out at 20°C for 20 min; different concentrations of lead ions and the nucleases were used as specified in the legends to the figures (Figures 1, 2 and 4). All reactions were terminated by adding an equal volume of stop solution (7.5 M urea, 20 mM EDTA and 0.02% xylene cyanol) and sample freezing. The products of the RNA cleavages were analyzed as described earlier (28). In order to assign the cleavage sites, the alkaline hydrolysis ladder, limited Cl3 and U2 ribonuclease digests (cytosine- and adenine-specific, respectively) of the same RNA molecule, were analyzed on the polyacrylamide gel. The alkaline hydrolysis ladder was generated by incubation of the labeled RNA in formamide containing 0.5 mM MgCl2 at 100°C for 15 min. Partial Cl3 ribonuclease digestion of RNAs was performed in denaturing conditions (10 mM Tris–HCl, pH 8.0 and 7.5 M urea) with 0.025 U of the enzyme. Partial U2 ribonuclease cleavage was carried out in semi-denaturing conditions (10 mM sodium citrate, pH 3.5 and 3.5 M urea) with 1 U of the enzyme. The reaction mixtures were incubated at 55°C for 15 min. The LKB UltroScan XL densitometer (LKB) and scintillation counter (Beckmann) were used for quantitative analyses.
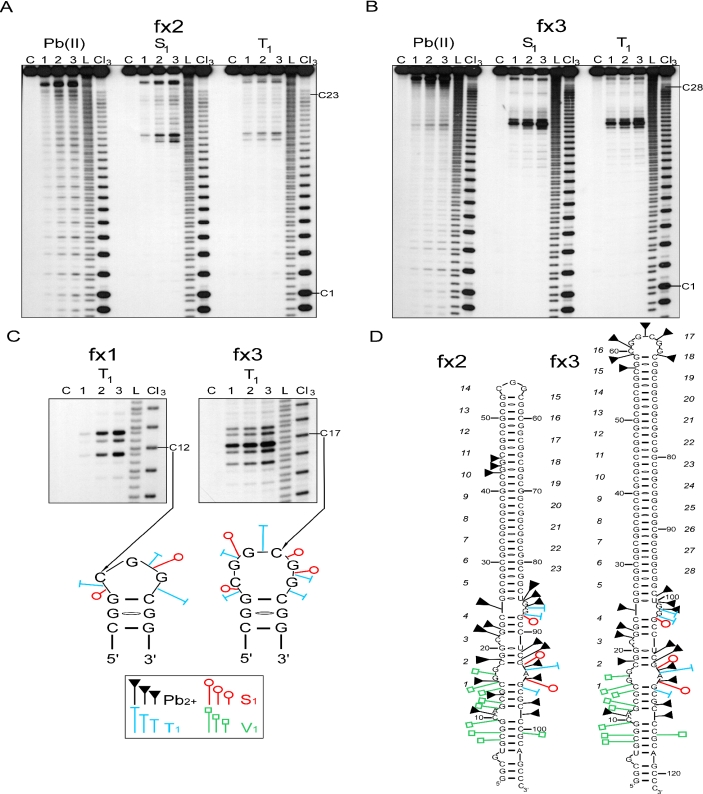
Figure 1.
Structure analysis of 5′UTR fragments of the FMR1 mRNA containing uninterrupted CGG repeats. (A) Cleavage patterns obtained for 5′ end labeled fx2 transcript harboring 23 CGG repeats treated with: Pb(II), lanes 1–3 correspond to 0.25, 0.5 and 1.0 mM lead acetate, respectively; S1, lanes 1–3 correspond to 0.675, 1.35 and 2.7 U/μl of S1 nuclease, respectively; T1, lanes 1–3 correspond to 0.1, 0.3 and 0.5 U/μl of T1 ribonuclease, respectively; lane C, incubation control; L, formamide ladder; Cl3, cytosine-specific ladder. The first and the last cytosine of the CGG tract are indicated. Electrophoresis was conducted in a 10% denaturing polyacrylamide gel. (B) Cleavage patterns obtained for 5′ end labeled fx3 transcript harboring 28 CGG repeats. The reaction conditions were the same as indicated in the panel A. (C) Cleavage patterns obtained for the terminal loops of the hairpin structures for fx1 transcript (odd number of CGG repeats) and fx3 transcript (even number of CGG repeats). Specificity and intensity of the cleavages induced by S1 and T1 nucleases are shown below. Size of the symbols corresponds to the relative cleavage intensity. (D) Proposed secondary structure model of the FMR1 5′UTR fragments containing 23 and 28 uninterrupted CGG repeats. It is noted that the regions flanking the CGG tract in both models, regardless of the number of repeats, adopted an identical secondary structure. The phosphodiester bonds susceptible to the lead-induced cleavages in the fx2 stem has only been shown for the 10th repeat of the hairpin for clarity; however, the same cleavage pattern was seen for all the remaining repeats of the CGG stem. The pattern of the lead ion cleavages observed in the terminal loop of fx3 is indicated. Phosphodiester bonds of the fx2 hairpin loop were very mildly cleaved by lead ions (repeats 13–15, not shown on the model). The sites of weakest cleavage induced by enzymatic probes in the CGG stem are not indicated. Numbers of the consecutive CGG repeats are italicized.
Figure 2.
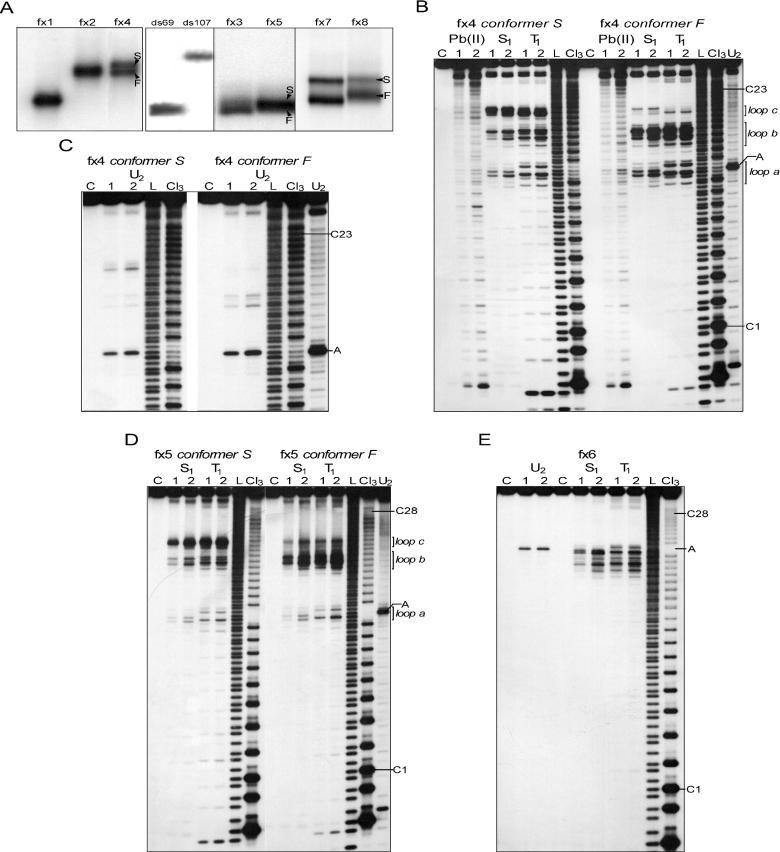
Structure analysis of 5′UTR fragments of the FMR1 mRNA containing CGG repeats with single AGG interruption. (A) Native polyacrylamide gel electrophoresis of the 5′ labeled transcripts. Arrowheads indicate coexisting stable slow migrating (S) and fast migrating (F) conformers. The ds69 and ds107 represent dsRNA migration standards (see Materials and Methods). (B–C) Cleavage patterns obtained for 5′ end labeled conformers S and F of the fx4 transcript treated with: Pb(II), lanes 1 and 2 correspond to 0.25 and 1.0 mM lead acetate, respectively; S1, lanes 1 and 2 correspond to 0.675 and 2.7 U/μl of S1 nuclease, respectively; T1, lanes 1 and 2 correspond to 0.1 and 0.5 U/μl of T1 ribonuclease, respectively; U2, lanes 1 and 2 correspond to 0.5 and 1 U/μl of U2 ribonuclease; lane C, incubation control; L, formamide ladder; Cl3, cytosine-specific ladder; U2, adenine-specific ladder. The first and the last cytosine of the CGG tract are indicated, as well as the adenine residue of the AGG interruption. The regions of the RNA molecules predicted to form single-stranded loops are shown. Electrophoresis was conducted in a 10% denaturing polyacrylamide gel. (D) Cleavage patterns obtained for 5′ end labeled conformers S and F of the fx5 transcript. The reaction conditions were the same as indicated in the panel B and C. (E) Results of the enzymatic probing of the 5′ end labeled fx6 transcript. The reaction conditions were the same as indicated in the panel B and C.
Figure 4.
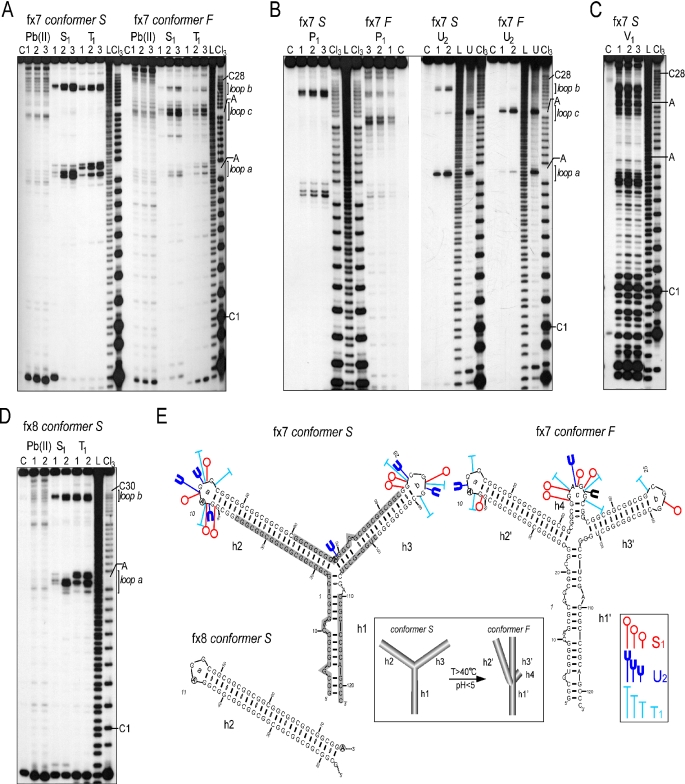
Structure analysis of 5′UTR fragments of the FMR1 mRNA containing CGG repeats with two AGG interruptions. (A) Cleavage patterns obtained for 5′ end labeled conformers S and F of the fx7 transcript harboring 28 CGG repeats with two AGG interruptions treated with Pb(II), S1 nuclease and T1 ribonuclease. The reaction conditions were the same as indicated in Figure 1. The first and the last cytosines of the CGG tract, as well as both the adenine residues of the AGG interruptions are indicated. The regions corresponding to the loops a, b and c are marked. Electrophoresis was conducted in a 10% denaturing polyacrylamide gel. (B) Cleavage patterns obtained for both the S and F conformers of the fx7 with: P1, lanes 1–3 correspond to 5, 7.5 and 10 μg/ml of P1 nuclease, respectively; U2, lanes 1 and 2 correspond to 0.5 and 1.0 U/μl of U2 ribonuclease, respectively. (C) Cleavage patterns obtained for conformer S of the fx7 transcript digested with V1 ribonuclease; lanes 1–3 correspond to 4.6, 9.1 and 18.2 μ/ml of V1 ribonuclease. (D) Results of the structure probing of the fx8 transcript (conformer S) with Pb(II), S1 nuclease and T1 ribonuclease. The reactions were carried out as described in the legend to the Figure 2. (E) Proposed secondary structure models of the FMR1 5′UTR fragments containing 28 and 30 CGG repeats with two AGG interruptions. Models of the conformers S and F are shown separately for the transcript fx7. The only difference between the structure of the fx7 and fx8 transcripts is the length of the helical region 2 (h2). Thus, for clarity, only the helix h2 terminated with the loop a is shown for the fx8 transcript conformer S. It is noted that the regions flanking the CGG tract in both the models, regardless of the number of repeats, adopted identical secondary structures. Specificity and intensity of the cleavages induced by S1, T1 and U2 ribonuclease are indicated. Size of the symbols corresponds to the relative cleavage intensity. Shaded area represents V1 ribonuclease-sensitive regions. Terminal loops of the helical regions h2 (h2′), h3 (h3′) and h4 are designated as a, b and c, respectively. Numbers indicating the trinucleotide repeats are italicized. Below, model for the temperature and low pH induced tertiary transition from the conformer S to the conformer F of the fx7 and fx8 transcripts (see Supplementary Figure 2).
Chemical modifications of RNAs with dimethyl sulfate
The RNA molecules labeled at the 3′ end were subjected to chemical modification with dimethyl sulfate (DMS, Aldrich) in both non-denaturing (10 min, 20°C in 200 mM HEPES, pH 8.0, 50 mM KCl and 10 mM MgCl2) and denaturing (1 min, 90°C in 200 mM HEPES, pH 8.0 and 1 mM EDTA) conditions. The reactions were conducted as described earlier (29).
RESULTS
RNA models and probes used for structure analyses
The structures of nine RNAs were analyzed in this study. All the transcripts harbored a tract of CGG repeats, which differed in the length of the repeating unit and/or their interruption status (Table 1). Three RNAs contained pure, uninterrupted CGG tracts composed of 19, 23 and 28 repeats (transcripts fx1–fx3, Figure 1). Three other transcripts harbored a single AGG interruption (fx4–fx6) and the remaining three RNAs had two AGG interruptions in the CGG region (fx7, fx8 and fx9; Table 1). All the transcripts represented different variants of the FMR1 5′UTR mRNA occurring in the general population. In order to properly select the length of the non-repeating FMR1 sequences flanking the repeats, the FMR1 mRNAs containing different length of CGG repeats were subjected to the extensive structure predictions using Mfold software (30) (Supplementary Figure 1A). The repeat flanking region that was always predicted to adopt identical structure, regardless of the length of the CGG tract, contained 14 nt at the 5′ side and 24 nt at the 3′ side of the repeats. Thus, this autonomous secondary structure module composed of 38 nt of total flanking sequences, in addition to the CGG repeating motif, was chosen for detailed structure analyses using different probing methods. In addition, this CGG repeats-containing module was also predicted to be capable of formation stable secondary structures by MatrixSS algorithm (31) (Supplementary Figure 1B). In order to preserve the natural FMR1 sequence and to avoid formation of transcripts with a heterogeneous sequence at the 5′ end (32,33), the in vitro transcription start site was selected at two consecutive G residues located 14 and 13 nt upstream of the CGG repeat tract. Length homogeneity and structure heterogeneity of the transcripts was determined using denaturing and non-denaturing polyacrylamide gel electrophoresis, respectively.
Uninterrupted CGG repeats form hairpin structures stabilized by flanking sequences
The results of non-denaturing polyacrylamide gel electrophoresis demonstrated that transcripts fx1, fx2 and fx3 formed single, stable conformers (Figure 2A; data not shown). The detailed structural analyses revealed that all three transcripts formed hairpin structures. It is evident that only one region of the repeating tract, in each of the transcripts, showed an enhanced reactivity to S1 and T1 nucleases (Figure 1). The strongest cleavages occurred at the 12th, 14th and at both the 16th and 17th CGG repeats for the fx1, fx2 and fx3 transcripts, respectively. The topography of the terminal loops cleavages, and the proposed loop structures are shown in Figure 1D. The number of phosphodiester bonds cleaved by the single-strand specific nucleases, as well as the cleavage intensity was very similar for fx1 and fx2 transcripts (odd number of CGG repeats), suggesting an identical terminal hairpin loop, composed of 3 nt (Figure 1D). The cleavage pattern observed for fx3, harboring an even number of CGGs, was significantly different and was consistent with a larger loop, containing 6 nt (Figure 1C and D).
The CGG repeats involved in the formation of the terminal hairpin loops in all three transcripts did not correspond to the center of symmetry of the repeated tract (repeats 10, 12, 14/15 for fx1, fx2 and fx3, respectively). This is possible only if part of the CGG repeats located at the 5′ end of the transcripts interact with the 3′-flanking, non-repeating sequences. Indeed, the CGG repeats 1–4 do not interact with the corresponding CGG repeats at the 3′ end of the transcripts, instead they stabilize hairpin structures of fx1, fx2 and fx3 by interacting with non-repeating flanking sequences (Figure 1D).
The second major difference, besides the size of the terminal hairpin loop, between transcripts harboring 19, 23 and 28 repeats, was the increasing stability of the hairpin stem with the lengthening of the repeating tract. This was evident from the reduced reactivity of the stem phosphodiester bonds (especially the most reactive CpG bond of the CGG motif) to lead ion induced hydrolysis (Figure 1A and B). The overall reactivity of the hairpin stem to lead ions suggested a relaxed structure of the hairpins stems, composed of two standard CG/GC base pairs, separated by GG mismatches. The enhanced reactivity of the CpG phosphodiester bond to lead ions, and a higher intensity of the S1 induced cleavage of the GpG bonds, supports the CGG alignment of the hairpin stems with the first G residue of each CGG motif involved in non Watson–Crick GG interactions.
Thus, we conclude that uninterrupted CGG repeats, in the context of the human FMR1 5′UTR sequence, formed stable hairpin structures. Unlike dCGG sequences (34), analyses of the CGG transcripts did not reveal the formation of tetraplex structures.
A single AGG interruption destabilizes CGG hairpins in a position-dependent manner
The RNA structure of the three transcripts harboring single C→A substitutions was analyzed. The fx6 transcript adopted a single stable structure, whereas fx4 and fx5 formed two different structures (Figure 2).
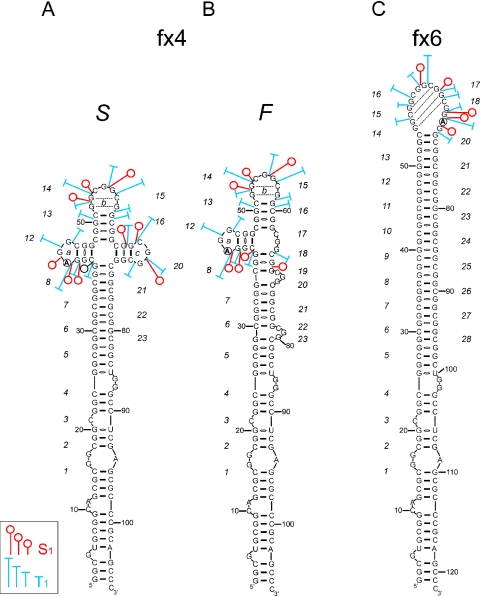
Transcript fx6 was the only RNA in the studied group harboring a single AGG interruption at the 3′ end of the CGG tract. Although the sequence of the fx6 transcript is similar to that of the fx5 RNA (both contain 28 CGG repeats with 1 AGG interruption), structurally, fx6 resembled the fx3 transcript (compare Figures 1B and 2E) much closer. Only one region of the repeat tract (repeats 14–19; Figure 3C) underwent an efficient cleavage by S1 and T1 nucleases. This region, encompassing 17 phosphodiester bonds, formed a large terminal loop of the proposed hairpin structure (Figure 3C). Uneven reactivity of the loop phosphodiester bonds to the single-strand specific probes suggested a dynamic structure stabilized by the presence of intra-loop interactions between specific residues of the 14th/15th and 17th/18th repeats (Figure 3C, dashed lines). In addition, efficient cleavage of the A69G70 phosphodiester bond by ribonuclease U2 confirmed the single-stranded status of the RNA region containing the AGG interruption (Figure 2E).
Figure 3.
Secondary structure models of the fx4 and the fx6 transcripts. Both FMR1 5′UTR fragments contain 23 CGG repeats with a single AGG interruption located at 10th (fx4) and 19th (fx6) repeat. For the fx4 transcript, the models of the S as well as the F conformer are shown. Size of the symbols corresponds to the relative cleavage intensity induced by S1 and T1 nucleases in the CGG region. Numbers indicating the consecutive CGG repeats are italicized.
Transcript fx4 is the shortest RNA harboring one AGG interruption analyzed. The single nucleotide substitution at position 42 that differentiates fx4 RNA from the fx2 transcript had significant structural consequences. The results of native polyacrylamide gel electrophoresis demonstrated that fx4 forms two stable coexisting conformers (Figure 2A). The ratio between F and S conformers did not depend on the RNA concentration (in the range of pg to μg, the same results obtained for fx4, fx5 and fx7–fx9). Thus, intermolecular interactions (i.e. dimer formation) in the case of a slow migrating conformer were highly unlikely.
The structures of the F (fast migration) and S (slow migration) conformers were analyzed separately. Structure probing analysis of the slower migrating conformer (fx4 S) revealed the presence of three regions within the CGG repeats tract that were susceptible to cleavages by S1 and T1 (Figure 2B). Loop a mapped at the AGG interruption (residues 42–44), loop b (residues of 13–15 repeat) and loop c (repeat 18) were easily accessible for these single-strand specific probes (Figures 2B and 3C). Results with U2 ribonuclease also corroborated these results (Figure 2C). In addition, quantitative analyses showed that lead ions induced a much stronger hydrolysis in the loop regions than in the rest of the repeating sequences.
The S1 and T1 cleavage patterns of the loop b revealed its similarity to the terminal hairpin loop of the fx2 transcript. The stem structure adjacent to the loop b was however, cleaved more efficiently than the fx2 stem region, perhaps due to the overall higher rigidity of the fx2 hairpin structure. On the other hand, the S1 and T1 digestion pattern of loop a (containing the AGG interruption) was different from the patterns obtained for CGG-containing loops (Figure 2B and D).
In addition to the distinct electrophoretic mobilities, the most apparent differences between the F and S fx4 conformers were the dramatic reduction in cleavages of loop c in fx4 F, accompanied by a minor increase of the reactivity at loops a and b (without change in the specificity of S1 and T1 cleavages, Figures 2B and 3A,B). In the secondary structure model of fx4 F shown in Figure 3B, we propose that the 3′ CGG region (repeats 16–23) forms a few small trinucleotide CGG bulges. Their location is random and can undergo dynamic changes (in the Figure 3B model, bulges formed by repeat 17, 20 and 23 are shown as one of several possible alignments). Hence, conformation F may represent a mixture of very similar slippery structures, all harboring two loops (a and b) and the alternatively aligned 3′ part of the CGG tract (repeats 16–23) with several 3 nt bulges, which are undetectable by S1 or T1 nuclease, possibly due to strong stacking interactions. The faster electrophoretic migration of fx4 F also supports its more rigid, hairpin-like structure when compared to the four-way junction in fx4 S RNA. Although this is the most plausible explanation of the structure probing data, we cannot entirely rule out the possibility of tertiary interactions between the loop c and other regions in the fx4 F transcript.
Interestingly, transcript fx5 harboring 28 CGG repeats with a single AGG interruption located close to the 5′ end of the repeats (Table 1), demonstrated similar structural properties to the fx4 RNA, i.e. formation of two stable conformers as well as very similar nuclease susceptibility patterns for the S and F conformers (Figure 2D). These data strongly suggest that fragments of the FMR1 5′UTR with a similar interruption arrangement will exhibit the same structural properties. Moreover, these results demonstrated that even single-nucleotide changes in the long stretch of the reiterative sequence can have a dramatic impact on the RNA secondary structure.
The CGG region of the most common FMR1 mRNA 5′UTR variants adopts a Y-shaped structure
Alleles harboring 28, 29 and 30 CGG repeats with two AGG interruptions comprise ∼65% of the different FMR1 5′UTR variants (11,12). The fx7 and fx8 transcripts formed two stable conformers, which differ significantly in their rate of migration during native PAGE (Figure 1A). Very slow migration of the fx7 S and fx8 S suggested that the structure of these conformers differs significantly from the transcripts described earlier. As demonstrated using P1, S1, V1, T1 and U2 nucleases, fx7 S and fx8 S adopt Y-shaped structures composed of 3 helical regions (h1, h2, h3) joint by a three-way junction, with CGG repeats located in helix 2 and 3 (Figure 4). The first AGG interruption formed a trinucleotide loop (loop a), structurally identical to loop a of fx4 or fx5 (compare Figures 4A,D and 2B,D), while the second AGG interruption was located at the three-way helical junction and was not cleaved by S1, T1 or P1 nucleases (Figure 4). Only lead ions were capable of detecting the structural changes induced by this three-way junction and hydrolyzing the phosphodiester bonds at the junction region (Figure 4A for fx7 S and 4d for fx8 S). It has been previously demonstrated that lead ions are often able to recognize and cleave more subtle structural elements than enzymatic probes (28,35,36). Lead ions also induced hydrolysis at the single-stranded regions of loop a, loop b, and internal loops of the helix 3 (Figure 4A and D). The relatively low reactivity of the fx7 S and fx8 S to the single-strand specific probes (except loops a and b) was accompanied by very high susceptibility to V1 nuclease—a double strand RNA-specific enzyme (Figure 4C). Regions flanking the CGG repeats, part of helix 2 (phosphodiester bonds p18–p37 in fx7 S) and part of helix 3 including the three-way junction (p61–p85 in fx7 S) were highly susceptible to the V1 cleavage, while the loops a and b were not recognized by this enzyme. Altogether, these results were consistent with the formation of a Y-shaped structure by the fx7 S RNA.
The results of the native gel electrophoresis indicated the presence of faster migrating fractions—conformers fx7 F and fx8 F. The structural polymorphism was independent of various electrophoresis conditions, including the concentration of Mg2+ ions (0.1–50 mM) or the presence of different monovalent ions such as Na+, K+ and NH4+. However, PAGE as well as S1 nuclease probing conducted at increasing temperatures indicated a much higher stability of the F conformer compared with S conformer (Supplementary Figure 2A and B). In addition, the pH of the incubation buffer had no influence on the stability of the F conformer, although a low pH (<4.5) readily induced a transformation from fx7 S to fx7 F (Supplementary Figure 2C).
Figure 4A and B shows the results of nuclease digestions and lead ion induced hydrolysis of the fx7 F transcript. It is important to note that all structure probing reactions were conducted at the same time for both S and F forms, and the reaction products were analyzed in the same polyacrylamide gel, therefore, enabling direct qualitative and quantitative comparison of the results. S1, P1 and T1 all cleaved the fx7 F conformer at precisely the same phosphodiester bonds, within loop a and loop b, as cleavages in the fx7 S conformer; however, the intensity of nuclease cuts was ∼5 times lower (Figure 4A and B). In addition, the central part of fx7 was hydrolyzed efficiently by single-strand specific endonucleases. This region encompassed nucleotides of the 15–21 repeats with phosphodiester bonds p68–p72, demonstrating the highest reactivity. Similarly, U2 ribonuclease cleaved the A42G43 bond very poorly in the fx7 F transcript compared with fx7 S. In contrast, the A69G70 phosphodiester bond (the second AGG interruption) was cut very efficiently in fx7 F (Figure 4B). This pattern of hydrolysis observed for fx7 F seems to be consistent with the presence of a hairpin structure that contains a large terminal loop spanning CGG repeats surrounding the second AGG interruption. Minor cuts detected in the loop a and b regions could, perhaps, result from a small amount of ‘contamination’ of fx7 F with the fx7 S conformer. However, two important facts argue against this hairpin model: first, the likelihood of cross contamination of the fx7 F with fx7 S has been experimentally excluded, since fx7 F demonstrated very high stability (the reverse process S→F is much more probable, Supplementary Figure 2); second, the transition process from conformer S to the hairpin structure would require the breaking of almost all hydrogen bonds in fx7 S and reestablishing entirely new interactions. A substantial input of energy would be necessary for such a dramatic structural change. This argues against a spontaneous S→F conversion, which can be effectively stimulated just by lowering the pH (Supplementary Figure 2C). Moreover, structural data obtained from S1 nuclease monitored thermal melting of the fx7 S, showed that the stability of the three-way junction played the most important role in the fx7 S→fx7 F transition (Supplementary Figure 2A). The high G+C contents of the FMR1 5′UTR implied the potential involvement of tetraplex structures. The data obtained from DMS modification experiments demonstrated that all the guanines of the fx7 F were susceptible to methylation, therefore ruling out the possibility of helices 2 and 3 adopting a tetraplex conformation (Supplementary Figure 3).
The secondary structure model of the fx7 F is presented in the Figure 4E. We propose, that the region of the second AGG interruption forms a third loop (loop c), stabilized by helix 4, resulting in a rearrangement of the three-way junction structure present in fx7 S into a four-way junction conformation. We hypothesize that the change in the type of junction influences the tertiary interactions between the helices (Figure 4E, inset).
Compared to fx7, transcript fx8 harbors 2 additional CGG repeats; one located at the 5′ region of the first AGG interruption and the other between the AGG interruptions. The only structural effect of the lengthening of this RNA molecule was the extension of helix 2 by one repetitive unit (CGG)·(CCG) (Figure 4E). All other properties of fx8 S and fx8 F were identical to those described above for fx7, again suggesting that the AGG interruption status will determine the structural properties of the FMR1 5′UTR. This conclusion was further corroborated by structural analyses of the fx9 transcript containing 47 repeats and the interruption pattern identical to the fx7 (Table 1, Supplementary Figure 4).
DISCUSSION
CGG repeat expansion in the FMR1 5′UTR causes four different clinical phenotypes: FXS, mild cognitive and behavioral deficits on the fragile-X spectrum, FXTAS and POF (8). Recently, it has become apparent that defects in FMR1 mRNA metabolism might be responsible for all the distinct phenotypes associated with the intermediate and premutation size alleles (40–200 CGG repeats). Several different processes in the FMR1 expression, including the elevated level of FMR1 transcription, changes in the transcription start sites (37), reduced translation of FMR1 mRNA (16,18,19), and interaction of the FMR1 mRNA with specific rCGG binding proteins (38), have been implicated in RNA-mediated pathogenesis of these disorders. We propose that the structure of the CGG region of FMR1 mRNA can contribute to most of the above mentioned RNA level defects found in the FMR1-related syndromes.
In this study, we have determined the structures of the trinucleotide repeat region in transcripts from a variety of normal FMR1 alleles containing uninterrupted CGG repeats, as well as those harboring 1 or 2 AGG interruptions. We intended to answer the question: could the CGG repeat region be protected from adopting stable RNA structures, similar to CUG hairpins known to affect normal functions of cells in myotonic dystrophy patients (28)? This question was particularly important from the perspective of the RNA-mediated pathogenesis of FXTAS, since a substantial portion of various FMR1 premutation alleles retain AGG interruptions as protective elements in their repeated sequences (12).
To evaluate the structural role of AGG interruptions in the CGG repeat tracts, we first determined the structure formed by pure repeats of various lengths and their non-repeating flanking sequences within the FMR1 5′UTR. The structure was a hairpin, more stable at its base due to the contribution of both the CGG repeats and the specific flanking sequences. The only imperfections present in its repeat portion were periodic GG mismatches formed within the hairpin stem. Although this arrangement of the repeated sequence was determined by the base-pairing occurring within the interacting flanking sequences, the CGG repeats were recently shown to align in a similar frame in shorter oligonucleotides in which they could fold without any constrains (23). Thus, the reference structure was established and its various distortions, caused by the presence of the AGG interruptions, were investigated in several FMR1 transcripts. Detailed analyses of the nature of these diverse structures demonstrated distinct positional effects of the AGG repeat interruptions.
From the perspective of RNA folding, the AGG interruptions within the CGG repeat tracts may be considered as either single or double C→A substitutions, occurring at specific positions or sequence intervals. In the established frames of the CGG repeats interacting within the stem structure, the A nucleotides would face the second G of the CGG repeat. This kind of interaction, however, is not observed in the analyzed transcripts. This is most likely because the AG oppositions, which are known to occur in different geometries in other RNA structures (39,40), are strongly disfavored in the repeated CGG sequence context. In fact, it has been demonstrated that the influence of GA mismatches on RNA helix stability strongly depends on the sequence context (41). In a pair of short CG rich complementary RNA oligonucleotides harboring two GA mismatches, the structural distortion introduced by GA mispairs could be so severe that, in some circumstances, it prevented the formation of a duplex RNA (41). In agreement, we observed a clear tendency of the A nucleotides to localize in the autonomous loop structures (e.g. loop a of fx4, fx7, fx8 and fx9). Among the structures analyzed, the only exception were the fx6 transcript in which the hairpin terminal loop was enlarged due to the appropriate A residue localization, and S conformers of the fx7 and fx8. In the latter two cases, the second A substitution lies within one of the double-stranded arms of the Y-shaped structure and pairs with the U residue of the 3′-flanking sequence in close vicinity of the three-way junction. However, in the F conformers of these transcripts, the second A residue was present within a newly formed loop c (Figure 4E). Since the S→F conformer transformation is pH-dependent and occurs more rapidly in acidic solution, it is tempting to speculate that the protonation of adenine and cytosine residues (pKa ∼ 4) at the critical three-way junction region (C18, C107 and A69) may be considered the driving force for the observed structural rearrangement. Protonation of A and C residues could lead to the destabilization of the canonical GC and AU base pairs (42) adjacent to the junction structure. Similar pH-induced conformational switch between two different RNA structures has been described for Escherichia coli 5S ribosomal RNA (43) and E.coli αmRNA (44).
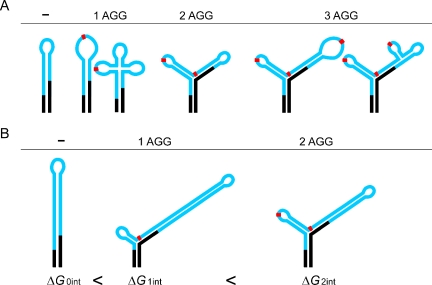
Biological role of the C→A substitutions present in the repeat tracts of transcripts from normal alleles of the FMR1 gene is to prevent the formation of long and stable CGG repeat hairpins (Figure 5), which may decrease the efficiency of the initiation of FMRP translation (19). Depending on the number and localization of these substitutions, the different hairpin structure destabilization strategies are carried out. They range from terminal loop enlargement as in fx6, to dissecting the repeats region into two or more smaller hairpins (Figure 5). A similar structural role seems to be played by the C→A substitutions in transcripts from the FMR1 premutation alleles (25). The repeat interruptions that occur in premutation alleles are biased toward those localizing at the 5′-end of the repeat (12). Therefore, they are likely to protect the premutation carriers from FXTAS or other RNA-related defects by shortening the effective length of hairpin composed of pure CGG repeats. The largest molecule analyzed in this study was just below the length threshold for the shortest premutation alleles (47 repeats). Therefore, we cannot entirely rule out that the elongated FMR1 mRNAs will adopt different structures after passing the pathogenic barrier of 50–55 repeats. We could only speculate that the lengthening of the transcripts by another 3–8 CGG repeats (from 47 to 50–55 repeats) will cause only minor structural differences if the AGG interruption status remains unchanged. This prediction is supported by our data on the structure of fx7 and fx9, which differ substantially in the total number of repeats (28 versus 47), but share virtually an identical structural plan (except for the elongation of the helix 3). Similarly, both fx1 and fx3 (19 and 28 repeats, respectively) formed hairpins and it would be highly unlikely to expect transcripts containing uninterrupted CGG tract in the premutation range to adopt different conformation. Thus, it seems to be justifiable to extrapolate our structural conclusions from the normal RNAs into the premutation size transcripts.
Figure 5.
Models for the interruption-mediated destabilization of the CGG tracts. Black regions represent flanking sequences, the CGG repeats in the 5′UTR of FMR1 mRNAs are shown in blue, and red rectangles represent AGG interruptions. (A) Comparison of different strategies of the AGG interruption induced CGG tract destabilization observed in the normal size FMR1 transcripts. Schematic models are based on experimental data and RNA structure prediction algorithms (30). (B) Influence of the interruptions on structure and free energy of the premutation size transcripts. Sequential loss of AGGs decreases ΔG of the RNA molecules and results in the formation of the most stable form —hairpin structure. Data based on secondary structure prediction of RNA transcripts harboring 60 repeats in the following configurations: (CGG)9AGG(CGG)9AGG(CGG)40, (CGG)9AGG(CGG)50 and (CGG)60.
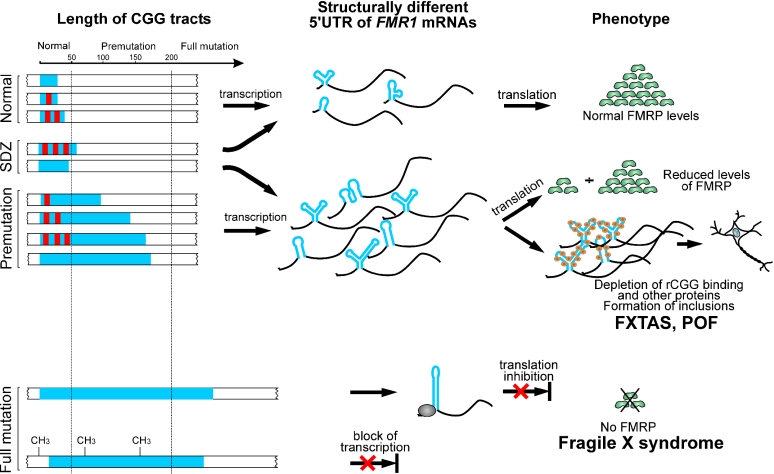
In the current model of RNA-mediated toxicity postulated for FXTAS, the total length of the CGG tract is essential (8). Herein, we propose that the AGG interruption status could influence the correlation between the repeat size and the clinical outcome. The role of RNA structure of the 5′UTR FMR1 mRNA may be particularly important for a phenotype in the carriers of the short premutation or long normal alleles. In the model (Figure 6), we propose the existence of the RNA structure-dependent zone (SDZ) encompassing both long normal and short premutation alleles. The fate of the transcripts falling into that zone primarily depends on the AGG interruption status, i.e. RNA structure of the FMR1 5′UTR, but not solely on the total length of the repeat tract (Figure 6, upper part). Therefore, long normal FMR1 transcripts harboring pure CGG repeats may lead to the FMR1-related premutation phenotypes (e.g. FXTAS), since long and very stable hairpin structures may influence RNA level processes such as aberrant RNA protein binding and RNA foci formation. On the other hand, individuals harboring premutation size FMR1 mRNAs but containing AGG interruptions (hence adopting less stable, ‘non-toxic’ RNA structure) can be completely asymptomatic. Thus, within the structure-dependent zone, RNA structure, not simply the alleles length, can determine the phenotypic outcome. It is also possible that the structural status of the FMR1 5′UTR influences the age of onset, phenotypic variability and/or severity of the FXTAS symptoms. It has already been demonstrated that only a fraction [17–75% depending on the age (45)] of male FMR1 premutation carriers will develop FXTAS. This incomplete penetrance of FXTAS may, at least in some cases, be caused by differences in the structure of the FMR1 mRNA leader. In order to verify this model, further clinical studies are necessary to correlate the incidence of FMR1-related phenotypes with the allelic variations in the premutation carriers and FMR1 mRNA structure.
Figure 6.
Postulated role of the 5′UTR structure in the pathogenesis of the FMR1-related CGG expansions diseases. Blue bars represent CGG tracts, while red rectangles symbolize AGG interruptions. The CGG region in the 5′UTR of FMR1 mRNAs is shown in blue. The remaining portion of FMR1 transcripts is shown in black. SDZ shows the RNA structure-dependent zone (see Discussion). Large CGG expansions (full mutations) in a vast majority of cases lead to the methylation-induced transcriptional silencing of the FMR1 gene; however, translation abnormalities (bottom) were also reported (17).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank Drs Robert D. Wells, Paul Hagerman, Richard Sinden and Vladimir Potaman for helpful discussions. This work was supported by the State Committee for Scientific Research, Grants No. 2P05A08826, PBZ/KBN/040/P04/12 and the Foundation for Polish Science, Grant No. 8/2000. Funding to pay the Open Access publication charges for this article was provided by the State Committee for Scientific Research.
REFERENCES
- 1.Webb T.P., Bundey S.E., Thake A.I., Todd J. Population incidence and segregation ratios in the Martin–Bell syndrome. Am. J. Med. Genet. 1986;23:573–580. doi: 10.1002/ajmg.1320230151. [DOI] [PubMed] [Google Scholar]
- 2.Verkerk A.J., Pieretti M., Sutcliffe J.S., Fu Y.H., Kuhl D.P., Pizzuti A., Reiner O., Richards S., Victoria M.F., Zhang F.P., et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 3.Fu Y.H., Kuhl D.P., Pizzuti A., Pieretti M., Sutcliffe J.S., Richards S., Verkerk A.J., Holden J.J., Fenwick R.G., Jr, Warren S.T., et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- 4.Kremer E.J., Pritchard M., Lynch M., Yu S., Holman K., Baker E., Warren S.T., Schlessinger D., Sutherland G.R., Richards R.I. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991;252:1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- 5.Oberle I., Rousseau F., Heitz D., Kretz C., Devys D., Hanauer A., Boue J., Bertheas M.F., Mandel J.L. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 6.Oostra B.A., Willemsen R. The X chromosome and fragile X mental retardation. Cytogenet. Genome Res. 2002;99:257–264. doi: 10.1159/000071602. [DOI] [PubMed] [Google Scholar]
- 7.Hagerman R.J., Hagerman P.J. The fragile X premutation: into the phenotypic fold. Curr. Opin. Genet. Dev. 2002;12:278–283. doi: 10.1016/s0959-437x(02)00299-x. [DOI] [PubMed] [Google Scholar]
- 8.Hagerman P.J., Hagerman R.J. The fragile-X premutation: a maturing perspective. Am. J. Hum. Genet. 2004;74:805–816. doi: 10.1086/386296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisch G.S., Snow K., Thibodeau S.N., Chalifaux M., Holden J.J., Nelson D.L., Howard-Peebles P.N., Maddalena A. The fragile X premutation in carriers and its effect on mutation size in offspring. Am. J. Hum. Genet. 1995;56:1147–1155. [PMC free article] [PubMed] [Google Scholar]
- 10.Heitz D., Devys D., Imbert G., Kretz C., Mandel J.L. Inheritance of the fragile X syndrome: size of the fragile X premutation is a major determinant of the transition to full mutation. J. Med. Genet. 1992;29:794–801. doi: 10.1136/jmg.29.11.794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichler E.E., Macpherson J.N., Murray A., Jacobs P.A., Chakravarti A., Nelson D.L. Haplotype and interspersion analysis of the FMR1 CGG repeat identifies two different mutational pathways for the origin of the fragile X syndrome. Hum. Mol. Genet. 1996;5:319–330. doi: 10.1093/hmg/5.3.319. [DOI] [PubMed] [Google Scholar]
- 12.Dombrowski C., Levesque S., Morel M.L., Rouillard P., Morgan K., Rousseau F. Premutation and intermediate-size FMR1 alleles in 10572 males from the general population: loss of an AGG interruption is a late event in the generation of fragile X syndrome alleles. Hum. Mol. Genet. 2002;11:371–378. doi: 10.1093/hmg/11.4.371. [DOI] [PubMed] [Google Scholar]
- 13.Eichler E.E., Holden J.J., Popovich B.W., Reiss A.L., Snow K., Thibodeau S.N., Richards C.S., Ward P.A., Nelson D.L. Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nature Genet. 1994;8:88–94. doi: 10.1038/ng0994-88. [DOI] [PubMed] [Google Scholar]
- 14.Tassone F., Hagerman R.J., Chamberlain W.D., Hagerman P.J. Transcription of the FMR1 gene in individuals with fragile X syndrome. Am. J. Med. Genet. 2000;97:195–203. doi: 10.1002/1096-8628(200023)97:3<195::AID-AJMG1037>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 15.Tassone F., Hagerman R.J., Taylor A.K., Gane L.W., Godfrey T.E., Hagerman P.J. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am. J. Hum. Genet. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kenneson A., Zhang F., Hagedorn C.H., Warren S.T. Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum. Mol. Genet. 2001;10:1449–1454. doi: 10.1093/hmg/10.14.1449. [DOI] [PubMed] [Google Scholar]
- 17.Feng Y., Zhang F., Lokey L.K., Chastain J.L., Lakkis L., Eberhart D., Warren S.T. Translational suppression by trinucleotide repeat expansion at FMR1. Science. 1995;268:731–734. doi: 10.1126/science.7732383. [DOI] [PubMed] [Google Scholar]
- 18.Primerano B., Tassone F., Hagerman R.J., Hagerman P., Amaldi F., Bagni C. Reduced FMR1 mRNA translation efficiency in fragile X patients with premutations. RNA. 2002;8:1482–1488. [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L.S., Tassone F., Sahota P., Hagerman P.J. The (CGG)n repeat element within the 5′ untranslated region of the FMR1 message provides both positive and negative cis effects on in vivo translation of a downstream reporter. Hum. Mol. Genet. 2003;12:3067–3074. doi: 10.1093/hmg/ddg331. [DOI] [PubMed] [Google Scholar]
- 20.Jin P., Zarnescu D.C., Zhang F., Pearson C.E., Lucchesi J.C., Moses K., Warren S.T. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron. 2003;39:739–747. doi: 10.1016/s0896-6273(03)00533-6. [DOI] [PubMed] [Google Scholar]
- 21.Tassone F., Hagerman R.J., Garcia-Arocena D., Khandjian E.W., Greco C.M., Hagerman P.J. Intranuclear inclusions in neural cells with premutation alleles in fragile X associated tremor/ataxia syndrome. J. Med. Genet. 2004;41:e43. doi: 10.1136/jmg.2003.012518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willemsen R., Hoogeveen-Westerveld M., Reis S., Holstege J., Severijnen L.A., Nieuwenhuizen I.M., Schrier M., van Unen L., Tassone F., Hoogeveen A.T., et al. The FMR1 CGG repeat mouse displays ubiquitin-positive intranuclear neuronal inclusions; implications for the cerebellar tremor/ataxia syndrome. Hum. Mol. Genet. 2003;12:949–959. doi: 10.1093/hmg/ddg114. [DOI] [PubMed] [Google Scholar]
- 23.Sobczak K., de Mezer M., Michlewski G., Krol J., Krzyzosiak W.J. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galvao R., Mendes-Soares L., Camara J., Jaco I., Carmo-Fonseca M. Triplet repeats, RNA secondary structure and toxic gain-of-function models for pathogenesis. Brain Res. Bull. 2001;56:191–201. doi: 10.1016/s0361-9230(01)00651-7. [DOI] [PubMed] [Google Scholar]
- 25.Handa V., Saha T., Usdin K. The fragile X syndrome repeats form RNA hairpins that do not activate the interferon-inducible protein kinase, PKR, but are cut by Dicer. Nucleic Acids Res. 2003;31:6243–6248. doi: 10.1093/nar/gkg818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jasinska A., Krzyzosiak W.J. Repetitive sequences that shape the human transcriptome. FEBS Lett. 2004;567:136–141. doi: 10.1016/j.febslet.2004.03.109. [DOI] [PubMed] [Google Scholar]
- 27.Bardoni B., Mandel J.L. Advances in understanding of fragile X pathogenesis and FMRP function, and in identification of X linked mental retardation genes. Curr. Opin. Genet. Dev. 2002;12:284–293. doi: 10.1016/s0959-437x(02)00300-3. [DOI] [PubMed] [Google Scholar]
- 28.Napierala M., Krzyzosiak W.J. CUG repeats present in myotonin kinase RNA form metastable ‘slippery’ hairpins. J. Biol. Chem. 1997;272:31079–31085. doi: 10.1074/jbc.272.49.31079. [DOI] [PubMed] [Google Scholar]
- 29.Peattie D.A., Gilbert W. Chemical probes for higher-order structure in RNA. Proc. Natl Acad. Sci. USA. 1980;77:4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Titov I.I., Ivanisenko V.A., Kolchanov N.A. FITness—a WWW-resource for RNA folding simulation based on genetic algorithm with local minimization. Comput. Techn. 2000;5:48–56. [Google Scholar]
- 32.Pleiss J.A., Derrick M.L., Uhlenbeck O.C. T7 RNA polymerase produces 5′ end heterogeneity during in vitro transcription from certain templates. RNA. 1998;4:1313–1317. doi: 10.1017/s135583829800106x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krzyzosiak W., Napierala M., Drozdz M. RNA structure modules with trinucleotide repeat motifs. In: Barciszewski J., Clark B.F.C., editors. RNA Biochemistry and Biotechnology. Dordrecht: Kluwer Academic Publishers; 1999. pp. 303–314. [Google Scholar]
- 34.Fry M., Loeb L.A. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl Acad. Sci. USA. 1994;91:4950–4954. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krzyzosiak W.J., Marciniec T., Wiewiorowski M., Romby P., Ebel J.P., Giege R. Characterization of the lead(II)-induced cleavages in tRNAs in solution and effect of the Y-base removal in yeast tRNAPhe. Biochemistry. 1988;27:5771–5777. doi: 10.1021/bi00415a056. [DOI] [PubMed] [Google Scholar]
- 36.Gornicki P., Baudin F., Romby P., Wiewiorowski M., Krzyzosiak W., Ebel J.P., Ehresmann C., Ehresmann B. Use of lead(II) to probe the structure of large RNA's. Conformation of the 3′ terminal domain of E.coli 16S rRNA and its involvement in building the tRNA binding sites. J. Biomol. Struct. Dyn. 1989;6:971–984. doi: 10.1080/07391102.1989.10506525. [DOI] [PubMed] [Google Scholar]
- 37.Beilina A., Tassone F., Schwartz P.H., Sahota P., Hagerman P.J. Redistribution of transcription start sites within the FMR1 promoter region with expansion of the downstream CGG-repeat element. Hum. Mol. Genet. 2004;13:543–549. doi: 10.1093/hmg/ddh053. [DOI] [PubMed] [Google Scholar]
- 38.Rosser T.C., Johnson T.R., Warren S.T. A cerebellar FMR1 riboCGG binding protein. Am. J. Hum. Genet. 2002;71:501. [Google Scholar]
- 39.Leontis N.B., Stombaugh J., Westhof E. The non-Watson–Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 2002;30:3497–3531. doi: 10.1093/nar/gkf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang H., Jossinet F., Leontis N., Chen L., Westbrook J., Berman H., Westhof E. Tools for the automatic identification and classification of RNA base pairs. Nucleic Acids Res. 2003;31:3450–3460. doi: 10.1093/nar/gkg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morse S.E., Draper D.E. Purine–purine mismatches in RNA helices: evidence for protonated GA pairs and next-nearest neighbor effects. Nucleic Acids Res. 1995;23:302–306. doi: 10.1093/nar/23.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voitiuk A.A., Blizniuk A.A. Effect of protonation of nucleic acid bases on the energy of formation of the Watson–Crick pairs. Mol. Biol. (Mosk) 1988;22:1080–1086. [PubMed] [Google Scholar]
- 43.Kao T.H., Crothers D.M. A proton-coupled conformational switch of Escherichia coli 5S ribosomal RNA. Proc. Natl Acad. Sci. USA. 1980;77:3360–3364. doi: 10.1073/pnas.77.6.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gluick T.C., Gerstner R.B., Draper D.E. Effects of Mg2+, K+, and H+ on an equilibrium between alternative conformations of an RNA pseudoknot. J. Mol. Biol. 1997;270:451–463. doi: 10.1006/jmbi.1997.1119. [DOI] [PubMed] [Google Scholar]
- 45.Jacquemont S., Hagerman R.J., Leehey M.A., Hall D.A., Levine R.A., Brunberg J.A., Zhang L., Jardini T., Gane L.W., Harris S.W., et al. Penetrance of the fragile X-associated tremor/ataxia syndrome in a premutation carrier population. JAMA. 2004;291:460–469. doi: 10.1001/jama.291.4.460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.