Abstract
The herpes simplex virus type 1 UL42 DNA polymerase processivity factor interacts physically with UL9 and enhances its ability to unwind short, partially duplex DNA. In this report, ATP hydrolysis during translocation of UL9 on single-stranded (ss) or partially duplex DNA was examined in the presence and absence of UL42 to determine the effect of UL42 on the catalytic function of UL9. Our studies reveal that a homodimer of UL9 is sufficient for DNA translocation coupled to ATP hydrolysis, and the steady-state ATPase catalytic rate was greater on partially duplex DNA than on ss DNA in the presence or absence of UL42. Although UL42 protein increased the steady-state rate for ATP hydrolysis by UL9 during translocation on either partially duplex or ss DNA, UL42 had no significant effect on the intrinsic ATPase activity of UL9. UL42 also had no effect on the catalytic rate of ATP hydrolysis when UL9 was not limiting but enhanced the steady-state ATPase rate at only subsaturating UL9 concentrations. At subsaturating UL9 to DNA ratios, stoichiometric concentrations of UL42 were shown to increase the amount of UL9 bound to ss DNA at equilibrium. These data support a model whereby UL42 increases the ability of UL9 to load onto DNA, thus increasing its ability to assemble into a functional complex capable of unwinding duplex DNA.
INTRODUCTION
The herpes simplex virus type 1 (HSV-1) genome possesses seven essential viral DNA replication genes which, when introduced into mammalian or insect cells, are necessary and sufficient for the amplification of plasmids containing a functional HSV-1 origin of replication (ori) (1–3). Six of these encode enzymatic activities that act at the replication fork, are conserved among all of the herpes viruses, and are sufficient in vitro to replicate single-stranded (ss) circular DNA without ori sequences (4,5). These activities include a heterodimeric DNA polymerase consisting of a catalytic subunit (pol) encoded by the UL30 gene and the pol processivity factor encoded by the UL42 gene (hereafter referred to as UL42), a ss DNA-binding protein called infected cell protein-8 (ICP8), encoded by the UL29 gene, and a heterotrimeric helicase-primase encoded by the UL5, UL52 and UL8 genes (3,6). Although not required in vitro for ori-independent replication, the seventh protein, which is encoded by the UL9 gene (hereafter referred to as UL9), binds specifically to HSV-1 ori sequences and possesses helicase and DNA-dependent ATPase activities (7–11). The sequence-specific DNA-binding domain resides in the C-terminal portion of the UL9 protein, while the helicase and ATPase activities are encoded by the N-terminal domain (12–14). Mutants with defects that destroy either the DNA-binding or the helicase activity of UL9 fail to synthesize viral DNA, suggesting that both represent essential functions for viral DNA replication (15–17). Moreover, most mutations in full-length UL9 that permit dimerization and binding of the protein to ori sequences, but which destroy the helicase and ATPase activities, have dominant-negative effects on virus replication in vivo (3,18,19).
Counterparts of the UL9 gene have been found for all of the alphaherpesviruses and for some of the betaherpesviruses, including human herpesviruses 6 and 7 (20–24). Based upon its ability to bind to cis-acting ori sequences (7), its co-localization with the other DNA replication proteins to replication compartments in infected cells (16), and its absolute requirement for the synthesis of viral DNA (25), UL9 is presumed to be involved in the initiation of HSV-1 DNA synthesis (3), although to date, no biochemical assay has demonstrated ori-dependent DNA replication in vitro. Forms consistent with unwinding of supercoiled plasmids containing ori sequences have been visualized by electron microscopy in the presence of UL9 protein and ICP8 (26), and together these two proteins can unwind short duplex DNA containing a minimal ori sequence (27). Also consistent with its role in initiation, UL9 physically interacts with other essential HSV DNA replication proteins, including ICP8 (28), UL42 (29) and the non-catalytic component of the helicase-primase complex encoded by UL8 (30). ICP8 binds to the extreme C-terminus of UL9 protein and enhances both its helicase and DNA-dependent ATPase activities (10,26–28,31–34). Although the mechanisms by which it does so have not been fully elucidated, removal of the C-terminal 27 amino acids of UL9 protein results in loss of the ability to bind to ICP8 and enhanced ability of UL9 protein to unwind DNA in vitro (31,34–36). Thus, the C-terminal portion of UL9 protein has been hypothesized to negatively regulate the activity of UL9 protein and binding of ICP8 to that region is proposed to abolish that regulation by masking that domain and/or by altering the conformation of the protein (33,35,36). Interestingly, the same C-terminal deletions in UL9 protein result in decreased ability to amplify ori-containing plasmids in transient assays (31), suggesting the importance of the ICP8–UL9 protein interaction for the replication of viral DNA.
Recently, we reported that the physical interaction between the UL9 and UL42 proteins led to enhancement of the helicase activity of UL9 on short non-specific partially duplex DNA substrates (37). However, UL42 enhanced the helicase activity of UL9 protein at only subsaturating concentrations of the latter with respect to DNA. In contrast, enhancement of UL9 helicase activity by ICP8 occurred at saturating and subsaturating concentrations of UL9 to DNA. In addition, ICP8, but not UL42, removed or dramatically reduced the lag period prior to unwinding by UL9 protein, which is thought to be due to a required assembly step (11,37). Thus, it appears likely that UL42 and ICP8 are both UL9 accessory proteins that facilitate unwinding by at least some distinct mechanisms. Uncovering the mechanism by which UL42 enhances UL9 unwinding is hampered by the fact that unwinding of DNA by UL9 is stoichiometric, requiring high molar ratios of UL9 to the DNA substrate. In contrast, hydrolysis of ATP, which drives the translocation of UL9 protein along DNA and is required for unwinding, can be described in catalytic terms. In the studies described in this report, we have investigated the impact of UL42 on the ATPase activity of UL9 to better understand the means by which UL42 enhances the function of UL9.
MATERIALS AND METHODS
Purification of proteins
The HSV-1 UL9 and UL42 proteins were expressed in Sf-9 insect cells using recombinant baculoviruses (gifts from Robert Lehman and Mark Charllberg, respectively). Proteins were purified to near homogeneity as described previously (37). UL9 protein was stored at 4°C, whereas UL42 was stored at 4°C or flash-frozen in liquid nitrogen and stored in aliquots at −80°C.
DNA substrates
Gel-purified synthetic oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA). For most experiments, the ss DNA substrate was a 38mer with the sequence 5′-TCTCCTCCTTGGCCTCTATCTCCTTTTTTTTTTTTTTT-3′. A 5′ biotinylated form of this sequence was used in equilibrium DNA-binding experiments. The partially duplex DNA substrate contained a 23mer with the sequence 5′-GGAGATAGAGGCCAAGGAGGAGA-3′ annealed to the above 38mer sequence to form a molecule with 23 bp of double-stranded (ds) DNA with a 3′ ss overhang of 15 nt. The fully duplex DNA substrate consisted of the above 38mer sequence annealed to the 38mer fully complementary sequence 5′-AAAAAAAAAAAAAAAGGAGATAGAGGCCAAGGAGGAGA-3′. Concentrations of oligonucleotides were determined spectrophotometrically. Partially duplex or fully ds DNA substrates were prepared by mixing a 1:1 molar ratio of the appropriate oligonucleotides in buffer containing 10 mM HEPES, pH 7.6, 50 mM NaCl and 5% glycerol, heating to 50°C for 3 min and cooling slowly to room temperature. Annealed substrates were stored at 4°C.
ATPase assays
Reactions were performed at 37°C in buffer containing 50 mM EPPS, pH 8.6, 25 mM NaCl, 4.5 mM MgCl2, 5 mM DTT, 0.1 mg BSA/ml, 10% glycerol, 8% DMSO, and non-radioactive ATP and [α-32P]ATP adjusted to a specific activity of ∼40 μCi/μmol. Except for ATP titration experiments, the final concentration of ATP in reactions was 3 mM. In experiments in which the ATP concentration was varied, the specific activity of the radioactively labeled ATP was held constant. Except as indicated, reactions were initiated by the addition of UL9 protein. However, in those experiments in which UL9 was preincubated with DNA, the reaction buffer also contained 2.5 mM EDTA and reactions were initiated by the addition of MgCl2 to a final concentration of 6 mM, with or without UL42 protein. Reactions were terminated by the addition of EDTA to a final concentration of 125 mM. The products were separated by ascending thin layer chromatography on polyethyleneimine–cellulose sheets using 1 M formic acid and 0.4 M LiCl as the developing buffer. The migration of non-radioactive nucleotide controls was determined under ultraviolet light and the products were quantified using a Molecular Dynamics phosphorImager and ImageQuant software. ATP hydrolyzed under condition n was determined as the amount of ADP as a proportion of total radioactivity in that lane, and was corrected for background presence of ADP in reactants (condition 0, no UL9) as follows:
| 1 |
The maximum observed rate of ATP hydrolysis under each condition was calculated based on kinetic profiles, and observed rate constants (kobs) were obtained by dividing the maximum rates by the concentration of UL9 used in that experiment. Plots of kobs versus concentration of ATP or DNA [Y] were fit to the Michaelis–Menten function
| 2 |
to estimate the catalytic rate constant (kcat) at infinite [Y] and the concentration of Y at half kcat (Km). All kinetic constants reported are apparent. To determine the efficiency of ATP hydrolysis by UL9 (Vmax/Km), the rate of ATP hydrolysis (V) was plotted as a function of UL9 concentration and fit to the Hill equation (38,39) with a coefficient (n) of 2:
| 3 |
where Vmax is the estimated maximum rate observed at infinite UL9 concentration [UL9] and Km is the concentration of UL9 at half Vmax. Statistical comparisons of kinetic constants in pairwise combinations were made using the Student's t-test with a 95% confidence interval.
Equilibrium DNA-binding assays
A fixed concentration of 38mer (200 nM), covalently linked to biotin at the 5′ end, was incubated with UL9 in 60 μl DNA-binding buffer (50 mM EPPS, pH 8.6, 25 mM NaCl, 5 mM DTT, 0.1 mg BSA/ml, 10% glycerol, 3 mM ATP and 2.5 mM EDTA) for at least 20 min at 0°C. For those experiments in which UL42 was added, UL9 was incubated with DNA for 10 min prior to the addition of UL42 and the entire mixture was incubated for an additional 20 min at 0°C. Protein–DNA complexes were cross-linked by the addition of formaldehyde to 1% and incubated on ice for 1 min. The biotinylated DNA and associated protein were pulled-down by the addition of 50 μl streptavidin beads (Sigma, St. Louis, MO), which had been washed and suspended in an equal volume of binding buffer. Samples were mixed for at least 10 min at 0°C and the beads were pelleted by low-speed centrifugation and washed in binding buffer. DNA and associated proteins were eluted by boiling in dissociation buffer (62.5 mM Tris–HCl, pH 6.8, 1% SDS, 5% 2-mercaptoethanol, 10% glycerol and 0.002% bromophenol blue) for 3 min followed by SDS–PAGE through 10% polyacrylamide gels. Proteins were transferred to nitrocellulose membrane followed by immunoblot analysis using polyclonal antibody generated in rabbits to purified UL9. [125I]Staphylococcus aureus protein A was used to detect antigen–antibody complexes as described previously (40) and the radioactive bands corresponding to a UL9 monomer (∼88 kDa) were quantified using a phosphorImager and ImageQuant software. The amount of UL9 bound to DNA was determined as the radioactivity in the band from reactions containing UL9 and DNA less than that from reactions containing UL9 but lacking DNA.
RESULTS
UL9 ATPase activity with standard helicase DNA substrate
Studies in our laboratory demonstrated that the HSV-1 polymerase accessory protein, UL42, enhanced the DNA helicase activity of UL9, but only at limiting concentrations of UL9 (37). To determine whether UL42 enhanced the DNA helicase activity of UL9 by altering its catalytic function, we examined the DNA-dependent ATPase activity of UL9. It is this activity that presumably drives the unidirectional (3′ to 5′) translocation of the UL9 protein as it unwinds DNA (8,9,11,37,41). Previous studies that have investigated the DNA-dependent ATPase activity of UL9 have employed high stoichiometries of the protein with respect to DNA (10,34,35). To better approximate the subsaturating conditions utilized in our DNA helicase assays, we employed a sensitive assay to measure the hydrolysis of [α-32P]ATP to [α-32P]ADP and inorganic phosphate by 50 nM UL9 in the presence of excess (500 nM) DNA facilitator. We began these studies using as the facilitator for translocation, the same DNA substrate we previously employed to study the UL9 helicase activity. This DNA was a 23/38mer partial duplex containing a 15 nt 3′ ss DNA overhang to provide a loading site for UL9.
To ensure that ATP concentrations were not limiting under our assay conditions, we determined the kinetics of ATP hydrolysis as a function of ATP concentration (0.3–5 mM). Following initiation of reactions with UL9, biphasic kinetics of ATP hydrolysis was observed, reflecting an initial lag period followed by a faster rate of hydrolysis thereafter (see Figure 1; data not shown). Rate constants were determined based on the maximum hydrolysis rate observed over a 30 min reaction period and were plotted as a function of ATP concentration. Concentrations of ATP above 5 mM were inhibitory, despite the addition of extra MgCl2 to compensate for the chelating activity of ATP (data not shown). The data fit well to the Michaelis–Menten function and estimated an apparent Km for ATP of 0.68 ± 0.11 mM and kcat of 360 ± 19 min−1 under these reaction conditions (Table 1). Thus, the use of 3 mM ATP, a concentration ∼5 times the Km value, in all subsequent reactions, ensured that ATP was not limiting.
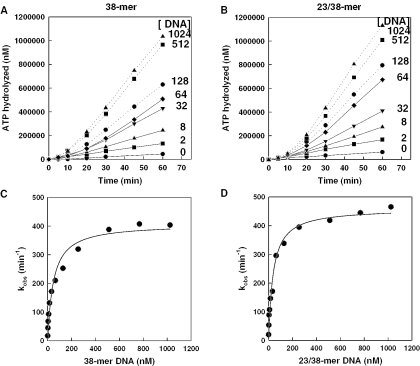
Figure 1.
DNA concentration dependence of UL9 ATPase activity. Kinetics of hydrolysis of a saturating concentration of ATP (3 mM) by 50 nM UL9 was determined in the presence of increasing concentrations of 38mer ss DNA (A) or partially duplex 23/38mer DNA (B). Observed rate constants (kobs) were calculated as the maximum rate observed divided by the concentration of UL9 and were plotted as a function of ss or partially duplex DNA concentration (C and D, respectively). The data in C and D were fit to the Michaelis–Menten function (Equation 2) to predict the apparent kinetic constants reported in Table 1.
Table 1.
Apparent kinetic constants for hydrolysis of ATP by UL9 during DNA translocation
| DNA | kcat (ATP) (min−1)a | Km (ATP) (mM)a | kcat (DNA) (min−1)b | Km (DNA) (nM)b | Vmax (UL9) (μM/min)c | Km (UL9) (nM)c |
|---|---|---|---|---|---|---|
| 38mer | n.d.d | n.d. | 407 ± 19 | 47 ± 9.7 | 17.8 ± 1.8 | 72.7 ± 14.7 |
| 23/38mer | 360 ± 19 | 0.68 ± 0.11 | 460 ± 16 | 38.0 ± 5.9 | 25.0 ± 2.2 | 94.2 ± 14.9 |
aA 50 nM fixed concentration of UL9 was incubated with excess (500 nM) DNA and increasing concentrations of ATP (0–5 mM). Observed ATP hydrolysis rate constants were plotted against ATP concentration and fit to the Michaelis–Menten function (Equation 2) to estimate apparent kcat and Km (±SD) for ATP as indicated in Materials and Methods.
bA 50 nM fixed concentration of UL9 was incubated with increasing concentrations of DNA (0–1024 nM) in the presence of excess (3 mM) ATP. Apparent kinetic constants were obtained by fitting plots of ATP hydrolysis rate constants as a function of DNA concentration to the Michaelis–Menten function.
cUL9 concentrations were varied (0–288 nM) and incubated with a fixed concentration of DNA (50 nM) and ATP (3 mM). Plots of ATP hydrolysis rates as a function of UL9 concentration were fit to the Hill equation (Equation 3) using a Hill coefficient of 2 to estimate the maximum rate at infinite UL9 concentration (Vmax) and the apparent Km for UL9 as indicated.
dNot done (n.d.).
Dependence of ATP hydrolysis by UL9 on type and concentration of DNA translocation substrate
Since translocation of UL9 was presumed to be along the 38mer strand in the 3′ to 5′ direction, we tested the ability of a fixed concentration of UL9 (50 nM) to hydrolyze ATP with the 38mer ss DNA as facilitator compared with the partially duplex 23/38mer. The kinetics of ATP hydrolysis by UL9 with increasing concentrations of the respective DNAs was monitored over a period of 1 h at 37°C. The kinetics of ATP hydrolysis with both ss and partially duplex DNA (Figure 1A and B, respectively) revealed a lag period from ∼10 to 20 min, depending upon the DNA concentration employed. The rate constants were calculated from the maximum observed rates and plotted as a function of DNA concentration (Figure 1C and D). Each plot fit well to the Michaelis–Menten function and estimated an apparent Km of 47 ± 9.7 nM for the ss 38mer and 38.0 ± 5.9 nM for the partial duplex (Table 1). Similar Km values were obtained in other experiments and suggested that the binding affinity of UL9 for partially duplex DNA translocation substrate was slightly higher than for ss DNA (P = 0.014). However, the catalytic rate constant for hydrolysis of ATP in the presence of the partially duplex DNA was significantly greater (P < 0.01) than with the ss 38mer, despite the fact that translocation of UL9 is along the 38mer strand in both cases. The faster catalytic rate of hydrolysis of ATP on partially duplex versus ss DNA was consistent in multiple experiments.
Kinetics of DNA-dependent ATP hydrolysis at increasing UL9 concentrations
The effect of UL9 concentration on the rate of ATP hydrolysis at a fixed concentration of each DNA translocation substrate was also examined in order to determine concentrations of protein, which were saturating or non-saturating with respect to DNA. For maximum sensitivity, we selected a DNA concentration of 50 nM, close to the apparent Km for DNA determined above using a fixed UL9 concentration of 50 nM. The kinetics of ATP hydrolysis over a 1 h period was monitored at concentrations of UL9 ranging from 6.3 to 288 nM. For both DNA facilitators, the data fit well to the Hill equation (Equation 3) with a coefficient of 2, but poorly to the Michaelis–Menten function (data not shown), suggesting that binding of UL9 for translocation along the ss and partially duplex DNA is cooperative (39). Because UL9 exists as a stable homodimer in solution (8,9), cooperativity presumably occurs between two homodimers of UL9. The apparent Kms for UL9 as it hydrolyzes ATP during translocation on the ss or partially duplex DNA were estimated from the Hill plots to be 72.7 ± 14.7 nM and 94.2 ± 14.9 nM, respectively (Table 1). This small difference was judged to be significant. The estimated maximum rate of hydrolysis of ATP (Table 1) was also significantly higher (P < 0.01) during translocation of UL9 on the partially duplex DNA (25.0 ± 2.2 μM/min) compared with that on ss DNA (17.8 ± 1.8 μM/min) for catalytic efficiencies of 265 min−1 and 245 min−1, respectively. These results suggest that some of the ATP hydrolysis on partially duplex substrates could be non-productive in terms of driving UL9 translocation or be used to facilitate the local separation of the duplex strands.
Effect of UL42 on kinetics of ATP hydrolysis by UL9
We next determined the effect of UL42 on the DNA-dependent ATPase activity of UL9 using a fixed concentration of UL9 and increasing concentrations of DNA translocation substrate. To facilitate complex formation between the UL9 and UL42 proteins, UL42 and UL9 were incubated at a 4:1 molar ratio, the optimum ratio previously determined for enhancement of UL9 unwinding activity (37), and reactions were initiated with the protein mixture to obtain final concentrations of 200 nM UL42 and 50 nM UL9. Figure 2 shows a plot of the observed rate constants for ATP hydrolysis at increasing concentrations of 38mer ss DNA translocation substrate. The results revealed a kcat (400 ± 17 min−1) and Km (55.0 ± 10.0 nM) for ss DNA not significantly different from the values obtained with 50 nM UL9 alone (compare Figure 2 with Figure 1C). The similar efficiencies (kcat/Km) for ATP hydrolysis during translocation of UL9 on ss DNA in the absence (7.28 per min per nM) compared with the presence of UL42 (8.59 per min per nM) suggests that UL42 does not have an impact on the coupling of ATP hydrolysis to DNA translocation. Studies to determine the effect of UL42 on the ATPase activity of UL9 during translocation on increasing concentrations of the partially duplex DNA substrate were also performed (data not shown). However, the Km for partially duplex DNA increased in the presence of UL42, most likely due to coating of the partially duplex DNA by the relatively high concentrations of UL42 that binds preferentially to ds DNA (42,43). Such coating likely interfered with the ability of UL9 to access the short ss portion of the 23/38mer required for UL9 to bind. Interference with UL9 binding to ss DNA has also been observed in the presence of ICP8, a protein that binds preferentially to ss DNA (35). To reduce the possibility of binding interference, we reduced the ratio of UL42 to UL9 to 2:1 and incubated the DNA with UL9 prior to the addition of UL42.
Figure 2.
Effect of UL42 on DNA dependence of UL9 ATPase activity. A 4:1 molar ratio of UL42 and UL9 were incubated together for 20 min at room temperature to facilitate complex formation. Reactions containing increasing concentrations of 38mer ss DNA were initiated by the addition of the protein mixture to achieve final concentrations of 50 nM UL9 and 200 nM UL42 and ATP hydrolysis was monitored over a 60 min period. The observed rate constants were calculated, plotted and fit to the Michaelis–Menten function as described in the legend to Figure 1 to estimate an apparent Km for DNA of 55 ± 10 nM and an apparent kcat of 400 ± 17 min−1 in the presence of UL42.
Effect of UL42 on steady-state rates of UL9 DNA-dependent ATP hydrolysis
We previously showed that UL42 enhanced the steady-state rate of unwinding of partially duplex DNA by UL9 at subsaturating but not the saturating concentrations of UL9 required for maximum helicase activity (37). To test the effect of UL42 on the DNA-dependent ATPase activity of UL9, we selected a concentration of UL9 (25 nM) well below the apparent Km for UL9 for ATP hydrolysis with a fixed DNA concentration of 50 nM (see Table 1). UL9 was incubated with ss or partially duplex DNA and ATP in the presence of 2.5 mM EDTA for at least 20 min to ensure equilibrium binding and assembly of UL9 on DNA. Reactions were then initiated by the addition of MgCl2 alone or together with UL42 (50 nM final concentration). In reactions containing UL9 as the only added protein, the lag period, previously observed in reactions that were initiated with UL9 at similar ratios to DNA, was virtually eliminated with the partially duplex DNA and substantially reduced with ss DNA (compare Figure 1A and B with Figure 3A and B). The kinetics of ATP hydrolysis was similar in reactions with or without UL42 for the first 20 min when either the ss or partially duplex DNA was used as the translocation substrate. However, 20 min after the addition of UL42, an increased rate of ATP hydrolysis by UL9 was observed in reactions containing both UL9 and UL42 compared with those containing UL9 only (Figure 3). The addition of UL42 increased the steady-state rate of ATP hydrolysis by UL9 by ∼40% with ss 38mer DNA as translocation substrate and ∼60% during translocation of UL9 on the 23/38mer partial duplex (Table 2). UL42 had no effect on steady-state ATP hydrolysis rates by UL9 in reactions in which no DNA facilitator was added or in those in which a duplex 38/38mer translocation substrate was provided (Table 2). Moreover, no significant ATP hydrolysis was observed in reactions that contained UL42 and no UL9 (data not shown). Thus, UL42 does not increase the intrinsic DNA-independent ATPase activity of UL9.
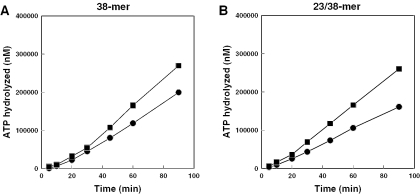
Figure 3.
Effect of UL42 on the steady-state rates of ATP hydrolysis at limiting UL9 concentration. A limiting concentration of UL9 (25 nM) was incubated to equilibrium with a fixed concentration (50 nM) of 38mer or 23/38mer DNA (A and B, respectively). Reactions were initiated by the addition of MgCl2 alone (filled circles) or with MgCl2 and UL42 to achieve a 50 nM final concentration (filled squares) and hydrolysis of ATP was monitored over a 90 min period at 37°C.
Table 2.
Effect of UL42 protein on UL9 ATPase activity
| Proteina | DNAb | ATPase rate (nM/min)c |
|---|---|---|
| UL9 | None | 212 |
| UL9 + UL42 | None | 0d |
| UL9 | 38mer | 2590 |
| UL9 + UL42 | 38mer | 3600 |
| UL9 | 23/38mer | 1960 |
| UL9 + UL42 | 23/38mer | 3180 |
| UL9 | 38/38mer | 469 |
| UL9 + UL42 | 38/38mer | 492 |
aWhere applicable, DNA was incubated with a subsaturating concentration of UL9 (25 nM) in reaction buffer containing 2.5 mM EDTA as described in Materials and Methods. Reactions were initiated by the addition of MgCl2 (6 mM) with or without UL42 (50 nM) to the final concentrations indicated.
bFinal concentration of DNA facilitator, as indicated, was 50 nM.
cReactions were performed at 37°C and portions were removed over a 90 min period and terminated by the addition of EDTA to 125 mM. Rates indicated are the maximum observed rate of hydrolysis.
dThe concentration of ADP observed did not exceed that present in reaction mixtures lacking UL9.
The 20 min lag period following the addition of UL42, which was required prior to an increased steady-state rate of hydrolysis of ATP by UL9, was reminiscent of the assembly lag observed following the addition UL9, particularly at low ratios of UL9:DNA (Figure 1A and 1B). The increased steady-state rate in the presence of UL42 was consistent with a greater occupancy of UL9 on the translocation substrates. If this interpretation is correct, UL42 should have no effect on the steady-state rate of ATP hydrolysis when saturating amounts of UL9 are bound to DNA at equilibrium. We employed a 12.5:1 ratio of UL9 to DNA, a ratio predicted to be saturating, with respect to ATPase activity, based on UL9 titration experiments (data not shown). Reactions containing 10 nM DNA were initiated by the addition of UL9 alone (125 nM final concentration) or by the addition of UL9 and UL42 (125 and 250 nM, respectively, final concentrations). The results (Table 3) demonstrate that UL42 had no significant effect on the rate of ATP hydrolysis by UL9 on either the ss or partially duplex DNA substrate, consistent with a mechanism that increases the occupancy of UL9 on DNA at limiting UL9 concentrations. Because UL42 does not affect the dissociation rate of UL9 from these short DNA substrates (37), these results are consistent with the hypothesis that UL42 enhances the ability of UL9 to load onto the DNA on which it translocates.
Table 3.
Effect of UL42 protein on UL9 DNA-dependent ATPase activity at saturating UL9:DNA ratios
| Proteina | DNAb | ATPase rate (nM/min)c |
|---|---|---|
| UL9 | 38mer | 4470 ± 600 |
| UL9 + UL42 | 38mer | 4450 ± 960 |
| UL9 | 23/38mer | 3510 ± 176 |
| UL9 + UL42 | 23/38mer | 3290 ± 160 |
aReactions were initiated by the addition of UL9 alone to achieve a final concentration of 125 nM or a pre-incubated mixture of a 1:2 molar ratio of UL9 and UL42 to achieve final concentrations of 125 and 250 nM, respectively.
bFinal concentration of DNA in reaction mixtures was 10 nM.
cAverages (±SD) of the maximum rate of conversion of ATP to ADP during steady-state reactions performed in triplicate.
Effect of UL42 on DNA-binding by UL9
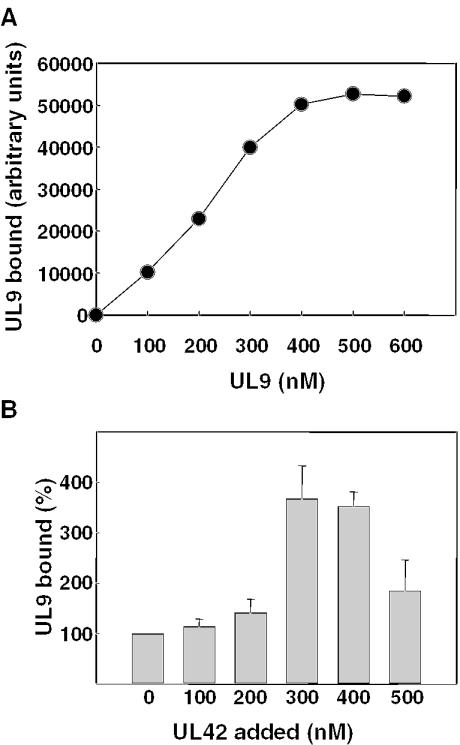
To further test this hypothesis, we performed equilibrium binding experiments of UL9 with 5′ biotinylated 38mer ss DNA. However, because UL9 dissociates rapidly from this short DNA, with a half-life of ∼2 min during translocation (37), bound UL9 was cross-linked to DNA with 1% formaldehyde prior to the isolation of DNA and DNA–protein complexes with streptavidin beads. Protein pulled down with the DNA was eluted, separated by SDS–PAGE, and the UL9-containing bands were detected by immunoblotting with specific polyclonal antibody to UL9 and [125I]protein A. Using a fixed concentration of DNA (200 nM), we demonstrated an approximately linear relationship between the amount of radioactivity in the UL9-containing bands and the concentration of UL9 added to DNA from 100 to 300 nM (Figure 4A). However, in the presence of higher concentrations of UL9 (400–600 nM), binding to DNA was saturated. These results confirm the ability of this assay to detect increases in the amount of UL9 capable of binding to DNA when incubated to equilibrium, provided that subsaturating amounts of UL9 are used. Therefore, we incubated a limiting concentration of UL9 (200 nM of monomer) with respect to DNA (200 nM) followed by the addition of UL42 or irrelevant protein (BSA) to assess the ability of UL42 to enhance UL9 loading onto DNA. DNA–protein complexes were cross-linked with formaldehyde, and the DNA and associated protein were isolated by incubation with streptavidin beads as described above. The amount of UL9 bound to the biotinylated 38mer increased significantly when stoichiometric concentrations of UL42 (300 or 400 nM) were added (Figure 4B). In contrast, addition of 200 or 300 nM BSA failed to increase the amount of UL9 bound to DNA (data not shown). With 500 nM UL42, UL9 binding was substantially reduced compared with that observed with 300 or 400 nM UL42, suggesting that large ratios of UL42 with respect to DNA and/or UL9 can interfere with the ability of UL9 to fully access this ss DNA. It is interesting to note that the ratios of UL42 to UL9 required to observe enhancement of UL9 binding to DNA were in the same range as those found to achieve significant enhancement of steady-state ATPase and helicase activities [(37) and this report]. Taken together, these results confirm that UL42 increases the ability of subsaturating amounts of UL9 to load onto ss DNA.
Figure 4.
Equilibrium binding of UL9 to ss DNA. (A) Increasing concentrations of UL9 were incubated with 200 nM 38mer biotinylated at the 5′ end, bound protein was cross-linked to DNA with 1% formaldehyde, and the DNA complexes were isolated using streptavidin beads as described in Materials and Methods. The amount of UL9 in the complexes was determined by phosphorimage analysis of immunoblots using UL9-specific antibody and [125I]protein A and plotted as a function of initial UL9 concentration. (B) A limiting concentration of UL9 (200 nM) was incubated with 200 nM biotinylated 38mer followed by the addition of increasing concentrations of UL42 as indicated. DNA–protein complexes formed at equilibrium were cross-linked, isolated, and the UL9 bound was quantified as indicated above. Within each experiment, the amount of UL9 bound to DNA in the presence of UL42 was normalized to that bound in the absence of UL42 (set at 100%). Results shown represent the mean normalized values obtained (± standard error) for three independent experiments.
DISCUSSION
Although it is hydrolysis of ATP during the unidirectional (3′ to 5′) translocation of UL9 along ss DNA that drives unwinding by UL9, UL9 hydrolyzes ATP catalytically but is required in stoichiometric quantities for efficient unwinding. In this report, we demonstrated that half saturation of 50 nM of the partial duplex by UL9 for ATP hydrolysis required 94 nM UL9, and 72 nM UL9 was required for half saturation of 50 nM ss 38mer DNA (Table 1), corresponding to 3–4 molecules of UL9 per DNA molecule. Given that UL9 exists in solution and binds DNA as a homodimer (8,9) and assuming an active protein concentration between 50 and 100%, this ratio suggests that DNA-dependent ATP hydrolysis is driven by translocation of a single homodimer of UL9 along DNA. Using 0.5 nM of a partially duplex 23/38mer DNA substrate, we previously observed that half-maximal unwinding required 9 nM UL9, indicating ∼36 UL9 molecules (monomeric) were required to unwind a single DNA partial duplex (37). The 10-fold increase in the amount of UL9 required for unwinding is likely to reflect the use in helicase assays of a 10-fold excess of ss DNA to trap unwound DNA molecules. Nevertheless, because little or no unwinding can be detected at low UL9:DNA in the absence of trap (11,37) (data not shown), it is difficult to determine how other proteins affect the activity or function of UL9 based upon the results of helicase assays alone.
We have extended our previous studies that demonstrated that UL42 enhanced unwinding of short partially duplex DNA by UL9 (37), and in this report, we examined the effect of the UL42 protein on the catalytic ATPase activity of UL9. The results demonstrate that UL42 increases the steady-state rate of ATP hydrolysis by UL9 in the presence of fully or partially ss DNA. However, the failure of UL42 to alter the ATPase activity of UL9 in the absence of DNA or in the presence of fully duplex DNA (Table 2) suggests that UL42 does not affect the global structure of the catalytic site on UL9 responsible for ATP hydrolysis. Because UL42 increased the steady-state rate of UL9 ATP hydrolysis with both ss and partially duplex DNA, enhancement of UL9 activity is not likely to be dependent upon the ds DNA-binding activity of UL42. In fact, when partially duplex DNA translocation substrate was provided, binding of high concentrations of UL42 to the DNA actually interfered with the ability of UL9 to bind the DNA, preventing an accurate assessment of Km for this DNA in the presence of UL42. Moderate interference of UL9 binding to DNA was also observed with high ratios of UL42 to ss DNA in direct DNA-binding experiments (Figure 4B).
We found that the kcat for ATP hydrolysis during translocation of UL9 along ss DNA did not differ in the presence or absence of UL42 (compare Figure 2 with Figure 1C). These results are in contrast to the effects of another UL9 accessory protein, ICP8, which was shown by others to increase the kcat for UL9 ATPase activity 4-fold (35). UL42 also appears to have no effect on the ability of UL9 to couple ATP hydrolysis to unwinding since we observed a similar ratio of kcat (∼1.4) for ATP hydrolysis on partially duplex compared with ss translocation substrate in the presence or absence of UL42 (Figures 1 and 2; data not shown).
Despite having no apparent effect on the catalytic activity of UL9 during translocation, UL42 did enhance the steady-state rate of hydrolysis of ATP by UL9 at subsaturating but not at saturating concentrations of UL9:DNA. In contrast, ICP8 has been reported to increase the DNA-dependent ATPase activity of UL9 at high UL9:DNA ratios (9,10,35). Enhancement of DNA-dependent UL9 ATPase activity by UL42 at limiting but not saturating concentrations of UL9 (Tables 2 and 3) is consistent with an increase in the number of UL9 molecules bound to DNA and/or an increase in the ability of UL9 to assemble into a functional translocation complex. In support of the former mechanism are results of steady-state experiments, in which UL9 was pre-loaded onto DNA prior to initiation. Pre-incubation of UL9 with DNA removed most of the lag period observed when reactions were initiated by the addition of UL9 (Figure 3), suggesting that fully assembled and functional UL9 was present prior to initiation. However, the addition of UL42 to the pre-bound UL9:DNA complexes resulted in an enhanced rate of ATP hydrolysis with ss or partially duplex DNA, but only after a lag period comparable with that observed following the addition of UL9 (compare Figure 3A and B with Figure 1A and B). The establishment of a new steady-state rate following the addition of UL42, but only after a lag period, was also observed for UL9 helicase activity (37). It is notable that despite the complex nature of the helicase assays, UL42 also enhanced the steady-state rate of unwinding by UL9 but only at concentrations of UL9:DNA which were limiting for that assay (37). More direct evidence in support for a model in which UL42 increases the ability of UL9 to load onto DNA was obtained in DNA-binding assays (Figure 4), which demonstrated that increasing concentrations of UL42 increased the amount of UL9 which could be cross-linked to ss DNA at equilibrium, but only when UL9 is present in limiting amounts with respect to DNA. Nevertheless, we cannot rule out the possibility that UL42 also moderately enhances the rate constant for assembly of productive UL9 complex onto DNA.
We believe it unlikely that the apparent increase in the UL9 bound to DNA in the presence of UL42 is due to decreased dissociation of UL9 from the DNA for three reasons. First, the length of DNA used (38 nt) was substantially shorter than the maximum length (60 nt) shown to enhance ATPase activity (10). Second, we have demonstrated that UL42 does not alter the half-life with which UL9 is associated with the partially duplex DNA during helicase assays (37). Third, UL42 increased UL9 binding to DNA even when dissociation of UL9 was prevented by formaldehyde cross-linking. Thus, UL42 is a UL9 loading protein that increases the affinity of UL9 for the DNA translocation substrate most likely by enhancing the rate of association of UL9 with DNA. Nevertheless, we observed no significant decrease in the apparent Km for ss DNA for hydrolysis of ATP by 50 nM UL9 in the presence or absence of UL42 (P = 0.10). Our failure to observe an effect on Km for DNA may have been due to the insensitivity of these determinations for moderate effects on UL9 loading or a relatively high error rate. Indeed, we observed an enhancement of 40% for steady-state ATPase rates on ss DNA, but the Km measurements in the presence or absence of UL42 had a standard deviation approximately equivalent to 20% of the determined values.
The finding that two different proteins can enhance the function of a DNA helicase involved in DNA replication is not unique. The essential T4 bacteriophage DNA helicase, gp41, requires both a loading protein, gp59, and the ss DNA-binding protein, gp32, to efficiently engage nascent replication forks (44,45). In contrast to UL42, gp59 has been shown to enhance both the intrinsic and DNA-dependent ATPase activity of its cognate helicase, gp41 (46), and gp59 is thought to alter the functional stoichiometry of gp41 helicase at the replication fork to facilitate unwinding (45,47). Like gp59, UL42 may increase the loading of its cognate helicase, UL9, onto DNA, thus facilitating its ability to assume the proper stoichiometry required for unwinding even when present at relatively low concentrations.
The results reported herein demonstrate that although UL42 enhances the loading of UL9 onto DNA, UL42 does not affect the catalytic activity of UL9. Therefore, we propose a model in which UL42 works synergistically with ICP8 in vivo to enhance UL9 function. It has been hypothesized that ICP8 may alter the conformation of UL9 and allow it to move from sequence-specific DNA-binding mode at ori sequences to an ss DNA-binding mode capable of translocation (35,36,48,49). However, there may be insufficient quantities of UL9 at the site to effect assembly and unwinding. Thus, at the nascent replication fork and under conditions in which UL9 concentrations are likely to be limiting, UL42 could be directed to the site of initiation as a result of its ability to physically interact with UL9. Once bound to UL9, UL42 could then direct the loading of additional UL9 homodimers to form a functional and efficient unwinding complex proximal to the origin, which in turn, could facilitate the entry of the other proteins required for DNA elongation. Studies are underway to examine the function of UL9 in the presence of both the UL42 and ICP8 accessory proteins to test this model.
Acknowledgments
We thank Shangling He for expert statistical analysis and the members of the Parris laboratory for helpful discussions. This work was supported in part by a grant from the National Institutes of Health (GM58809) to D.S.P. During part of this work, Y.Z. was supported by a training grant from the National Cancer Institute (CA 09338). Funding to pay the Open Access publication charges for this article was provided by the Department of Molecular Virology, Immunology and Medical Genetics, and OhioLink.
REFERENCES
- 1.Wu C.A., Nelson N.J., McGeoch D.J., Challberg M.D. Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J. Virol. 1988;62:435–443. doi: 10.1128/jvi.62.2.435-443.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stow N.D. Herpes simplex virus type 1 origin-dependent DNA replication in insect cells using recombinant baculoviruses. J. Gen. Virol. 1992;73:313–321. doi: 10.1099/0022-1317-73-2-313. [DOI] [PubMed] [Google Scholar]
- 3.Marintcheva B., Weller S.K. A tale of two HSV-1 helicases: roles of phage and animal virus helicases in DNA replication and recombination. Prog. Nucleic Acid Res. Mol. Biol. 2001;70:77–118. doi: 10.1016/s0079-6603(01)70014-1. [DOI] [PubMed] [Google Scholar]
- 4.Skaliter R., Lehman I.R. Rolling circle DNA replication in vitro by a complex of herpes simplex virus type 1-encoded enzymes. Proc. Natl Acad. Sci. USA. 1994;91:10665–10669. doi: 10.1073/pnas.91.22.10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falkenberg M., Lehman I.R., Elias P. Leading strand and lagging strand DNA synthesis in vitro by a reconstituted herpes simplex virus type 1 replisome. Proc. Natl Acad. Sci. USA. 2000;97:3896–3900. doi: 10.1073/pnas.97.8.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boehmer P.E., Lehman I.R. Herpes simplex virus DNA replication. Annu. Rev. Biochem. 1997;66:347–384. doi: 10.1146/annurev.biochem.66.1.347. [DOI] [PubMed] [Google Scholar]
- 7.Elias P., Lehman I.R. Interaction of origin binding protein with an origin of replication of herpes simplex virus 1. Proc. Natl Acad. Sci. USA. 1988;85:2959–2963. doi: 10.1073/pnas.85.9.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruckner R.C., Crute J.J., Dodson M.S., Lehman I.R. The herpes simplex virus 1 origin binding protein: a DNA helicase. J. Biol. Chem. 1991;266:2669–2674. [PubMed] [Google Scholar]
- 9.Fierer D.S., Challberg M.D. Purification and characterization of UL9, the herpes simplex virus type 1 origin-binding protein. J. Virol. 1992;66:3986–3995. doi: 10.1128/jvi.66.7.3986-3995.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodson M.S., Lehman I.R. The herpes simplex virus type 1 origin binding protein: DNA-dependent nucleoside triphosphatase activity. J. Biol. Chem. 1993;268:1213–1219. [PubMed] [Google Scholar]
- 11.Boehmer P.E., Dodson M.S., Lehman I.R. The herpes simplex virus type-1 origin binding protein: DNA helicase activity. J. Biol. Chem. 1993;268:1220–1226. [PubMed] [Google Scholar]
- 12.Deb D., Deb S.P. A 269-amino-acid segment with a pseudo-leucine zipper and a helix–turn–helix motif codes for the sequence-specific DNA-binding domain of herpes simplex virus type 1 origin-binding protein. J. Virol. 1991;65:2829–2838. doi: 10.1128/jvi.65.6.2829-2838.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elias P., Gustafsson C.M., Hammarsten O., Stow N.D. Structural elements required for the cooperative binding of the herpes simplex virus origin binding protein to oriS reside in the N-terminal part of the protein. J. Biol. Chem. 1992;267:17424–17429. [PubMed] [Google Scholar]
- 14.Abbotts A.P., Stow N.D. The origin-binding domain of the herpes simplex virus type 1 UL9 protein is not required for DNA helicase activity. J. Gen. Virol. 1995;76:3125–3130. doi: 10.1099/0022-1317-76-12-3125. [DOI] [PubMed] [Google Scholar]
- 15.Martinez R., Shao L., Weller S.K. The conserved helicase motifs of the herpes simplex virus type 1 origin-binding protein UL9 are important for function. J. Virol. 1992;66:6735–6746. doi: 10.1128/jvi.66.11.6735-6746.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malik A.K., Shao L., Shanley J.D., Weller S.K. Intracellular localization of the herpes simplex virus type-1 origin binding protein, UL9. Virology. 1996;224:380–389. doi: 10.1006/viro.1996.0545. [DOI] [PubMed] [Google Scholar]
- 17.Marintcheva B., Weller S.K. Residues within the conserved helicase motifs of UL9, the origin-binding protein of herpes simplex virus-1, are essential for helicase activity but not for dimerization or origin binding activity. J. Biol. Chem. 2001;276:6605–6615. doi: 10.1074/jbc.M007743200. [DOI] [PubMed] [Google Scholar]
- 18.Stow N.D., Hammarsten O., Arbuckle M.I., Elias P. Inhibition of herpes simplex virus type 1 DNA replication by mutant forms of the origin-binding protein. Virology. 1993;196:413–418. doi: 10.1006/viro.1993.1496. [DOI] [PubMed] [Google Scholar]
- 19.Malik A.K., Weller S.K. Use of transdominant mutants of the origin-binding protein (UL9) of herpes simplex virus type 1 to define functional domains. J. Virol. 1996;70:7859–7866. doi: 10.1128/jvi.70.11.7859-7866.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGeoch D.J., Dalrymple M.A., Davison A.J., Dolan A., Frame M.C., McNab D., Perry L.J., Scott J.E., Taylor P. The complete DNA sequence of the long unique region of herpes simplex virus type 1. J. Gen. Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 21.Davison A.J., Scott J.E. The complete DNA sequence of varicella zoster virus. J. Gen. Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 22.Telford E.A., Watson M.S., McBride K., Davison A.J. The DNA sequence of equine herpesvirus-1. Virology. 1992;189:303–316. doi: 10.1016/0042-6822(92)90706-u. [DOI] [PubMed] [Google Scholar]
- 23.Wu T.F., Sun W., Boussaha M., Southwick R., Coussens P.M. Cloning and sequence analysis of a Marek's disease virus origin binding protein (OBP) reveals strict conservation of structural motifs among OBPs of divergent alphaherpesviruses. Virus Genes. 1996;13:143–157. doi: 10.1007/BF00568907. [DOI] [PubMed] [Google Scholar]
- 24.Lindquester G.J., Greenamoyer C.A., Anton E.D., O'Brian J.J., Pellett P.E., Dambaugh T.R. Comparison of a 20 kb region of human herpesvirus 6B with other human beta herpesviruses reveals conserved replication genes and adjacent divergent open reading frames. Arch. Virol. 1997;142:193–204. doi: 10.1007/s007050050070. [DOI] [PubMed] [Google Scholar]
- 25.Malik A.K., Martinez R., Muncy L., Carmichael E.P., Weller S.K. Genetic analysis of the herpes simplex virus type 1 UL9 gene: isolation of a lacZ insertion mutant and expression in eukaryotic cells. Virology. 1992;190:702–715. doi: 10.1016/0042-6822(92)90908-8. [DOI] [PubMed] [Google Scholar]
- 26.Makhov A.M., Lee S.S.-K., Lehman I.R., Griffith J.D. Origin-specific unwinding of herpes simplex virus 1 DNA by the viral UL9 and ICP8 proteins: visualization of a specific preunwinding complex. Proc. Natl Acad. Sci. USA. 2003;100:898–903. doi: 10.1073/pnas.0237171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aslani A., Olsson M., Elias P. ATP-dependent unwinding of a minimal origin of DNA replication by the origin-binding protein and the single-strand DNA-binding protein ICP8 from herpes simplex virus type 1. J. Biol. Chem. 2002;277:41204–41212. doi: 10.1074/jbc.M208270200. [DOI] [PubMed] [Google Scholar]
- 28.Boehmer P.E., Lehman I.R. Physical interaction between the herpes simplex virus 1 origin binding protein and single-stranded DNA-binding protein ICP8. Proc. Natl Acad. Sci. USA. 1993;90:8444–8448. doi: 10.1073/pnas.90.18.8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monahan S.J., Grinstead L.A., Olivieri W., Parris D.S. Interaction between the herpes simplex virus type 1 origin-binding and DNA polymerase accessory proteins. Virology. 1998;241:122–130. doi: 10.1006/viro.1997.8953. [DOI] [PubMed] [Google Scholar]
- 30.McLean G.W., Abbotts A.P., Parry M.E., Marsden H.S., Stow N.D. The herpes simplex virus type 1 origin-binding protein interacts specifically with the viral UL8 protein. J. Gen. Virol. 1994;75:2699–2706. doi: 10.1099/0022-1317-75-10-2699. [DOI] [PubMed] [Google Scholar]
- 31.Boehmer P.E., Craigie M.C., Stow N.D., Lehman I.R. Association of origin binding protein and single strand DNA-binding protein, ICP8, during herpes simplex virus type 1 DNA replication in vivo. J. Biol. Chem. 1994;269:29329–29334. [PubMed] [Google Scholar]
- 32.Makhov A.M., Boehmer P.E., Lehman I.R., Grittith J.D. Visualization of the unwinding of long DNA chains by the herpes simplex virus type 1 UL9 protein and ICP8. J. Mol. Biol. 1996;258:789–799. doi: 10.1006/jmbi.1996.0287. [DOI] [PubMed] [Google Scholar]
- 33.Lee S.S.K., Lehman I.R. The interaction of herpes simplex type 1 virus origin-binding protein (UL9 protein) with box I, the high affinity element of the viral origin of DNA replication. J. Biol. Chem. 1999;274:18613–18617. doi: 10.1074/jbc.274.26.18613. [DOI] [PubMed] [Google Scholar]
- 34.Boehmer P.E. The herpes simplex virus type-1 single strand DNA-binding protein, ICP8, increases the processivity of the UL9 protein DNA helicase. J. Biol. Chem. 1998;273:2676–2683. doi: 10.1074/jbc.273.5.2676. [DOI] [PubMed] [Google Scholar]
- 35.Arana M.E., Haq B., Le Gac N.T., Boehmer P.E. Modulation of the herpes simplex virus type 1 UL9 DNA helicase by its cognate single-strand DNA-binding protein, ICP8. J. Biol. Chem. 2001;276:6840–6845. doi: 10.1074/jbc.M007219200. [DOI] [PubMed] [Google Scholar]
- 36.Murata L.B., Dodson M.S. The herpes simplex virus type 1 origin binding protein: sequence specific activation of adenosine triphosphatase activity by a double-stranded DNA containing box I. J. Biol. Chem. 1999;274:37079–37086. doi: 10.1074/jbc.274.52.37079. [DOI] [PubMed] [Google Scholar]
- 37.Trego K.S., Parris D.S. Functional interaction between the herpes simplex virus type 1 polymerase processivity factor and origin-binding proteins: enhancement of UL9 helicase activity. J. Virol. 2003;77:12646–12659. doi: 10.1128/JVI.77.23.12646-12659.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bell J.E., Bell E.T. Proteins and Enzymes. Englewood Cliffs, NJ: Prentice-Hall, Inc.; 1988. pp. 465–467. [Google Scholar]
- 39.Neet K.E. Cooperativity in enzyme function: equilibrium and kinetic aspects. Methods Enzymol. 1980;64:139–192. doi: 10.1016/s0076-6879(80)64009-9. [DOI] [PubMed] [Google Scholar]
- 40.Goodrich L.D., Rixon F.J., Parris D.S. Kinetics of expression of the gene encoding the 65-kilodalton DNA-binding protein of herpes simplex virus type 1. J. Virol. 1989;63:137–147. doi: 10.1128/jvi.63.1.137-147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levin M.K., Patel S.S. Helicases as molecular motors. In: Schliwa M., editor. Molecular Motors. Weinheim, Germany: Wiley–VCH Verlag GmbH; 2003. pp. 170–198. [Google Scholar]
- 42.Gallo M.L., Jackwood D.H., Murphy M., Marsden H.S., Parris D.S. Purification of the herpes simplex virus type 1 65-kilodalton DNA-binding protein: properties of the protein and evidence of its association with the virus-encoded DNA polymerase. J. Virol. 1988;62:2874–2883. doi: 10.1128/jvi.62.8.2874-2883.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gottlieb J., Challberg M.D. Interaction of herpes simplex virus type 1 DNA polymerase and the UL42 accessory protein with a model primer template. J. Virol. 1994;68:4937–4945. doi: 10.1128/jvi.68.8.4937-4945.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barry J., Alberts B. Purification and characterization of bacteriophage T4 gene 59 protein. A DNA helicase assembly protein involved in DNA replication. J. Biol. Chem. 1994;269:33049–33062. [PubMed] [Google Scholar]
- 45.Ma Y., Wang T., Villemain J.L., Giedroc D.P., Morrical S.W. Dual functions of single-stranded DNA-binding protein in helicase loading at the bacteriophage T4 DNA replication fork. J. Biol. Chem. 2004;279:19035–19045. doi: 10.1074/jbc.M311738200. [DOI] [PubMed] [Google Scholar]
- 46.Morrical S.W., Hempstead K., Morrical M.D. The gene 59 protein of bacteriophage T4 modulates the intrinsic and single-stranded DNA-stimulated ATPase activities of gene 41 protein, the T4 replicative DNA helicase. J. Biol. Chem. 1994;269:33069–33081. [PubMed] [Google Scholar]
- 47.Raney K.D., Carver T.E., Benkovic S.J. Stoichiometry and DNA unwinding by the bacteriophage T4 41:59 helicase. J. Biol. Chem. 1996;271:14074–14081. doi: 10.1074/jbc.271.24.14074. [DOI] [PubMed] [Google Scholar]
- 48.Aslani A., Macao B., Simonsson S., Elias P. Complementary intrastrand base pairing during initiation of herpes simplex virus type 1 DNA replication. Proc. Natl Acad. Sci. USA. 2001;98:7194–7199. doi: 10.1073/pnas.121177198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He X., Lehman I.R. Unwinding of a herpes simplex virus type 1 origin of replication (oris) by a complex of the viral origin binding protein and the single-stranded DNA binding protein. J. Virol. 2000;74:5726–5728. doi: 10.1128/jvi.74.12.5726-5728.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]