Abstract
To generate data for comparative analyses of zygomycete mitochondrial gene expression, we sequenced mtDNAs of three distantly related zygomycetes, Rhizopus oryzae, Mortierella verticillata and Smittium culisetae. They all contain the standard fungal mitochondrial gene set, plus rnpB, the gene encoding the RNA subunit of the mitochondrial RNase P (mtP-RNA) and rps3, encoding ribosomal protein S3 (the latter lacking in R.oryzae). The mtP-RNAs of R.oryzae and of additional zygomycete relatives have the most eubacteria-like RNA structures among fungi. Precise mapping of the 5′ and 3′ termini of the R.oryzae and M.verticillata mtP-RNAs confirms their expression and processing at the exact sites predicted by secondary structure modeling. The 3′ RNA processing of zygomycete mitochondrial mRNAs, SSU-rRNA and mtP-RNA occurs at the C-rich sequence motifs similar to those identified in fission yeast and basidiomycete mtDNAs. The C-rich motifs are included in the mature transcripts, and are likely generated by exonucleolytic trimming of RNA 3′ termini. Zygomycete mtDNAs feature a variety of insertion elements: (i) mtDNAs of R.oryzae and M.verticillata were subject to invasions by double hairpin elements; (ii) genes of all three species contain numerous mobile group I introns, including one that is closest to an intron that invaded angiosperm mtDNAs; and (iii) at least one additional case of a mobile element, characterized by a homing endonuclease insertion between partially duplicated genes [Paquin,B., Laforest,M.J., Forget,L., Roewer,I., Wang,Z., Longcore,J. and Lang,B.F. (1997) Curr. Genet., 31, 380–395]. The combined mtDNA-encoded proteins contain insufficient phylogenetic signal to demonstrate monophyly of zygomycetes.
INTRODUCTION
Fungi constitute a huge group of highly diverse organisms, including some of the most-studied and best-understood eukaryotic model systems: ‘baker's yeast’ (Saccharomyces cerevisiae), fission yeast (Schizosaccharomyces pombe), and the filamentous euascomycetes Neurospora crassa and Aspergillus nidulans. These species all belong to the Ascomycota. Substantially fewer scientific studies have been performed in members of the sister phylum Basidiomycota and very little is known in the remaining two phyla, Zygomycota and Chytridiomycota, often classified as ‘lower fungi’. This expression is a taxonomically vague concept borrowed from Aristotle's philosophy, visioning directed evolution from the simple (primitive, low) to the highly complex. The misnomer is most evident in the ‘higher’ ascomycetes and basidiomycetes, which evolve toward microscopic, unicellular and genetically simplified yeast-like organisms in some lineages, and toward morphologically complex, gene-rich and biochemically versatile multicellular organisms in others.
Although the availability of complete nuclear and mitochondrial sequences of more than a dozen ascomycetes provides a strong basis for biochemical investigations, only a few complete mitochondrial sequences are known from chytrids, and none from zygomycetes, a situation that has motivated the work presented here. In fact, the number of zygomycete nuclear gene sequences (mostly rRNA sequences) is so limited that it is impossible to determine with confidence whether or not Zygomycota is a monophyletic taxon (1–3). The lack of resolution in these analyses is consistent with estimates that even the combined LSU- and SSU-rRNA would contain far too little information to resolve many fungal phylogenetic relationships with confidence (4). This dataset is at most sufficient for resolving fungal inferences below the phylum level (5,6).
Sequences of complete mtDNAs from several zygomycetes might be a first step in remediating this situation. Mitochondrial phylogenies can be based on up to 13 protein sequences, and have been shown to resolve deep divergences in the fungal and animal lineages (7–9). For instance, Alexopolous et al. (10) indicate that ‘additional study is needed to determine whether the class (Trichomycetes) is a monophyletic group belonging to Zygomycota, or merely a collection of orders grouped together on the basis of a unique shared habitat’. Molecular phylogenies based on rRNA sequences were successful in moving the order Amoebidiales away from zygomycetes, although they were then placed with mesomycetozoan rotists, whose phylogenetic affiliation was unresolved and controversial (11). Only subsequent analysis with multiple mitochondrial proteins placed this group at the base of the animal divergence, with high confidence (9). This example clearly illustrates the requirement of multi-gene datasets with at least several thousand amino acid positions, for resolution of trees at the kingdom level.
Furthermore, zygomycete mtDNAs are of considerable interest for comparative gene expression studies: our preliminary data indicated the presence of a mitochondrial rnpB gene, which encodes the RNA subunit of RNase P, the enzymatically active part of an endonuclease (ribonucleo-protein) responsible for tRNA maturation. In mitochondria, the size and sequence of the RNA subunit varies substantially, which has considerably complicated its identification. The gene is apparently absent from all completely sequenced basidiomycete and chytridiomycete mtDNAs, and has a patchy distribution in ascomycetes (12); to date, there are no published data on zygomycete mitochondrial RNase P (mtP-RNA).
To help remediate the lack of data for phylogenetic inferences, and to facilitate biochemical investigations and comparative mitochondrial genome analyses, we have sequenced mtDNAs from three distantly related zygomycetes, the Mucorales Rhizopus oryzae, the Mortierellales Mortierella verticillata and the Harpellales Smittium culisetae. In this article, we compare their mitochondrial genomes (gene content, gene organization, genetic code and widely conserved 3′ RNA processing sites). We will then present secondary structure models and expression data for seven newly identified zygomycete mtP-RNAs. Finally, we will test whether or not zygomycetes are monophyletic, and provide evidence that the group I introns invasion of cox1 gene in angiosperms originated in a zygomycete close to Rhizopus.
MATERIALS AND METHODS
Strains and culture conditions
The various zygomycete strains were obtained from Kerry O'Donnell (National Center for Agricultural Utilization Research, Peoria, IL; NRRL), R.W. Lichtwardt (Department of Botany, University of Kansas, Laurence, KS; RWL) and Carolyn Babcock (Canadian Collection of Fungal Cultures, Ottawa; DAOM). All strains, R.oryzae (DAOM 148428, previously designated as Rhizopus stolonifer), R.stolonifer (DAOM 194667), Rhizopus oligosporus (NRRL 2710), M.verticillata (NRRL 6337), Radiomyces spectabilis (NRRL 2753), Mucor mucedo (NRRL 3635) and S.culisetae (strain 18-3; R.W. Lichtwardt), were grown in YG medium consisting of 0.5% yeast extract and 3% glycerol. Liquid cultures of 500 ml in 2 L Erlenmeyer flasks were grown at room temperature under gentle shaking (∼100 r.p.m.).
DNA and RNA extractions
For mtDNA and RNA extractions, the cells were broken mechanically, and a mitochondrial fraction was isolated by differential centrifugation (8). This fraction was lysed in the presence of 1% SDS and 100 μg/ml proteinase K at 50°C for 1 h, and after phenol–chloroform extraction, the nucleic acids were precipitated with ethanol. For RNA purifications, the high molecular weight RNA fraction was precipitated with 2 M LiCl, redissolved in RNase-free water and ethanol-precipitated. MtDNAs from all zygomycete strains were purified from total cellular nucleic acids by Cesium chloride/bisbenzimide density gradient centrifugation.
Cloning and sequencing of complete mtDNAs
Library construction and DNA sequencing followed previously published protocols (8). Briefly, mtDNAs were physically sheared by nebulization (13), and a size fraction of 1300–4000 bp was recovered after agarose gel electrophoresis. The DNA was incubated with a mixture of T7 DNA polymerase and Escherichia coli DNA polymerase I (the Klenow fragment) to generate blunt ends, and then cloned into the EcoRV cloning site of the phagemid pBFL6 (B. F. Lang, unpublished data). Recombinant plasmids containing mtDNA inserts were identified by colony hybridization using mtDNA as a probe. Clones contained in the random libraries were sequenced to ∼8-fold coverage, and remaining gaps were closed by primer walking or sequencing of PCR-amplified DNA fragments. The expected quality of the sequenced mtDNAs is <1 error in 10 000 bp.
The mtDNA sequences of R.oryzae, M.verticillata and S.culisetae have been deposited in GenBank (accession nos AY863212, AY863211 and AY8632133, respectively).
PCR amplification of rnpB genes
Mitochondrial rnpB genes of R.stolonifer, M.mucedo, R.spectabilis and R.oligosporus were PCR-amplified from ∼100 ng of the respective mtDNAs in a 50 μl reaction mixture [200 μM dNTP, 2.5 mM MgCl2, 2 nM of primers, 5 μl of 10× buffer and 3 U of DNA polymerases mixture from the Expand high fidelity kit (Roche Catalog no. 1732650) and degenerate primers]. The annealing temperature of the PCR amplification was 50°C. Sequences of the degenerate primers are 5′-GTAATGGCAGCATACTAGACTCAT-3′ and 5′-TTGAACTCCCAAGTTTTATGTATG-3′. The amplification products were cloned and sequenced. The rnpB sequences of R.stolonifer, R.oligosporus, M.mucedo and R.spectabilis have been deposited in GenBank (accession nos AY861439, AY861440, AY861441 and AY861442, respectively).
RNA circularization by ligation and RT–PCR
RNA ligation of mtP-RNAs, followed by RT–PCR amplification, was performed to determine the precise 5′ and 3′ termini of RNase P RNAs according to the previously published protocols (12). The primers are 5′-CTCTTATAGGATAATACAAAGTTG-3′ and 5′-GGCCGAAGAATAAAGAGGGA-3′ for M.verticillata, and 5′-ACCCTAATTTTCATTAGATATTT-3′ and 5′-AATCCTTAGTAAGGATAGCTT-3′ for R.oryzae.
RT–PCR of mtP-RNA from R.oryzae
Mitochondrial RNA of R.oryzae was treated with DNase I and extracted with phenol/chloroform, until no genomic DNA could be amplified by PCR using the mtP-RNA-specific primers 5′-TTCTTAGAGTTAAATAAGCC-3′ and 5′-TTGGAGGAAAGTCCGGG-3′. Following these treatments, we amplified the mtP-RNA by RT–PCR as described above using a sample without reverse transcription as negative control. Following amplification and separation on a 0.8% agarose gel, the resulting DNA fragment was cloned and sequenced.
S1 mapping
DNA oligonucleotides and a 10 bp DNA ladder (Invitrogen 10821-015) were labeled at their 3′ termini with ddATP-32P (Amersham PB10235) and terminal deoxynucleotidyl transferase (MBI Fermentas EP0161) according to the manufacturer's recommendations.
A total of 100 000 c.p.m. of gel-purified labeled oligonucleotides were hybridized to ∼10 μg of total RNA, as described in the protocol by Hahn and Breeden (http://www.fhcrc.org/labs/hahn/methods/mol_bio_meth/s1_oligo_probe.html), and S1 nuclease digestions were carried out at 37°C for 30 min, after the addition of 20 U of S1 nuclease and the buffer provided by the manufacturer (MBI Fermentas EN0321). The product was then ethanol-precipitated and dissolved in 4 μl of RNase-free water. An aliquot of 2 μl of the product was mixed with 2 μl of the dye solution provided with the 10 bp ladder (Invitrogen 10821-015), denatured at 75°C and loaded on a 7% polyacrylamide denaturing sequencing gel. The following oligonucleotides were used:
SSU-rRNA, 5′-AAATAAAGGGTTTAATATATTGGGAGGGACTTATTGTCCCCCCGGTAATAACCATTCAGCCACTCGTTCCCGAACGGCT-3′
cox1 mRNA, 5′-CAGATTCTAAGGGGGTGTTATATTATTAATTTAATTAAGATTGAACTGGTAATGAATTTACAGTATGGA-3′.
Phylogenetic inference
Mitochondrial protein sequences from all completely sequenced zygomycetes, chytridiomycetes and basidiomycetes were included in phylogenetic inference (for species names and GenBank accession nos, see legend of Figure 5). Protein sequences from apocytochrome b (Cob), as well as 7 subunits of NADH dehydrogenase (Nad), 3 cytochrome c oxidase (Cox) subunits and 2 ATP synthetase (Atp) subunits were aligned with Muscle (14), concatenated and trimmed with Gblocks [using default parameters (15)] to remove ambiguously aligned regions. The resulting alignment contained 2890 aligned positions. Maximum likelihood (ML) phylogenies were inferred from this alignment using both PHYML (16) and IQPNNI (17); ML bootstrap support was determined based on 100 replicates, using both programs. All phylogenies were inferred using the JTT amino acid substitution model and gamma distribution (Γ) correction for variation of rates across sites.
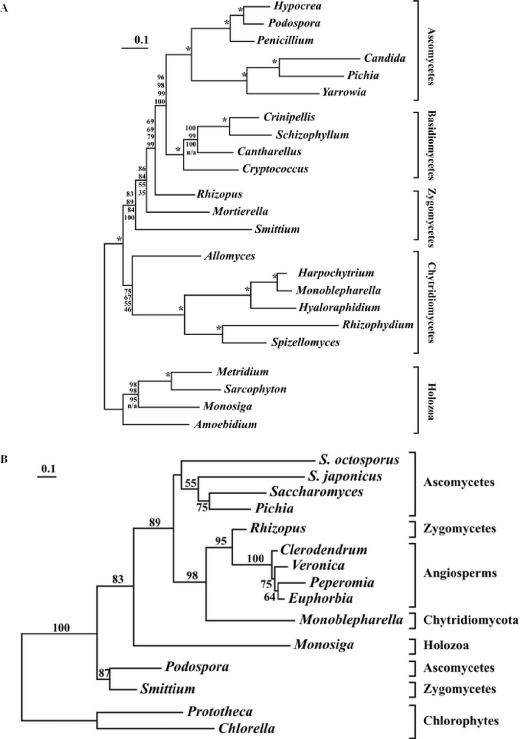
Figure 5.
(A) Fungal phylogeny based on multiple proteins. Mitochondrion-encoded protein sequences from Harpochytrium sp. 94, Crinipellis perniciosa, Cryptococcus neoformans, Hypocrea jecorina, Amoebidium parasiticum, Spizellomyces punctatus, Yarrowia lipolytica, Monosiga brevicolis, R.oryzae, Rhizophydium sp. JEL136, Penicillium marneffei, Pichia canadensis, Cantharellus cibarius, Sarcophyton glaucum, S.culisetae, Monoblepharella15, Metridium senile, A.macrogynus, Hyaloraphidium curvatum, Candida albicans, P.anserina, S.commune and M.verticillata were aligned, concatenated and trimmed. Phylogenies were inferred from the resulting 2890 character alignment using four different methods. Shown here is the ML tree inferred using IQPNNI, along with bootstrap support values from PHYML, IQPNNI, MrBayes and separate ML analysis, in order from top to bottom, based on 100 replicates. Nodes with 100% bootstrap support using all methods are indicated by an ‘asterisk’. Clearly, both the position of A.macrogynus and the branching order of the zygomycetes remain unclear, although the topology is robust overall. (B) Phylogeny of intronic ORFs. Sequences of intronic ORFs inserted in cox1 genes were obtained from the following species: ORF305 from R.oryzae, ORF248 from S.culisetae, ORF318 from Monoblepharella15 (NP_803527), ORF333 from Schizosaccharomyces japonicus (NP_705621), ORF317 from S.octosporus (NP_700369), ORF313 from P.anserina (NP_074934), ORF319 from Pichia canadensis (NP_038209), ia4 from S.cerevisiae (AAB21126), ORF251 Chlorella vulgaris (T07187), ORF234.2 Prototheca wickerhamii (NP_042245), ORF280 from Peperomia obtusifolia (AAB86934), ORF279 from Veronica catenata (CAA11340) and ORF277 from Maranta leuconeura (CAA11350). Sequences were aligned as described in Materials and Methods, and a phylogeny was inferred by ML. Only bootstrap support values >50% are shown. This tree robustly supports the monophyly between sequences from Monoblepharella15, R.oryzae, and the angiosperms, strongly suggesting horizontal intron transfer between these two groups.
Phylogenetic inference based on concatenated datasets can lead to tree reconstruction artefacts, such as long-branch attraction, resulting from differences in relative evolutionary rates between genes [sometimes referred to as the covarion-like behavior of different genes (18)]. Ideally, during the ML tree search procedure, nuisance parameters, such as branch lengths and the Γ distribution shape parameter (α), could be optimized separately for each gene; however, this method results in the estimation of many more parameters, potentially more than can be statistically justified. For this reason, we partitioned the dataset into four functional categories: Atp (253 positions), Cob (361 positions), Cox (876 positions) and Nad (1400 positions). The additional parameters included in this partitioned dataset were justified using the χ2 test (P < 0.0001). Using this partitioned dataset, a topology was inferred from this alignment using MrBayes, with branch lengths and α parameter unlinked across partitions (19). In addition, an ML tree was determined from this partitioned dataset using an adaptation of the method of (18). All possible tree topologies were generated, with constraints of groups that received at least 95% bootstrap support under ML, using both IQPNNI and PHYML (Supplementary Figure 1). Log-likelihoods were calculated separately for each partition using PHYML, under each of the topologies, and the sum over all partitions was calculated for each tree. The tree found to maximize the sum log-likelihood of the dataset was taken to be the ML tree (this method is referred to henceforth as separate analysis). Bootstrap support was also calculated using both of these methods, based on 100 replicates. Likelihood ratio tests were performed using genewise optimized site likelihoods, given the 99 tree topologies described in Supplementary Figure 1, and using Tree-Puzzle to generate sitewise likelihoods (20) along with CONSEL (21).
Additionally, phylogenetic analysis of closely related fungal, green algal and plant intronic open reading frames (ORFs) of the same cox1 intron (for species names and GenBank accession nos, see legend of Figure 5) were carried out. Sequences were aligned using Muscle and trimmed with Gblocks, and the resulting alignment was manually refined. An ML phylogeny was inferred using IQPNNI, and ML bootstrap support was determined based on 100 replicates.
RESULTS AND DISCUSSION
Genes in addition to the standard fungal set, rnpB and rps3
The mitochondrial genomes of the zygomycetes R.oryzae (previously listed incorrectly as R.stolonifer), M.verticillata and S.culisetae were completely sequenced. Like most other fungal mtDNAs, they map as circular molecules (Figure 1), although they are most likely organized as linear multimeric concatamers in vivo, as in other fungi (22). mtDNAs of the three species carry the basic fungal set of genes (Table 1), and encode a full set of tRNAs [only trnI(cau) is absent in R.oryzae], the RNA component of mitochondrial RNase P (rnpB) and a ribosomal protein (rps3; lacking in R.oryzae). MtDNA-encoded rps3 has previously been identified in several ascomycetes and in one chytridiomycete (Allomyces macrogynus; Blastocladiales), but not in other chytridiomycete orders, including Monoblepharidales, Spizellomycetales and Chytridiales. It has been proposed that this gene has been lost independently three times from opisthokont mitochondrial genomes: in the chytridiomycete lineage, in the animal lineage (23) and now also in one of the three zygomycetes presented here. We assume that, in all cases, rps3 has been transferred to the nuclear genome, like other ribosomal protein genes missing in fungal mitochondrial genomes (24).
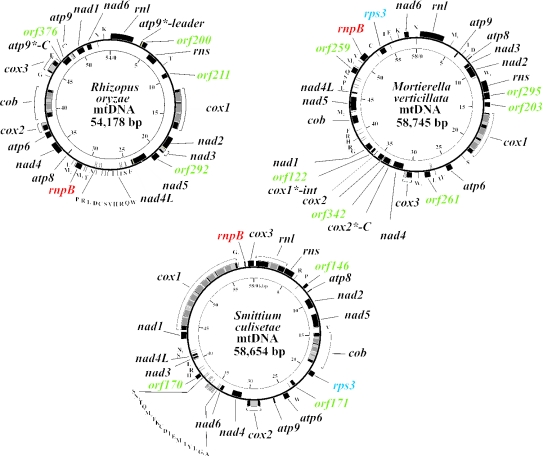
Figure 1.
Genomic maps of the mtDNAs of R.oryzae, M.verticillata and S.culisetae. The inner circle gives a scale in kilo base pair. The outer circle indicates the location of genes, exons (black) and introns plus intronic ORFs (gray). Names of ORFs, rps3 and rnpB are colored to distinguish them from standard fungal genes (black).
Table 1.
Overview of gene, ORF and intron content in zygomycete mtDNAs
| Genes and introns | R.oryzae | M.verticillata | S.culisetae |
|---|---|---|---|
| rns, rnl | ▪ | ▪ | ▪ |
| atp6,8,9 | ▪ | ▪ | ▪ |
| cob, cox1,2,3 | ▪ | ▪ | ▪ |
| nad1-6,4L | ▪ | ▪ | ▪ |
| trnA-W | 24 [trnI (cau) missing] | 26 | 27 |
| rnpB | ▪ | ▪ | ▪ |
| rps3 | □ | ▪ | ▪ |
| Group I introns (intronic ORFs) | 9(5) | 4(3) | 14(13) |
| Intron locations (number) | cox1(3), cox2(1) cox3(1), cob(2) nad3(1), atp9(1) | cox1(3), cox3(1) | rnl(1), cox1(9) cox2(1), cob(3) |
| Other ORFs | 4 | 7 | 3 |
The sizes of zygomycete mtDNAs are within a close range of 54–58 kb (Figure 1). Genes are encoded on both strands, but are not as tightly packed as in animals and some ascomycetes: only 40.6% in R.oryzae, 43.1% in M.verticillata and 35.3% in S.culisetae are coding. Nonetheless, the coding regions of nad2/nad3 and nad4L/nad5 of R.oryzae, respectively, overlap by 1 nt (i.e. the last nucleotide position of the UAA stop codon of the upstream gene is the first nucleotide of the AUG start codon of the downstream gene; Figure 1). No conservation of mitochondrial gene order is observed between these species.
First steps toward a derived genetic code in zygomycetes
Both R.oryzae and the fast-evolving S.culisetae have retained the standard translation code for protein coding genes, a trait inherited from the eubacterial ancestors of mitochondria. However, M.verticillata reassigns two UGA ‘stop’ codons as tryptophan, once each in nad3 and nad4. UGA(Trp) codons are also present in the S.culisetae intronic ORF283 and ORF248, both encoding group I introns homing endonucleases of the LAGLI-DADG type. UGA(Trp) at amino acid position 237 of ORF248 is part of a distinctive, highly conserved sequence motif of this class of endonuclease, strongly suggesting its translation as tryptophan. It is possible that the presence of this UGA(Trp) is a vestige of horizontal intron transfer from a fungus adapted to this translation code. In fact, according to our phylogenetic analyses of intron endonucleases (Figure 5A), S.culisetae ORF248 is closely related to ORF313 of Podospora anserina, which makes preferential use of UGA(Trp) in its genes and intronic ORFs. Like in the fission yeast S.pombe and the basidiomycete Schizophyllum commune, the mtDNAs of S.culisetae and M.verticillata do not encode trnW(uca), which would efficiently recognize both UGA and UGG tryptophan codons. We assume that in all these cases, UGA codons are (albeit inefficiently) decoded by trnW(cca) (25,26). However, it cannot be excluded that, alternatively, the C in the wobble position of the anticodon is either modified or partially edited to allow efficient recognition of UGA(Trp) codons.
The zygomycete mtDNAs described here encode complete sets of tRNAs sufficient to recognize all encountered codons (for codon usage, see Supplementary Table 1S). R.oryzae does not have trnI(cau); however, ATA(Ile) codons are absent in standard mitochondrial genes, although they occur in intronic ORFs. Incidentally, a strikingly similar scenario exists in Schizosaccharomyces octosporus. It has been suggested (27) that either (i) the tRNA required for translation of ATA(Ile) is imported from the cytoplasm to recognize these codons or (ii) the intronic ORFs are neither translated nor required for intron splicing. A further explanation is that these codon positions are recognized by other tRNAs at low efficiency, resulting in amino acid misincorporation, which might be permissible at poorly conserved amino acid positions of proteins.
Whatever the mechanism of codon recognition, we suggest that such unexpected codon usage in intronic ORFs reflects horizontal intron transfer from species that are adapted to the use of UGA(Trp) and/or ATA(Ile). This codon usage is common in fungi and several other eukaryotic lineages.
Eubacteria-like mtP-RNAs in zygomycete mitochondria
Mitochondrial rnpB genes (encoding the mitochondrial RNA subunit of RNase P, mtP-RNA) were identified by in silico analysis in all three zygomycetes using the previously described procedures (12), and their RNA secondary structures were modeled by phylogenetic comparative analysis (Figure 2) (see also http://megasun.bch.umontreal.ca/People/lang/rnpB/). The presence of this gene in all three zygomycetes is striking, because outside fungi, rnpB is only present in the green alga Nephroselmis olivacea (28) and in various jakobids (B. F. Lang and E. Seif, unpublished data), including Reclinomonas americana (29). Within fungi, it is only present in some ascomycetes, but absent in basidiomycetes and chytridiomycetes (12).
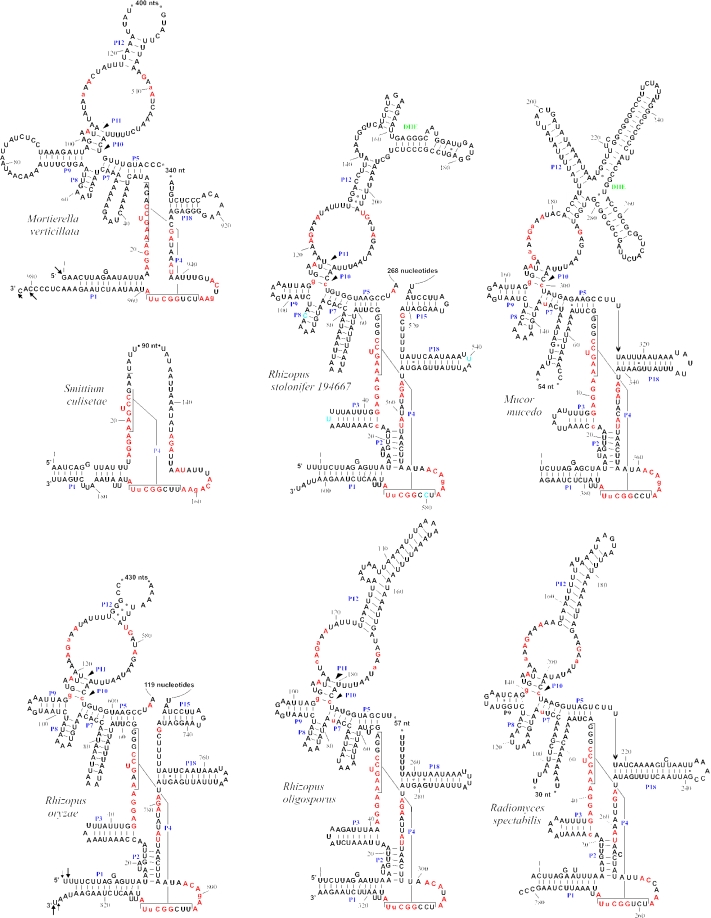
Figure 2.
Secondary structure models for mtP-RNAs from R.oryzae, R.stolonifer 194667, R.oligosporus, R.spectabilis, M.mucedo, S.culisetae and M.verticillata. Positions in red are invariant in the minimum bacterial consensus (32); uppercase letters in the mtP-RNAs indicate 100%, lowercase 90%, conservation of the minimum bacterial consensus sequence. The arrows pinpoint experimentally determined termini; arrow length is proportional to the percentage of molecules ending at a defined position. Double hairpin elements are named in green. The few nucleotides colored blue in the R.stolonifer mtP-RNA model are different in its close relative R.oryzae.
The inferred size of the S.culisetae mtP-RNA is 145 nt, close to the shortest known example (140 nt, in Saccharomycopsis fibuligera) (30). Most remarkably, the highly reduced mtP-RNA structures of S.culisetae and budding yeasts are almost identical (Figure 2), perfectly matching the minimum consensus secondary structure of fungal mtP-RNAs (12). In contrast, rnpB from M.verticillata and R.oryzae are the largest genes of this class ever identified (980 and 830 bp, respectively), even larger than rnpB genes studied in bacteria (31). In addition, the zygomycete mitochondrial RNA secondary structures are the most bacteria-like among fungi.
To verify the expression of the R.oryzae and M.verticillata genes, we determined their precise 5′ and 3′ ends by sequencing RT–PCR products of circularized mtP-RNAs (Figure 2). The 3′ end of M.verticillata is 9 nt longer than anticipated, elongated by a cytidine-rich stretch of sequence. A similar extension is located at the 3′ terminus of Schizosaccharomyces octosporus mtP-RNA (12) and downstream of protein coding genes in a variety of fungi (see below; Figure 3). The 5′ end of M.verticillata, and both the 5′ and 3′ termini of R.oryzae mtP-RNAs, match the proposed secondary structure model and reveal little heterogeneity of mtP-RNA termini (Figure 2).
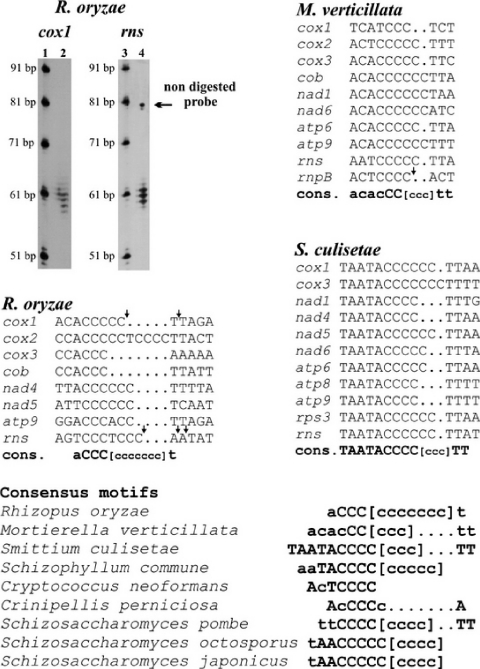
Figure 3.
3′ RNA processing motifs in zygomycetes, basidiomycetes and fission yeasts. The 3′ termini of the R.oryzae mitochondrial SSU rRNA and of cox1 mRNA were determined by nuclease S1 assays and run on a sequencing gel against a commercial 10 bp ladder (Invitrogen 10821-015) that was 3′ labeled with ddATP (32P). For experimental details see Materials and Methods. The positions of 3′ termini for both molecules are indicated in the derived consensus sequences by arrows. A small fraction of the undigested form of the SSU rRNA probe is apparent on the gel. In the lower part of the figure, additional, similar motifs in fissions yeasts and basidiomycetes are presented. Uppercase letters indicate 100% conservation and lowercase letters correspond to at least 60% nucleotide conservation. Lowercase Cs between brackets indicate the C-clusters of variable length.
In evolutionary terms, zygomycete mtP-RNA structures cover an unprecedented wide range of intermediate stages in loss of RNA structural elements. The mtP-RNAs from R.oryzae and closely related species have the most bacteria-like secondary structures, containing almost all structural elements of the bacterial minimum secondary structure consensus (32). They are followed by M.verticillata, whose structure closely resembles the more derived mtP-RNA of the ascomycete Taphrina deformans (12). Finally, the tiny, yeast-like mtP-RNA molecule of S.culisetae has no P2 helix, which is otherwise almost omnipresent in mtP-RNAs. Note that this helix is also absent in M.verticillata, potentially indicating its loss in a common ancestor.
The most bacteria-like fungal mtP-RNA secondary structure is that of R.oryzae, only lacking P13, P14 and P19, which are otherwise only present in the protist mtP-RNAs of N.olivacea and R.americana. The large size of the R.oryzae and M.verticillata mtP-RNAs is due to insertions at the J5-15 and J5-18 junctions, respectively, and in the P12 helix. In order to determine whether these regions are conserved structural elements or more variable insertion elements or introns, we amplified the cDNA sequence of the R.oryzae mtP-RNA, and the genomic sequences from the closely related Mucorales M.mucedo, R.spectabilis, R.oligosporus and R.stolonifer. Figure 2 shows that the insertion sequences can be folded into double hairpin structures [DHEs (33)]. Because the cDNA sequence of the R.oryzae mtP-RNA is identical to the genomic sequence, these variable regions are not introns. Furthermore, the insertion points and sizes of these regions vary substantially, indicating that they have been acquired recently and independently. Their presence in mtP-RNAs pinpoints structural regions that are likely not critical for RNase P activity. An analogous situation has been described in some cyanobacteria, where P-RNAs contain short tandem repeats that increase the length of helix P12. Site-directed mutagenesis experiments have shown that this helix is not required for catalytic activity in vitro, implying that it is also unlikely to be crucial for the in vivo activity (34).
Conserved C-rich motifs in mRNAs and SSU-rRNA
Small C-rich clusters are present downstream of mitochondrial protein- and SSU-rRNA coding regions, in all three species (Figure 3). The consensus sequence of this motif varies only slightly among zygomycetes. It also exists in basidiomycetes and in fission yeasts, pointing to a shared function. Mapping of the 3′ end of the M.verticillata mtP-RNA (which also terminates with this motif as discussed above; Figure 2), of cox1 and the SSU-rRNA of R.oryzae shows that these C-rich motifs are the site of 3′ RNA processing, and are retained in the mature RNA molecules. The presence of ragged 3′ ends (Figure 3) indicates that these are generated by an exonuclease trimming mechanism. Similar observations have been made in fission yeasts (12,27). This mechanism resembles that of nuclear and viral RNAs terminating in polyuridine motifs that serve as a binding site for the La protein implicated in RNA protection against exonucleases [reviewed in (35); homologs of La are known from a range of fungi and animals].
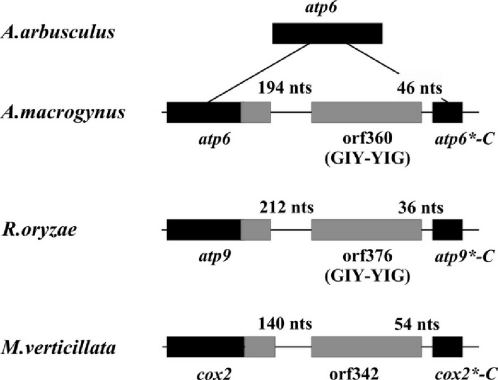
More instances of mobile endonuclease elements?
As reported earlier, the chytridiomycete fungus A.macrogynus has a novel mtDNA insertion element whose sequence is absent in a close relative, Allomyces arbusculus. This element consists of a duplicate C-terminus of a foreign atp6 gene, plus an ORF encoding an endonuclease that is responsible for its mobility (36). The inserted atp6 portion is fused in phase with the resident gene (Figure 4), reconstituting an obviously functional hybrid gene of standard length and amino acid conservation. In fact, it has been shown that homing endonucleases can be mobile even independent of introns and genes (37), and that they are capable of carrying genetic material from one site to another when they migrate.
Figure 4.
Schematic view of atp6 regions of A.macrogynus and A.arbusculus (36), atp9 of R.oryzae and cox2 of M.verticillata. Coding sequences are enclosed in boxes and intergenic spacers are represented by a thick line. Black boxes indicate sequences present before the invasion by the corresponding ORF. Gray boxes represent ORFs and newly acquired sequences.
Intriguingly, similar gene hybrids are present in the mtDNAs of R.oryzae (atp9*-C) and M.verticillata (cox2*-C), including ORF376 (R.oryzae) and ORF342 (M.verticillata) (Figure 3). ORF342 has no significant similarity to known endonucleases, but ORF376, like the mobile element endonuclease of A.macrogynus (36), encodes a protein related to homing endonucleases of the GIY-YIG type (38,39). The same structural organization is seen in the atp6-ORF360-atp6*-C gene region in A.macrogynus, atp9-ORF376-atp9*-C in R.oryzae and cox2-ORF342-cox2*-C in M.verticillata (Figure 3), indicating the presence of similar mobile elements. However, contrary to the A.macrogynus case, we currently do not have biochemical evidence for the endonucleolytic activity. Both the cox2*-C and atp9*-C fragments of the two zygomycetes encode C-terminal ends that are 100% identical at the amino acid level, suggesting that the source of potential transfers are closely related zygomycete species.
Lateral transfer of a group I intron from zygomycete to angiosperm mitochondria
A significant portion of the genomes described here is occupied by introns (R.oryzae 15.8%; M.verticillata 8.6%; and S.culisetae 27.4%). With 14 introns, the mtDNA of S.culisetae contains the largest number, 9 of which are located in the cox1 gene (Table 1). Here, all the identified zygomycete introns are of group I and 22 contain intronic ORFs: sixteen of the LAGLI-DADG type and six of the GIY-YIG type. In R.oryzae, we identified one intron [cox1-i1(ORF305)], which is most similar to introns inserted at the same positions of angiosperm cox1 genes (highest BLAST expect value of e−114 with Philodendron oxycardium, Lamium sp. and Malpighia glabra). Because this is the only group I intron in vascular plant mtDNAs, it has most likely been acquired by lateral transfer. The hypothesis of recent horizontal transfer of this intron from a fungal donor to flowering plants (40) is further based on the incongruence between the intron and organismal phylogenies, and its closer phylogenetic relationship to a fungal intron than to those of Marchantia and Prototheca (40–43). However, because of the high mobility of this intron and the possibility of multiple lateral transfers, the published phylogenetic inferences have to be interpreted with caution.
The presence of additional, highly similar introns in three fungi, R.oryzae, S.culisetae (this paper) and Monoblepharella15 (8), allows us to more rigorously test the hypothesis of Vaughn et al. by phylogenetic analysis (Figure 5A). Our analysis reveals that ORFs from R.oryzae, Monoblepharella15 and the three angiosperms group together with high support (98%). ORF305 from R.oryzae is the closest and most similar relative of the angiosperm ORFs, suggesting that the fungal donor of the group I intron and its resident ORF was a zygomycete. This scenario is biologically meaningful, because symbiotic mycorrhizal zygomycetes live in close association with most plants. However, note that ORF248 from S.culisetae branches with ORF313 from P.anserina. This observation, along with the presence of a UGA codon specifying tryptophan in ORF248 (see above), suggests a second case of lateral transfer, this time from a euascomycete close to P.anserina, into a zygomycete.
Phylogenetic analysis with standard mitochondrial proteins: are zygomycetes paraphyletic?
The availability of complete mtDNAs from three distant zygomycetes provides an opportunity for testing the monophyly of Zygomycota. Tree topologies were inferred from a 2890 position concatenated protein alignment using two standard ML-based methods, MrBayes and separate ML analysis. The results of these analyses are summarized in Figure 5B, which presents the tree produced by IQPNNI, along with bootstrap values from all methods. All methods produced similar results, except that separate analysis recovered monophyly of S.culisetae and M.verticillata (81% bootstrap support). The latter topology was also found in the ‘credible set’ in three independent runs of MrBayes (5.3, 44.7 and 99.0% posterior probability, respectively; MrBayes bootstrap support for this grouping was 45%). Although these data support both of these topologies, it is interesting that the monophyly of S.culisetae and M.verticillata is better supported under more sophisticated (yet statistically valid) models.
Although this phylogeny is generally robust, bootstrap support values indicate two major areas of uncertainty: the position of A.macrogynus and the relative branching positions of the zygomycetes. Indeed, also the approximately unbiased likelihood ratio test suggests these same problems. When sitewise likelihoods are calculated separately for each functional class, the confidence set contained a total of 17 tree topologies that failed to reject the data (P < 0.05). Among these topologies, A.macrogynus branches immediately above, immediately below or monophyletic with the chytridiomycetes, and R.oryzae and M.verticillata are found to be either monophyletic or paraphyletic (with either species branching more deeply than the other). Although S.culisetae and M.verticillata are monophyletic in the best tree under this model of separately optimized functional classes, all topologies in which all three zygomycetes are monophyletic are rejected. It is worth noting that several obvious wrong positions for S.culisetae were also observed (e.g. as most ancestral among fungi, or among the chytridiomycetes), suggesting that the accelerated evolutionary rate in this species causes a long-branch attraction artefact (44). Similar results are obtained when sitewise likelihoods were calculated from the fully concatenated alignment. Clearly, these data are insufficient to resolve the phylogeny of the zygomycetes, most likely because these three species diverge deeply within fungi, and at relatively short distance from each other. In such a situation, two strategies can be used to resolve the dilemma, addition of more zygomycete and neighboring fungal lineages or addition of more sequence per species. As our protein dataset is already based on complete mtDNAs, this latter strategy implies resorting to expressed sequence tag and/or nuclear genome sequences, a currently ongoing project.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank Gertraud Burger (Université de Montréal) for helpful comments on the manuscript, Michael W. Gray and Murray Schnare for help in the analysis of the genes for small and large rRNAs, Kerry O'Donnell (National Center for Agricultural Utilization Research, Peoria, IL), R. W. Lichtwardt (Department of Botany, University of Kansas, Laurence, KS) and Carolyn Babcock (Canadian Collection of Fungal Cultures, Ottawa) for supply of fungal strains. We further acknowledge the excellent technical assistance of Z. Wang. Salary and interaction support from the Canadian Institutes of Health Research (MOP 42475; B.F.L.), the Canadian Institute for Advanced Research (CIAR; B.F.L.), the Nova Scotia Health Research Foundation (J.L.), and the ‘Bourses d'Excellence biT’ (CIHR; Y.L.) and supply of laboratory equipment and informatics infrastructure by Genome Quebec/Canada is gratefully acknowledged. Funding to pay the Open Access publication charges for this article was provided by the CIHR.
REFERENCES
- 1.Voigt K., Cigelnik E., O'Donnell K. Phylogeny and PCR identification of clinically important Zygomycetes based on nuclear ribosomal–DNA sequence data. J. Clin. Microbiol. 1999;37:3957–3964. doi: 10.1128/jcm.37.12.3957-3964.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanabe Y., O'Donnell K., Saikawa M., Sugiyama J. Molecular phylogeny of parasitic Zygomycota (Dimargaritales, zoopagales) based on nuclear small subunit ribosomal DNA sequences. Mol. Phylogenet. Evol. 2000;16:253–262. doi: 10.1006/mpev.2000.0775. [DOI] [PubMed] [Google Scholar]
- 3.Voigt K., Wostemeyer J. Reliable amplification of actin genes facilitates deep-level phylogeny. Microbiol. Res. 2000;155:179–195. doi: 10.1016/S0944-5013(00)80031-2. [DOI] [PubMed] [Google Scholar]
- 4.Berbee M.L., Carmean D.A., Winka K. Ribosomal DNA and resolution of branching order among the ascomycota: how many nucleotides are enough? Mol. Phylogenet. Evol. 2000;17:337–344. doi: 10.1006/mpev.2000.0835. [DOI] [PubMed] [Google Scholar]
- 5.Kurtzman C.P. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res. 2003;4:233–245. doi: 10.1016/S1567-1356(03)00175-2. [DOI] [PubMed] [Google Scholar]
- 6.James T.Y., Porter D., Leander C.A., Vilgalys R., Longcore J.E. Molecular phylogenetics of the Chytridiomycota supports the utility of ultrastructural data in chytrid systematics. Can. J. Bot. 2000;78:226–350. [Google Scholar]
- 7.Leigh J., Seif E., Rodriguez N., Jacob Y., Lang B.F. Fungal evolution meets fungal genomics. In: Arora D.K., editor. Handbook of Fungal Biotechnology. 2nd edn. NY: Marcel Dekker Inc.; 2003. pp. 145–161. [Google Scholar]
- 8.Bullerwell C.E., Forget L., Lang B.F. Evolution of monoblepharidalean fungi based on complete mitochondrial genome sequences. Nucleic Acids Res. 2003;31:1614–1623. doi: 10.1093/nar/gkg264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lang B.F., O'Kelly C., Nerad T., Gray M.W., Burger G. The closest unicellular relatives of animals. Curr. Biol. 2002;12:1773–1778. doi: 10.1016/s0960-9822(02)01187-9. [DOI] [PubMed] [Google Scholar]
- 10.Alexopolous C.J., Mims C.W., Blackwell M. Introductory Mycology. NY: John Wiley & Sons; 1996. [Google Scholar]
- 11.Ustinova I., Krienitz L., Huss V.A. Hyaloraphidium curvatum is not a green alga, but a lower fungus; Amoebidium parasiticum is not a fungus, but a member of the DRIPs. Protist. 2000;151:253–262. doi: 10.1078/1434-4610-00023. [DOI] [PubMed] [Google Scholar]
- 12.Seif E.R., Forget L., Martin N.C., Lang B.F. Mitochondrial RNase P RNAs in ascomycete fungi: lineage-specific variations in RNA secondary structure. RNA. 2003;9:1073–1083. doi: 10.1261/rna.5880403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okpodu C.M., Robertson D., Boss W.F., Togasaki R.K., Surzycki S.J. Rapid isolation of nuclei from carrot suspension culture cells using a BioNebulizer. BioTechniques. 1994;16:154–159. [PubMed] [Google Scholar]
- 14.Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 16.Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 17.Vinh le S., Von Haeseler A. IQPNNI: moving fast through tree space and stopping in time. Mol. Biol. Evol. 2004;21:1565–1571. doi: 10.1093/molbev/msh176. [DOI] [PubMed] [Google Scholar]
- 18.Bapteste E., Brinkmann H., Lee J.A., Moore D.V., Sensen C.W., Gordon P., Durufle L., Gaasterland T., Lopez P., Muller M., et al. The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba. Proc. Natl Acad. Sci. USA. 2002;99:1414–1419. doi: 10.1073/pnas.032662799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt H.A., Strimmer K., Vingron M., von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 21.Shimodaira H., Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- 22.Bendich A.J. Structural analysis of mitochondrial DNA molecules from fungi and plants using moving pictures and pulsed-field gel electrophoresis. J. Mol. Biol. 1996;255:564–588. doi: 10.1006/jmbi.1996.0048. [DOI] [PubMed] [Google Scholar]
- 23.Bullerwell C.E., Burger G., Lang B.F. A novel motif for identifying rps3 homologs in fungal mitochondrial genomes. Trends Biochem. Sci. 2000;25:363–365. doi: 10.1016/s0968-0004(00)01612-1. [DOI] [PubMed] [Google Scholar]
- 24.Lang B.F., Gray M.W., Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- 25.Bullerwell C.E., Leigh J., Seif E., Longcore J.E., Lang B.F. Evolution of the fungi and their mitochondrial genomes. In: Arora D.K., Khachatourians G.G., editors. Applied Mycology and Biotechnology. Vol. 3. Amsterdam: Elsevier Science; 2003. pp. 133–159. [Google Scholar]
- 26.Paquin B., Laforest M.J., Forget L., Roewer I., Wang Z., Longcore J., Lang B.F. The fungal mitochondrial genome project: evolution of fungal mitochondrial genomes and their gene expression. Curr. Genet. 1997;31:380–395. doi: 10.1007/s002940050220. [DOI] [PubMed] [Google Scholar]
- 27.Bullerwell C.E., Leigh J., Forget L., Lang B.F. A comparison of three fission yeast mitochondrial genomes. Nucleic Acids Res. 2003;31:759–768. doi: 10.1093/nar/gkg134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turmel M., Lemieux C., Burger G., Lang B.F., Otis C., Plante I., Gray M.W. The complete mitochondrial DNA sequences of Nephroselmis olivacea and Pedinomonas minor. Two radically different evolutionary patterns within green algae. Plant Cell. 1999;11:1717–1730. doi: 10.1105/tpc.11.9.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lang B.F., Burger G., O'Kelly C.J., Cedergren R., Golding G.B., Lemieux C., Sankoff D., Turmel M., Gray M.W. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 30.Wise C.A., Martin N.C. Dramatic size variation of yeast mitochondrial RNAs suggests that RNase P RNAs can be quite small. J. Biol. Chem. 1991;266:19154–19157. [PubMed] [Google Scholar]
- 31.Brown J.W. The Ribonuclease P Database. Nucleic Acids Res. 1999;27:314. doi: 10.1093/nar/27.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siegel R.W., Banta A.B., Haas E.S., Brown J.W., Pace N.R. Mycoplasma fermentans simplifies our view of the catalytic core of ribonuclease P RNA. RNA. 1996;2:452–462. [PMC free article] [PubMed] [Google Scholar]
- 33.Paquin B., Laforest M.J., Lang B.F. Double-hairpin elements in the mitochondrial DNA of Allomyces: evidence for mobility. Mol. Biol. Evol. 2000;17:1760–1768. doi: 10.1093/oxfordjournals.molbev.a026274. [DOI] [PubMed] [Google Scholar]
- 34.Vioque A. The RNase P RNA from cyanobacteria: short tandemly repeated repetitive (STRR) sequences are present within the RNase P RNA gene in heterocyst-forming cyanobacteria. Nucleic Acids Res. 1997;25:3471–3477. doi: 10.1093/nar/25.17.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolin S.L., Cedervall T. The La protein. Annu. Rev. Biochem. 2002;71:375–403. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- 36.Paquin B., Laforest M.J., Lang B.F. Interspecific transfer of mitochondrial genes in fungi and creation of a homologous hybrid gene. Proc. Natl Acad. Sci. USA. 1994;91:11807–11810. doi: 10.1073/pnas.91.25.11807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eddy S.R., Gold L. Artificial mobile DNA element constructed from the EcoRI endonuclease gene. Proc. Natl Acad. Sci. USA. 1992;89:1544–1547. doi: 10.1073/pnas.89.5.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burger G., Werner S. The mitochondrial URF1 gene in Neurospora crassa has an intron that contains a novel type of URF. J. Mol. Biol. 1985;186:231–242. doi: 10.1016/0022-2836(85)90100-7. [DOI] [PubMed] [Google Scholar]
- 39.Michel F., Cummings D.J. Analysis of class I introns in a mitochondrial plasmid associated with senescence of Podospora anserina reveals extraordinary resemblance to the Tetrahymena ribosomal intron. Curr. Genet. 1985;10:69–79. doi: 10.1007/BF00418495. [DOI] [PubMed] [Google Scholar]
- 40.Vaughn J.C., Mason M.T., Sper-Whitis G.L., Kuhlman P., Palmer J.D. Fungal origin by horizontal transfer of a plant mitochondrial group I intron in the chimeric CoxI gene of Peperomia. J. Mol. Evol. 1995;41:563–572. doi: 10.1007/BF00175814. [DOI] [PubMed] [Google Scholar]
- 41.Adams K.L., Clements M.J., Vaughn J.C. The Peperomia mitochondrial coxI group I intron: timing of horizontal transfer and subsequent evolution of the intron. J. Mol. Evol. 1998;46:689–696. doi: 10.1007/pl00006349. [DOI] [PubMed] [Google Scholar]
- 42.Cho Y., Qiu Y.L., Kuhlman P., Palmer J.D. Explosive invasion of plant mitochondria by a group I intron. Proc. Natl Acad. Sci. USA. 1998;95:14244–14249. doi: 10.1073/pnas.95.24.14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer J.D., Adams K.L., Cho Y., Parkinson C.L., Qiu Y.L., Song K. Dynamic evolution of plant mitochondrial genomes: mobile genes and introns and highly variable mutation rates. Proc. Natl Acad. Sci. USA. 2000;97:6960–6966. doi: 10.1073/pnas.97.13.6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Felsenstein J. Cases in which parsimony and compatibility methods will be positively misleading. Syst. Zool. 1978;27:401–410. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.