Abstract
The RNA-binding ability of ribosomal protein L1 is of profound interest since the protein has a dual function as a ribosomal protein binding rRNA and as a translational repressor binding its mRNA. Here, we report the crystal structure of ribosomal protein L1 in complex with a specific fragment of its mRNA and compare it with the structure of L1 in complex with a specific fragment of 23S rRNA determined earlier. In both complexes, a strongly conserved RNA structural motif is involved in L1 binding through a conserved network of RNA–protein H-bonds inaccessible to the solvent. These interactions should be responsible for specific recognition between the protein and RNA. A large number of additional non-conserved RNA–protein H-bonds stabilizes both complexes. The added contribution of these non-conserved H-bonds makes the ribosomal complex much more stable than the regulatory one.
INTRODUCTION
L1, one of the largest ribosomal proteins, is located on the side protuberance, opposite the L7/L12 stalk of the 50S ribosomal subunit. In the isolated state L1 proteins have two different conformations: a closed conformation in the case of bacterial L1 from Thermus thermophilus (1) and an open one in the case of its archaeal homologues from Methanococcus jannaschii (2) or Methanococcus thermolithotrophicus (3). L1 is a primary RNA-binding ribosomal protein, which associates independently, specifically and strongly with 23S rRNA (4). We have recently determined the crystal structure of L1 in complex with a specific 55 nt fragment of 23S rRNA (5). It is known that L1 proteins in Escherichia coli and in Methanococcus species regulate their own expression by binding to their mRNAs, thereby acting as translational repressors (6–10). L1 from Escherichia coli (EcoL1) mediates autogenous regulation of translation by binding to a region within the leader sequence, close to the Shine–Dalgarno sequence, of the mRNA of the L11 operon coding for ribosomal proteins L1 and L11 (6). L1 from M.vannielii (MvaL1) was shown to be an auto-regulator of the MvaL1 operon encoding ribosomal proteins L1, L10 and L12 (8). It was also shown that EcoL1 can inhibit the in vitro translation of MvaL1 polycistronic mRNA and, conversely, that MvaL1 can inhibit the synthesis of both L11 and L1 proteins of E.coli (9). However, there are no structural data available on the conformation of the mRNA to which L1 binds, either in isolation or with L1 bound.
In bacteria and archaea, the L1 regulatory target site exhibits high similarity in both sequence and secondary structure to the L1 binding site on the 23S rRNA. L1 proteins from mesophilic and thermophilic bacteria and archaea bind to the specific site on 23S rRNA with 5- to 10-fold higher affinity than to their regulatory binding site on the mRNA (10). This difference fits the requirements of classical regulation of ribosomal synthesis (feedback inhibition) based on direct competition between the two binding sites. Direct comparison of the structures of the complexes formed by regulatory ribosomal proteins with their targets on rRNA and mRNA is an attractive task. Only one such investigation has been made recently for ribosomal protein S8 (11).
We prepared and crystallized several complexes between bacterial and archaeal L1 proteins and specific fragments of their mRNAs (S. Tishchenko, unpublished). The structure of one of these complexes, M.jannaschii L1 (MjaL1) with a fragment of its mRNA, has been determined and compared with the structure of L1–rRNA complex. The comparison revealed a network of strongly conserved H-bonds, which are inaccessible to the solvent. Atoms involved in these H-bonds must be responsible for the specific RNA–protein recognition.
MATERIALS AND METHODS
Identification of a MjaL1mRNA fragment suitable for MjaL1–mRNA complex formation
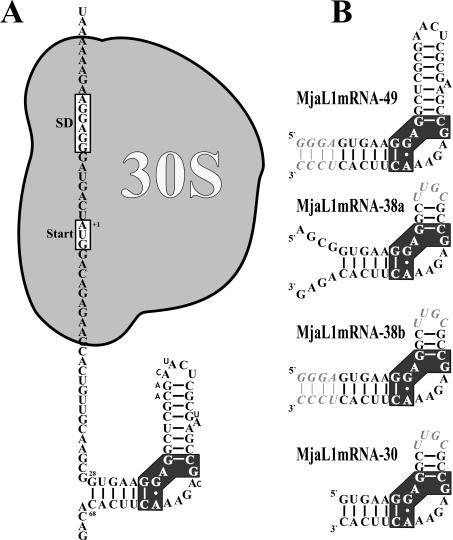
The first aim was to design a ‘minimal’ mRNA fragment, an RNA fragment as short as possible which still would retain the full affinity for L1. From the structure of the L1–rRNA complex (5), we concluded that, apart from the nucleotides essential for L1 recognition (shown with black background in Figure 1), the two helices flanking the asymmetric loop might be essential for L1 binding. Comparison of the L1 binding sites on the mRNA from M.jannaschii with that of the closely related M.vannielii revealed that the 7 bp helix (nucleotides +28 to +34 and +62 to +68) flanking the asymmetric loop was identical in both species, whereas the sequence of the upper part of the stem–loop structure (comprising nucleotides +40 to +51) was only poorly conserved between the two species (Figure 1A). Thus, it was a reasonable assumption that this stem–loop structure was not essential for L1 binding.
Figure 1.
Secondary structure of the regulatory L1-binding site on the L1mRNA of M.jannaschii and derivatives thereof used in binding experiments and crystallization trials. (A) Localization of the L1-binding site and of the 30S ribosomal subunit as part of the translation initiation complex on the L1mRNA. Nucleotides different in the M.vannielii L1mRNA are shown with smaller sized letters. (B) Fragment MjaL1mRNA-49 comprising nucleotides +28 to +68 with four additional base pairs (italic) at the 3′ and the 5′ end. Fragment MjaL1mRNA-38a comprising nucleotides +24 to +38 and +54 to +72 is closed by the tetraloop UUGC (italic). Fragment MjaL1mRNA-38b is similar to MjaL1mRNA-38a, but the free 3′ and 5′ ends have been replaced by an extended helix as in MjaL1mRNA-49. Fragment MjaL1mRNA-30 comprising nucleotides +28 to +38 and +54 to +68 is closed by a tetraloop as in MjaL1mRNA-38. The cluster of nucleotides conserved in all 23S rRNA and mRNA binding sites is shown with black background.
Several RNA constructs that included the regulatory MjaL1 binding site were synthesized (Figure 1B) and tested for their MjaL1-binding activity using filter binding assays. As Table 1 shows, fragments MjaL1mRNA-49, MjaL1mRNA-38a and MjaL1mRNA-38b (Figure 1B) bind L1 with virtually the same (full) affinity as a 250 nt MjaL1mRNA fragment, confirming that the stem–loop structure distal to the asymmetric loop does not contribute to the L1–mRNA interaction, and can be shortened to a 3 bp helix closed by a tetraloop without any loss of affinity for L1. Unexpectedly, the 30 nt fragment MjaL1mRNA-30 (Figure 1B) does not exhibit any specific affinity for MjaL1, indicating that the 7 bp helix alone is not sufficient for L1 binding. The apparent dissociation constant determined for MjaL1 in complex with the MjaL1mRNA-30 (Kd of 10−7 M) is virtually the same as for MjaL1 in complex with a 16S rRNA fragment, which was used as a negative control. MjaL1, in general, shows a rather high unspecific affinity for any RNA. L1 from the closely related M.vannielii, which does not show such an unspecific affinity for RNA, does not bind to MjaL1mRNA-30, but exhibits the full affinity for the other three short mRNA fragments tested (data not shown).
Table 1.
Binding of MjaL1 to different RNA fragments
| RNA | Description | Kd [M] |
|---|---|---|
| MjaL1mRNA-250 | 250 nt fragment of the MjaL1mRNA (22), comprising nucleotides −29 to +220, used as positive control | 5.0 × 10−10 |
| MjaL1mRNA-49 | Figure 1B | 5.2 × 10−10 |
| MjaL1mRNA-38a | Figure 1B | 5.8 × 10−10 |
| MjaL1mRNA-38b | Figure 1B | 4.9 × 10−10 |
| MjaL1mRNA-30 | Figure 1B | 1.0 × 10−7 |
| Mja16SrRNA | 145 nt fragment of 16S rRNA (23) used as negative control | 2.5 × 10−7 |
Binding was carried out in the presence of 350 mM KCl as described in Materials and Methods.
All three transcripts exhibiting the full affinity for L1 were used to prepare MjaL1–mRNA complexes for crystallization; so far the best crystals were obtained with the 49 nt fragment. Large single crystals of this complex containing SeMetL1 were used for the data collection and for structural studies.
Protein and RNA preparation
The fragment MjaL1mRNA-49 was obtained by transcription from linearized plasmid DNA using T7 RNA polymerase. RNA was purified on denaturing (8 M urea) 15% (w/v) acrylamide (19:1, acrylamide/bis-acrylamide) gels, using 90 mM Tris-borate, pH 8.2, 1 mM EDTA, as running buffer. RNA was eluted by 50 mM Tris–HCl buffer, pH 7.5 (25°C), 1 mM EDTA, purified by anion-exchange (DEAE-Sepharose) chromatography, precipitated by ethanol and dissolved in 1 mM sodium citrate, pH 6.4. The gene for MjaL1 was cloned and overexpressed in E.coli strain BL21(DE3) as a host (12). To avoid the potential misincorporation of amino acids [e.g. lysine instead of arginine, as in (13)] the host strain was co-transformed with pUBS520, a plasmid carrying the gene for (14). For MAD phasing, selenomethionine-MjaL1 was produced by essentially the same procedure but with E.coli strain B834(DE3) and minimal media containing selenomethionine (15). Cells were suspended in 100 mM Tris–HCl buffer (pH 7.5) (25°C), 0.8 M NaCl, 100 mM MgCl2, 0.2 mM EDTA, 5 mM β-mercaptoethanol, 0.1 mM PMSF and disrupted by brief sonication. The cell debris and the ribosomes were removed by centrifugation. The main contaminating proteins in the supernatant were then precipitated by heating for 10 min at 65°C and removed by centrifugation. The supernatant was diluted with 50 mM sodium acetate (pH 5.5) to adjust the concentration of NaCl to 0.15 M and the protein was purified by cation-exchange (CM-Sepharose) chromatography. A linear gradient from 0.15 M to 0.8 M NaCl was used for elution. The protein preparation was dialyzed into 10 mM sodium cacodylate buffer, pH 6.0, containing 100 mM NaCl.
Filter-binding assay
Uniformly 32P-labeled RNA fragments were synthesized in vitro from templates linearized by SmaI in the presence of [α-32P]UTP (800 Ci/mmol; New England Nuclear Corp.) using the MAXIscript T7 Kit (Ambion Inc., Austin, TX). Unincorporated nucleotides were removed with QIAquick Nucleotide Removal Kit (QIAGEN Inc.). The purity and integrity of the transcripts were confirmed by electrophoresis on 12.5% polyacrylamide gels containing 8 M urea. The affinity of MjaL1 to RNA-binding fragments was determined by nitrocellulose filter-binding assay as described previously (10). The binding buffer contained 50 mM Tris–HCl, pH 7.6, 20 mM MgCl2, 350 mM KCl, 1 mM β-mercaptoethanol and 0.04% BSA.
Crystallization
The RNA fragment and the protein were mixed in equimolar amounts (3 mg/ml for RNA and 4.5 mg/ml for MjaL1). Crystals were grown by the vapor diffusion method at 22°C. Hanging drops were made by mixing MjaL1–mRNA complex with 3% PEG 10 K, 300 mM KCl, 50 mM sodium cacodylate buffer, pH 6.0 and 6% 2-methyl-2,4-pentandiol, 120 mM KCl in a 6:1:1 volume ratio, respectively, and the well solution was made from 30% PEG 10 K in 100 mM sodium cacodylate buffer, pH 6.0. Crystals appear within five days. Before freezing in liquid nitrogen, the crystals were transferred to the solution composed of 21% butane-2,3-diol, 1.5% PEG 10 K, 60 mM KCl, 50 mM sodium cacodylate, pH 6.0.
Data collection and structure determination
Data were collected from a single SeMet crystal of MjaL1–mRNA at the MPG/GBF beamline BW6, DESY (Hamburg, Germany) using a MAR CCD detector and were processed and merged with the XDS program suite (16). The crystal structure of the regulatory complex MjaL1–mRNA was initially solved by the MAD method in space group I222. An electron density map of a workable quality was obtained and allowed us to build models of both mRNA and L1 molecules within the complex. However, we could not refine this structure to an R-free value less than 40%. Detailed analysis of experimental data shows that crystals of the L1–mRNA complex are pseudohemihedrally twinned and belong to space group C2 with unit cell parameters a = 212.3 Å, b = 68.9 Å, c = 115.9 Å and β = 123.0°, and two monomers in the asymmetric unit. Because for these crystals a × cosβ is almost equal to −c, the cell can be indexed as I222 with a = 68.9, b = 116.1 and c = 178.5 Å, where orthorhombic a-axes are parallel to a* axes of the monoclinic reciprocal unit. The cumulative intensity distribution calculated with TRUNCATE of the CCP4 program suite (17) indicated pseudohemihedral twinning in space group C2. The twin law h + 2l, −k, −l describes a real space rotation about an axis perpendicular to the crystallographic 2-fold. The twin fraction was estimated to be ∼0.35 by the Britton plot (18). The location of the complex in the correct unit cell was done by the molecular replacement method using the obtained structure as a model. The molecular replacement yielded an unambiguous solution with a correlation coefficient of 70.7% and R-factor of 41.2%. Unfortunately, the electron density map calculated with detwinned structure-factor amplitudes and model phases was of lower quality than with twinned data. Therefore, we used twinned data for the model building and refinement. The final model, refined to an R-factor of 27.5% and an R-free of 31.4% at 3.4 Å resolution, includes 214 amino acid residues and 49 nt. Data and refinement statistics are summarized in Table 2. Determination of heavy atom positions, initial phasing, density modification and refinement were executed using CNS (19). The map interpretation and model building were performed with O (20). NCS restraints were used during the early stages of refinement, but the two molecules in the asymmetric unit were finally refined separately. The structural data and the coordinates have been deposited in the Protein Data Bank (accession code 1U63).
Table 2.
Data collection and refinement statistics
| Crystallographic dataa | |
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 212.3, b = 68.9, c = 115.9, α = 90°, β = 123.0°, γ = 90° |
| Wavelength (Å) | 1.05 |
| Resolution (Å) | 30–3.40 (3.61–3.40) |
| Number of reflections | 33 549 (5012) |
| Number of unique reflections | 17 645 (2718) |
| Completeness (%) | 89.4 (83.9) |
| Averaged redundancy | 1.9 (1.84) |
| I/σi(I) | 9.68 (2.79) |
| Rsym(I) (%)b | 6.2 (31.8) |
| Refinement statistics | |
| Resolution range (Å) | 8–3.4 |
| Reflections | 16 761 (2066) |
| R-factor (%) | 27.5 (38.5) |
| Free R-factor (%) | 31.4 (43.0) |
| r.m.s. deviation | |
| Bond lengths (Å) | 0.0104 |
| Bond angles (°) | 1.87561 |
| Improper angles (°) | 4.56 |
| Mean B value (overall, Å2) | 70.4 |
aValues in parenthesis are statistics for the highest resolution shell.
bRsym = ∑|I − 〈I〉|/∑〈I〉, where I is the measured intensity of each reflection and 〈I〉 is the intensity averaged from several observations of symmetry-related reflections.
RESULTS AND DISCUSSION
Structure of L1–mRNA complex
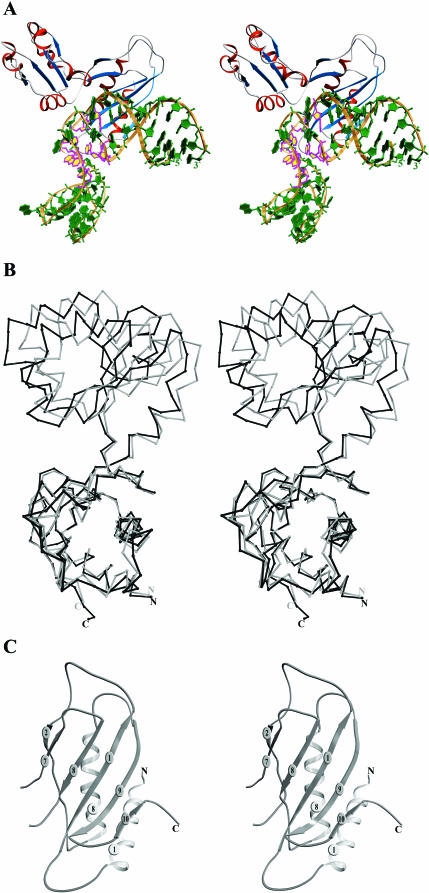
Here, we report the crystal structure of ribosomal protein L1 from M.jannaschii in complex with the 49 nt fragment of the M.jannaschii L1 binding site on its mRNA. A stereo view of the complex is shown in Figure 2A. L1 is an elongated molecule with two domains connected by a hinge region. The overall three-dimensional structure of MjaL1 bound with mRNA is close to that of MjaL1 in the isolated state described earlier (2). In both cases, the protein is in the ‘open’ conformation and the structure of each domain does not undergo essential conformational change upon complex formation. However, MjaL1 in the mRNA-bound form displays a small (about 2 Å) closing of the cavity between the two domains (Figure 2B), which might be dependent on the crystal packing. The protein interacts with RNA mostly through domain I, a ribbon representation of which is given in Figure 2C.
Figure 2.
(A) Stereo view of the L1–mRNA complex. The ribose-phosphate backbone is in gold, bases are in green, β-strands in blue and α-helices in red. Conserved nucleotides are shown in magenta and yellow. (B) Superposition of the structures of the isolated MjaL1 protein (gray) and MjaL1 from the present complex (black) with least squares minimization of differences in Cα atom coordinates of domain I. (C) Stereo view of the MjaL1 domain I.
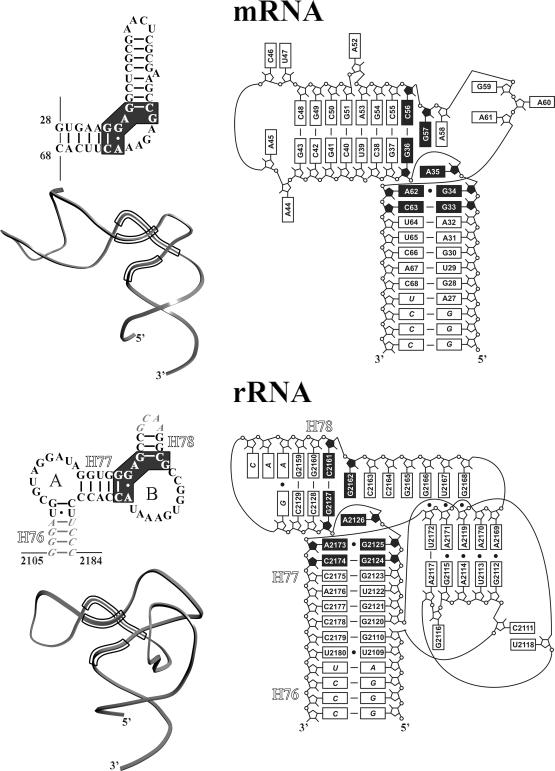
The fragment of mRNA is made of two regular double helices separated by a sharp turn (Figures 2 and 3). The first of these helices is terminated by the non-canonical A62–G34 base pair, the second one contains a bulged nucleotide A52 and terminates with a tetraloop (the RNA is numbered from the A of the AUG codon, Figure 1). Chain A27–G43 is bent through 90° at position G36 whereas chain C48–C68 forms a loop A58–A61 in the middle part of the fragment. As a result, ribose-phosphate groups of G37 and C63 are brought into proximity. Phosphate groups of nucleotides G34–C38 and U65–C66 are approximately in the same plane and surround a slightly concave region of about 14 × 16 Å2 through which mRNA mainly interacts with L1. The surface of the first helix of the mRNA fragment is complementary to the surface of the β-sheet of domain I.
Figure 3.
Secondary structures, ribbon representation and schematic illustration of three-dimensional structures of the mRNA and rRNA fragments. Nucleotide replacements, which were introduced to the RNA fragments to facilitate the complex crystallization, are in italic. Conserved nucleotides are shown with black background.
Two monomers in the asymmetric unit are connected by a non-crystallographic 2-fold axis and have slightly different conformations at the regions of two flexible loops β1–β2 and α8–β9 (Figure 2C), not involved in interactions with mRNA and located in domain I. Superposition of domains I yields an r.m.s. deviation of 0.89 Å for all Cα atoms; that of domain II is 0.56 Å. The two mRNA molecules have similar conformations with r.m.s. deviation between P atoms of about 0.57 Å, whereas those nucleotides that interact with the protein coincide much better (r.m.s. deviation of 0.27 Å). Two complexes in the asymmetric unit form a tightly associated dimer, where RNA molecules contact each other through the interconnecting loop and the bulged nucleotide A52. In the crystal, symmetry-related dimers interact through the residues of domain II or making a head-to-tail RNA arrangement.
Comparison of the structures of the L1–mRNA and L1–rRNA complexes
Earlier, we have published the crystal structure of ribosomal protein L1 from Sulfolobus acidocaldarius in complex with a specific 55 nt fragment of 23S rRNA from T.thermophilus (5). Now it is possible to compare the structure of the ribosomal complex L1–rRNA and the structure of the present regulatory complex L1–mRNA (Figures 3 and 4). The L1 proteins in both complexes have similar overall three-dimensional structures and demonstrate an ‘open’ conformation.
Figure 4.
Stereo ribbon representation of the L1–mRNA and L1–rRNA complexes in the same orientation.
Schemes of the secondary and spatial structures of the mRNA and rRNA fragments as well as their ribbon models are presented in Figure 3. It is seen that the rRNA molecule has a more complicated three-dimensional structure, because of two loops interacting with each other. In mRNA, one of these loops (A), is absent, the other (B) is six residues shorter. In spite of this difference, both RNAs have the same unique region in the junction of two helices. In the rRNA fragment, helices 76 and 77 form a co-axial helix that is perpendicular to helix 78, similar to the structure formed by the two perpendicular helices of the mRNA binding site. In both RNAs, the junction of the two helices contains nucleotides strictly conserved in all L1-binding sites in large rRNAs as well as in sites specific for L1 in mRNAs from those bacteria and archaea, for which feedback regulation has been experimentally proved. These nucleotides are connected by a network of conserved hydrogen bonds, most of which are inaccessible to the solvent. This network strongly stabilizes the unique three-dimensional structure of the region. Comparison of these unique sites in both kinds of L1–RNA complexes shows that they are structurally conserved with an r.m.s. deviation between P atoms of ∼0.50 Å.
Comparison of the protein–RNA interactions in the complexes
In the ribosomal and regulatory complexes, protein L1 interacts with RNA through both domains. Domain I contacts RNA via the slightly concave surface formed by the inner face of the β-sheet and two spatially adjacent loops, containing residues identical in all known L1 sequences. The number of residues that contact the RNA is substantially less in domain II than in domain I, particularly in the L1–mRNA complex.
The contact area between L1 protein and RNA is more extensive in the ribosomal complex than in the regulatory one due to differences in the spatial organization of the two RNAs. In each complex there are two sites of interaction with L1. In the mRNA, one site includes mainly nucleotides of the first helix and of the junction region. On the rRNA surface, the analogous site is formed by nucleotides of helix 77 and one strand of helix 78. Both RNA molecules interact through this site mainly with domain I of the protein. Highly conserved nucleotides of this site (G33, G34, G36, C63 in the mRNA or G2124, G2125, G2127, C2174 in the rRNA) form hydrogen bonds with strictly conserved residues located in the beginning of strand β1 and spatially adjacent loop β9–β10.
The second region of interaction is formed by nucleotides of loop B in the rRNA or the interconnecting loop in the mRNA. Nucleotides of this site interact with L1 residues of domain II. The more than 2-fold shortening of the interconnecting loop in the mRNA relative to loop B in the rRNA decreases significantly the number of RNA–protein contacts in the regulatory complex.
It should be noted that nucleotides at 3′ and 5′ ends, which form 4 bp in the first helix of MjaL1mRNA-49 fragment, are not involved in interaction with L1. Consequently, we can suggest that the absence of L1-binding capacity in the 30 nt fragment of MjaL1mRNA may be associated with some deformation of the unique RNA structure in this short mRNA fragment. Probably, these 4 bp are needed for retention of this unique spatial structure of L1-binding site on RNA.
RNA–protein recognition
Binding sites on the protein and RNA surfaces include regions responsible for RNA–protein recognition (we call them ‘recognition modules’) and regions, which form additional intermolecular contacts (21). It is known that many ribosomal proteins are structurally and functionally interchangeable between ribosomes from different species. These data suggest that the structure of recognition modules on the proteins and rRNAs should be highly conserved and complementary to each other. The polar atoms of recognition modules usually form a network of intermolecular RNA–protein hydrogen bonds, some of which are inaccessible to the solvent. We have recently suggested that atoms involved in these hydrogen bonds are responsible for RNA–protein recognition (21).
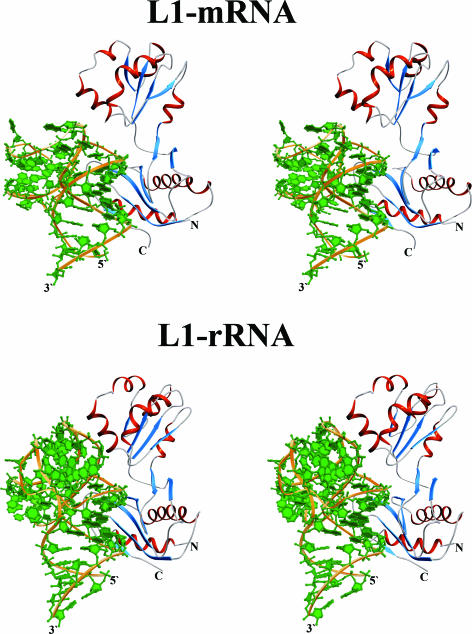
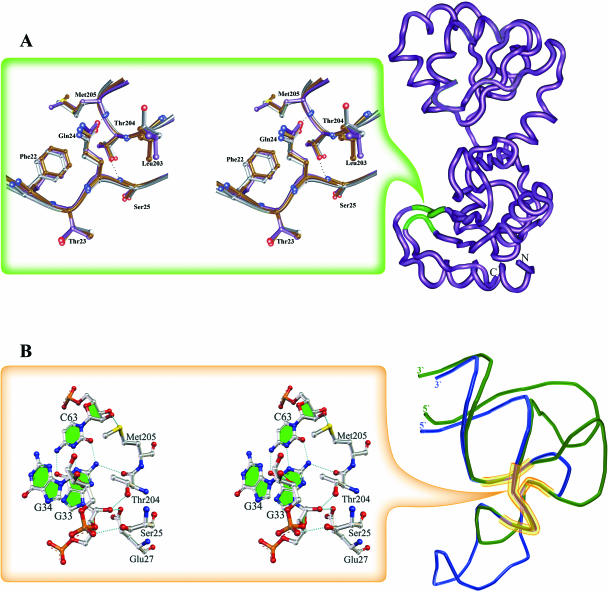
Detailed analysis of all known crystal structures of L1 proteins from different organisms revealed a structurally invariant region in domain I. This region contains a cluster of strictly conserved amino acid residues; their relative positions and side-chain conformations are stabilized by intramolecular hydrogen bonds. It is noteworthy that there is no difference in the conformation of this region in the isolated L1 molecule and in L1 bound to rRNA or mRNA (Figure 5A). It seems that the structure of this site is preformed to bind RNA and does not undergo sufficient conformational changes upon binding. These data enable us to consider this region of the protein as its RNA recognition module. It should be mentioned that we suggested a key role of this region in RNA binding previously (3), based solely on the analysis of crystal structures of isolated L1 proteins. The comparison between the two L1–RNA complexes proves this suggestion to be correct.
Figure 5.
Invariant regions in L1 proteins and in the rRNA and mRNA fragments. Conserved H-bonds are shown with dotted lines. (A) The location of the invariant structure on the surface of L1 proteins is shown in green on the MjaL1 model (right). Superposition of the corresponding regions in L1 proteins in isolated and RNA-bound forms is shown on the left. Isolated MjaL1 is in gray, MjaL1 complexed with mRNA in magenta and SacL1 complexed with rRNA in brown. (B) Superposition of the mRNA (blue) and rRNA (green) fragments; the conserved unique structure is outlined (right).
In two L1–RNA complexes, the rRNA and mRNA fragments also possess practically identical structures in and around the junction of two corresponding helices (Figure 5B). These regions contain highly conserved nucleotides and form the unique conformation strongly stabilized by intramolecular hydrogen bonds.
In the L1–rRNA and L1–mRNA complexes, polar atoms of the RNAs and L1 invariant regions form five conserved RNA–protein hydrogen bonds inaccessible to the solvent (Figure 5B). These bonds are formed by the atoms of amino acid residues Ser25, Glu27, Thr204, Met205 and of nucleotides G33, G34, C63. We have made three point mutations, Thr204Gly, Met205Gly and Met205Asp, and characterized them with respect to their RNA-binding potency by a filter-binding assay. The analysis revealed that the above listed mutants have decreased rRNA-binding ability compared with the wild-type protein and do not bind the mRNA at all. Structural modelling shows that these mutant proteins lose one of the conserved hydrogen bonds with RNA. Thus, in both cases conserved RNA–protein hydrogen bonds play a crucial role in L1–RNA recognition and binding. However, the binding of mRNA to L1 turned out to be more sensitive to point mutations than the binding of rRNA. This may be the result of considerably fewer contacts between L1 and mRNA than between L1 and rRNA.
Acknowledgments
This work was supported by the Russian Academy of Sciences, the Russian Foundation for Basic Research (No. 03-04-48327 and No. 04-04-49634), the Program of RAS on Molecular and Cellular Biology and the Program of the RF President on support of outstanding scientific schools. The research of M.G. was supported in part by International Research Scholar's award from the Howard Hughes Medical Institute. The research of W.P. was supported by the Austrian Science Fund (FWF, P17164-B10). Funding to pay the Open Access publication charges for this article was provided by the Austrian Science Fund.
REFERENCES
- 1.Nikonov S., Nevskaya N., Eliseikina I., Fomenkova N., Nikulin A., Ossina N., Garber M., Jonsson B.-H, Briand C., Svensson A., Aevarsson A., Liljas A. Crystal structure of the RNA-binding ribosomal protein L1 from Thermus thermophilus. EMBO J. 1996;15:1350–1359. [PMC free article] [PubMed] [Google Scholar]
- 2.Nevskaya N., Tishchenko S., Fedorov R., Al-Karadaghi S., Liljas A., Kraft A., Piendl W., Garber M., Nikonov S. Archaeal ribosomal protein L1: the structure provides new insights into RNA binding of the L1 protein family. Structure. 2000;8:363–371. doi: 10.1016/s0969-2126(00)00116-7. [DOI] [PubMed] [Google Scholar]
- 3.Nevskaya N., Tishchenko S., Paveliev M., Smolinskaya Y., Fedorov R., Piendl W., Nakamura Y., Toyoda T., Garber M., Nikonov S. Structure of ribosomal protein L1 from Methanococcus thermolithotrophicus. Functionally important structural invariants on the L1 surface. Acta Crystallogr. 2002;D58:1023–1029. doi: 10.1107/s0907444902006157. [DOI] [PubMed] [Google Scholar]
- 4.Zimmermann R.A. Interactions among protein and RNA components of the ribosome. In: Chambliss G., Craven G., Davies J., Davies K., Kahan L., Nomura M., editors. Ribosomes. Structure, Function and Genetics. Baltimore: University Park Press; 1980. pp. 135–169. [Google Scholar]
- 5.Nikulin A., Eliseikina I., Tishchenko S., Nevskaya N., Davydova N., Platonova O., Piendl W., Selmer M., Liljas A., Drygin D., Zimmermann R., Garber M., Nikonov S. Structure of the L1 protuberance in the ribosome. Nature Struct. Biol. 2003;10:104–108. doi: 10.1038/nsb886. [DOI] [PubMed] [Google Scholar]
- 6.Gourse R., Sharrock R., Nomura M. Control of ribosome synthesis in Escherihia coli. In: Hardesty B., Kramer G., editors. Structure, Function, and Genetics in Ribosomes. New York: Springer-Verlag; 1986. pp. 766–788. [Google Scholar]
- 7.Baier G., Piendl W., Redl B., Stöffler G. Structure, organization and evolution of L1 equivalent ribosomal protein gene of the archaebacterium Methanococcus vannielii. Nucleic Acids Res. 1990;18:719–724. doi: 10.1093/nar/18.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayer C., Köhrer C., Gröbner P., Piendl W. MvaL1 autoregulates the synthesis of the three ribosomal proteins encoded on the MvaL1 operon of the archaeon Methanococcus vannielii by inhibiting its own translation before or at the formation of the fist peptide bond. Mol. Microbiol. 1998;27:455–468. doi: 10.1046/j.1365-2958.1998.00693.x. [DOI] [PubMed] [Google Scholar]
- 9.Hanner M., Mayer C., Köhrer C., Golderer G., Gröbner P., Piendl W. Autogenous translational regulation of the ribosomal MvaL1 operon in the archaebacterium Methanococcus vannielii. J. Bacteriol. 1994;176:409–418. doi: 10.1128/jb.176.2.409-418.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Köhrer C., Mayer C., Neumair O., Gröbner P., Piendl W. Interaction of ribosomal L1 proteins from mesophilic and thermophilic Archaea and Bacteria with specific L1-binding sites on 23SrRNA and mRNA. Eur. J. Biochem. 1998;256:97–105. doi: 10.1046/j.1432-1327.1998.2560097.x. [DOI] [PubMed] [Google Scholar]
- 11.Merianos H.J., Wang J., Moore P.B. The structure of a ribosomal protein S8/spc operon mRNA complex. RNA. 2004;10:954–964. doi: 10.1261/rna.7030704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kraft A., Lutz C., Lingenhel A., Gröbner P., Piendl W. Control of ribosomal protein L1 synthesis in mesophilic and thermophilic archaea. Genetics. 1999;152:1363–1372. doi: 10.1093/genetics/152.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calderone T.L., Stevens R.D., Oas T.G. High-level misincorporation of lysine for arginine at AGA codons in a fusion protein expressed in Escherichia coli. J. Mol. Biol. 1996;262:407–412. doi: 10.1006/jmbi.1996.0524. [DOI] [PubMed] [Google Scholar]
- 14.Brinkmann U., Mattes R.E., Buckel P. High-level expression of recombinant genes in Escherichia coli is dependent on the availability of the dnaY gene product. Gene. 1989;85:109–114. doi: 10.1016/0378-1119(89)90470-8. [DOI] [PubMed] [Google Scholar]
- 15.Leahy D.S., Hendrickson W.A., Aukhil I., Erickson H.P. Structure of a fibronectin type III domain from tenascin phased by MAD analysis of the selenomethionyl protein. Science. 1992;258:987. doi: 10.1126/science.1279805. [DOI] [PubMed] [Google Scholar]
- 16.Kabsch W. Integration, scaling, space-group assignment, post refinement. In: Rossmann M.G., Arnold E., editors. International Tables for Crystallography. Vol. F. Dordrecht: Kluwer Academic Publishers; 2001. [Google Scholar]
- 17.Bailey S. The CCP4 suit: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 18.Fisher R.G., Sweet R.M. Treatment of diffraction data from crystals twinned by merohedry. Acta Crystallogr. 1980;A36:755–760. [Google Scholar]
- 19.Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.-S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 20.Jones T.A., Zhou J.Y., Cowan S.W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 21.Nevskaya N.A., Nikonov O.S., Revtovich S.V., Garber M.B., Nikonov S.V. Identification of RNA-recognizing modules on the surface of ribosomal proteins. Mol. Biol. (Moscow) 2004;38:789–798. [PubMed] [Google Scholar]
- 22.Kraft A. 2000. Interaction of ribosomal L1 proteins from thermophilic Methanoccoccus species with their specific binding sites on 23S rRNA and mRNA. PhD thesis, Universit of Innsbruck Austria.
- 23.Gruber T., Köhrer C., Lung B., Shcherbakov D., Piendl W. Affinity for ribosomal protein S8 from mesophilic and (hyper)thermophilic archaea and bacteria for 16S rRNA correlates with the growth temperatures of the organisms. FEBS Lett. 2003;549:123–128. doi: 10.1016/s0014-5793(03)00760-9. [DOI] [PubMed] [Google Scholar]