Abstract
Computational evaluation of protein–DNA interaction is important for the identification of DNA-binding sites and genome annotation. It could validate the predicted binding motifs by sequence-based approaches through the calculation of the binding affinity between a protein and DNA. Such an evaluation should take into account structural information to deal with the complicated effects from DNA structural deformation, distance-dependent multi-body interactions and solvation contributions. In this paper, we present a knowledge-based potential built on interactions between protein residues and DNA tri-nucleotides. The potential, which explicitly considers the distance-dependent two-body, three-body and four-body interactions between protein residues and DNA nucleotides, has been optimized in terms of a Z-score. We have applied this knowledge-based potential to evaluate the binding affinities of zinc-finger protein–DNA complexes. The predicted binding affinities are in good agreement with the experimental data (with a correlation coefficient of 0.950). On a larger test set containing 48 protein–DNA complexes with known experimental binding free energies, our potential has achieved a high correlation coefficient of 0.800, when compared with the experimental data. We have also used this potential to identify binding motifs in DNA sequences of transcription factors (TF). The TFs in 79.4% of the known TF–DNA complexes have accurately found their native binding sequences from a large pool of DNA sequences. When tested in a genome-scale search for TF-binding motifs of the cyclic AMP regulatory protein (CRP) of Escherichia coli, this potential ranks all known binding motifs of CRP in the top 15% of all candidate sequences.
INTRODUCTION
Protein–DNA interactions are involved in the regulations of many important cellular processes, such as transcription, replication, recombination and translation. Hence, accurate prediction of protein–DNA interactions is essential to our understanding of many cellular regulations, including transcriptional regulation. It is also important for genome annotation, where there is probably not a ‘recognition code’ in the regulator–DNA binding with obvious sequence preferences (1). Typically, binding motifs of transcription factors (TFs) are predicted through the identification of conserved sequence fragments using various approaches, such as Gibbs sampling (2,3), Hidden Markov Model (4–7) and combinatorial optimization techniques (8–12). Biophysical approaches have also been used to search for binding sites of TFs in DNA sequences, based on estimations of sequence-specific binding energy and the chemical potential of TFs (13). While these methods perform well in identifying potential binding motifs, they clearly lack the capabilities for validating their predictions. We believe that a validation capability could be and need to be developed through the application of protein structural information.
The rapid growth in the number of solved protein–DNA complex structures provides a rich source of information, from which residue–nucleotide interaction potentials could be derived. We know that the knowledge-based potential, based on the theory of mean force field, has proven to be a powerful tool for estimating binding affinities using structural data. Margalit and co-workers (14,15) have developed a set of scoring parameters for assessing amino acid–nucleotide interactions. These parameters have been used to make a quantitative measure on the binding affinity between sequence variants of zinc-finger protein zif268 and their DNA-binding sites. Sarai and co-workers (16–19) systematically studied various aspects of interactions between regulatory proteins and their DNA-binding motifs, which include sequence composition, structural symmetry, conformational distribution in thermodynamics and physiochemical properties, such as solvent density and long-range dipole field distribution. They proposed a position-dependent knowledge-based potential at the residue/nucleotide level and employed a ‘threading’-like procedure to predict DNA-binding sites recognized by a regulatory protein (20). Recently, Olson and co-workers (21–25) found that protein–DNA interactions often lead to the conformational deformation of DNA. They also discovered that specific interaction environments around the contacting amino acids and nucleotides contribute significantly to the binding affinity. This suggests that the simple amino acid–nucleotide interaction model, which is widely used in the statistical analysis in studies of mean force field, could not deal with the conformational changes adequately. More sophisticated multi-body interactions and well-thought details of the interaction environments need to be incorporated into the knowledge-based potential.
The rapid increase in the number of experimental structures has made it possible to use distance-dependent knowledge-based potentials in protein folding, and predictions of protein–ligand and protein–protein interactions (26–30). In most of these potentials, the reference states have been carefully defined in order to eliminate the background noise of interactions. Among the existing models for reference states, the ‘independent’ reference state was the simplest one (31,32). It simulates the ideal gas state and assumes that there are no interactions between any units (atoms, amino acids or nucleotides). Though this model has been widely used in earlier predictions, it has low accuracy in quantitative predictions of binding affinities.
The ‘uniform density’ reference state is a more general model and its revised forms are currently being widely used (32–36). In this type of reference model, the density of pairwise interactions in each distance bin is assumed to be the average densities of those of all folded structures. In such a model, all pairwise interactions are treated uniformly, and it is assumed that there are no interactions between the interaction pairs. While promising, most of the reference states constructed in this way did not have a well-behaved uniform density distribution across all distance bins, due to reasons, such as finiteness of structural volume and the small sampling size of complex structures, which does not reflect well the continuity of physical forces. For example, there were often fluctuations in the counts of interactions among adjacent distance bins, especially for those in the distance bins beyond interaction cutoff or without interactions. Hence, various corrections were proposed in the reference state. Zhou and co-workers (26,27) revised the ‘uniform density’ reference state by considering the interactions between interaction pairs. By introducing an adjustable distance parameter, their reference state could represent a distance-scaled finite-gas reference state that is quite similar to that of a stable protein structure. Muegge and Martin (28) introduced an accurate volume correction to get a consistent reference state with respect to distance bins. Shakhnovich and Ishchenko (30) adopted two exponential parameters for counts of the interacting units to correct for the reference state, which could potentially consider the influence of interaction distances or volume sizes and the effects between different interaction pairs. All these efforts led to more sensible reference states, and improved the scoring accuracy for protein folding and prediction of protein–ligand interaction (32–36).
In this paper, we use ‘DNA tri-nucleotides’ (triplets) as an interaction unit to facilitate the representation of multi-body interactions between a protein and DNA, which can handle the effects of DNA structural deformation and local interaction environments. A distance-dependent knowledge-based potential is developed based on a statistical analysis of interactions between protein–residues and DNA triplets in known protein–DNA complex structures. On the basis of the ‘uniform density’ reference state, we proposed a strategy for distance-normalization, where a distribution of interactions is considered as the ‘distance-normalized’ if it fits the ideal uniform density reference state. The corrections are conducted in two ways. One is to correct for the effect of interaction distances using our consistent reference state. The other is to correct for the effects caused by the triplet size and the number of occurrences of single interaction units (protein residue or DNA triplet). Then, the potential parameters are further optimized using a Z-score optimization. The details of each of these steps are explained in Methods. Our distance-dependent potential has been thoroughly tested on a large set of test data. The test results are highly encouraging, as described in the Results Section.
METHODS
Structural sets
Through a systematic search of the PDB (release January 26, 2004) (37), we have found 571 protein–DNA complex structures, of which 228 (all containing double-stranded DNA) are involved in transcription regulation. In this study, we only consider the 186 TF–DNA structures that were solved using X-ray crystallography with a resolution better than 3.5 Å. Out of these complexes, 141 have diverse DNA-binding sequences (sequence similarity <75.0%) consisting of at least three different consecutive base pairs (bp) (Table 1). The protein structures involved in these complexes represent 48 non-redundant protein domains, based on the SCOP classification (38). We have used these 141 complexes as the structural set to derive our distance-dependent knowledge-based potential.
Table 1.
Data set of protein–DNA complexes
| Structural set (141 complexes) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1a02 | 1a0a | 1a1g | 1a1h | 1a1k | 1a3q | 1akh | 1am9 | 1an2 | 1an4 | 1apl | 1au7 |
| 1b01 | 1b3t | 1b72 | 1b8i | 1bc8 | 1bdt | 1bf5 | 1bl0 | 1by4 | 1c0w | 1c9b | 1cdw |
| 1cez | 1cf7 | 1cgp | 1cit | 1d3u | 1d5y | 1ddn | 1dh3 | 1du0 | 1dux | 1e3o | 1ea4 |
| 1efa | 1egw | 1f2i | 1f5t | 1fjl | 1fos | 1fzp | 1g2f | 1gd2 | 1gji | 1gt0 | 1gu4 |
| 1gu5 | 1gxp | 1h6f | 1h8a | 1h9d | 1h9t | 1hbx | 1hcq | 1hlo | 1hlz | 1hw2 | 1hwt |
| 1ic8 | 1if1 | 1ig7 | 1ign | 1imh | 1io4 | 1j59 | 1je8 | 1jfi | 1jgg | 1jj4 | 1jk1 |
| 1jk2 | 1jnm | 1jt0 | 1k6o | 1k78 | 1k79 | 1k7a | 1kb2 | 1kb4 | 1kb6 | 1ku7 | 1l3l |
| 1lat | 1lb2 | 1le5 | 1le9 | 1llm | 1lmb | 1lq1 | 1mdy | 1mhd | 1mjm | 1mjo | 1mm8 |
| 1mnm | 1mnn | 1mur | 1n6j | 1ngm | 1nkp | 1nvp | 1nwq | 1oct | 1odh | 1owf | 1p47 |
| 1p7h | 1pdn | 1per | 1pp7 | 1pp8 | 1pue | 1puf | 1pyi | 1pzu | 1r0o | 1r4o | 1r4r |
| 1ram | 1rio | 1rpe | 1run | 1skn | 1tf6 | 1tgh | 1tsr | 1ubd | 1yrn | 1ysa | 1ytb |
| 1ytf | 2cgp | 2drp | 2gli | 2hap | 2hdd | 2or1 | 6cro | 6pax | |||
DNA triplet representation
DNA has a compact structure with high density of hydrogen bonds. For a double-stranded DNA structure, its conformational deformation has a significant effect on the thermodynamic stability and the binding affinity to a protein (21–25). In some complexes, such as the zinc-finger protein–DNA complex, it has been shown that the triplet can form a binding core and is one of the principal binding modes in protein–DNA interaction (39–42). Hence, to evaluate the protein–DNA association accurately, it is critical to consider the local DNA interaction environment and the multi-body effects at the protein–DNA interaction interface.
For each amino acid α at the protein–DNA interaction interface, we consider its interactions with all DNA nucleotides within 15 Å (see details later). We consider (i) two-body interactions between α and each of these nearby nucleotides, (ii) three-body interactions between α and any combination of two of these nucleotides and (iii) four-body interactions between α and any combination of three of these nucleotides. The total interaction energy is defined as the sum of these individual interaction energy terms. To simplify our discussion, we introduce a unified triplet representation for the DNA involved in the interactions, which could be used to represent two-, three- and four-body interactions discussed above.
We consider an alphabet of 5 nucleotides in DNA structures, four native nucleotides (nt) A, T, C, G and a pseudo nucleotide O that does not have any structural and energy contributions but just as a placeholder. A two-body interaction between the residue α and a nucleotide, says A, is represented as an interaction between α and triplet AOO (which is termed type-1 triplet and represented as [100]). Similarly, a three-body interaction involving α and nucleotides A, C will be represented as an interaction between α and triplet ACO (which is termed type-2 triplet and represented as [110]). Hence, an interaction between α and triplet ACG (which is termed type-3 triplet and represented as [111]) represents a four-body interaction between α and nucleotides A, C, G. In this representation, the sequential order of nucleotides is insignificant. In the following, a ‘triplet’ could mean 3 nt, 2 nt plus an O, or 1 nt plus two Os. We require that any two real nucleotides, if existing, in a triplet have their distance less than a cutoff distance (termed triplet size R), which is set to be 8 Å in this study (see Discussion). It is easy to see that there are 34 types of triplets (4 of [100], 10 of [110] and 20 of [111]). The position of each nucleotide is represented as that of its glycosidic nitrogen atom connecting the base and the deoxyribose of the nucleotide. The nitrogen atoms are N1 in C and T, and N9 in A and G. The coordinate of the triplet will be the geometric center of the three corresponding nucleotides, while the pseudo nucleotide O is not included.
Clearly, the triplet representation allows representing both the preference of individual nucleotides (triplet [100]) and the local environment around the nucleotides (triplet [110] and [111]). In addition, when DNA binds to protein, its conformational deformation can be implicitly reflected by the change of the coordinates of the corresponding DNA triplets. Those multi-body effects are mainly contributed from DNA nucleotides with only one body from interaction protein. Clearly, more complicated combinations of DNA nucleotides, such as quadruplet, could provide more DNA local environment information. However, the weakness of such representation is apparent, knowing that a greater number of possible combinations of quadruplets and the amount of data will be needed to estimate the relevant parameters.
In DNA structures, the observed numbers (K) of the aforementioned three types of triplets have different correlations with the triplet size. When different triplet sizes are used in deriving the potential function, they will greatly affect the distributions of the interactions involving the three triplet types, and affect the stability of the potential. Therefore, the effect of triplet size on distribution of interactions between protein residue and DNA triplets should be corrected. In an ideal model where a nucleotide is treated as a point and the nucleotides are distributed uniformly in an infinite space, the number of the observed neighboring nucleotides (L) of a given nucleotide within the cutoff of triplet size (R) is proportional to the volume of the corresponding neighborhood, which is proportional to the cube of R (L ∝ R3). Under such an ideal situation, it will provide an upper limit of the correlation between the number K of the observed triplets and R. First note that for a given nucleotide t and a triplet size R, the number of triplets of type [100] involving t is apparently 1. The number of triplets of type [110] involving t is proportional to the cube of R (K = L ∝ R3). Similarly, the number of triplets of type [111] involving t is proportional to the square of R3 {K = [L (L − 1)/2] ∝ R6}. However, in any real protein–DNA complex, it will not reach the ideal correlation between the numbers of the observed triplets (K) and R, because the complex has a finite volume and nucleotides are not strictly uniformly distributed in a DNA structure. We have systematically analyzed the three types of DNA triplets in our training complex structures without distinguishing the types of the nucleotides. Based on this analysis, we have derived a correction function between the number of triplets (K) and R (Equation 1), where T is the DNA triplet type and K(T, R) is the number of the observed triplets normalized by the actual number of triplets of type [100]. This function will be used in our development of the potential to correct for the effect of triplet size.
| 1 |
Derivation of a consistent reference state
In deriving both distance-independent and distance-dependent knowledge-based potentials, researchers often use a distance cutoff. It is generally assumed that the region beyond the distance cutoff has no significant contributions to the interaction energy. Such a distance cutoff is often set in an arbitrary manner. Moreover, it is known that short-distance pairwise interactions are stronger, and therefore should have higher statistical significance compared with longer-distance interactions. However, the observed frequencies for such short-distance interactions are often lower than the true counts due to the distance or volume effect. Such distance effect has reduced the statistical significance of short-distance interactions, and greatly impaired the potential's performance especially those distance-independent potentials. Removing such distance effects poses a challenge for a finite protein–DNA system. Based on the assumption of ‘uniform density’ reference states, we propose a strategy of distance-normalization to overcome the distance effect, through an application of a consistent reference background (CRB) derived through fitting the interaction background using a distance function. After the normalization and additional corrections, the system will be well consistent with the ‘uniform density’ reference state.
First note that in each distance bin, the sum of the probabilities of all interactions is 1. Generally, the smaller the distance bin is, the more accurate the potential will be, assuming that we have sufficient amount of data. In practice, when the distributions are stable and the bins are small enough, the variation of bin-length will not affect the potential's performance in general.
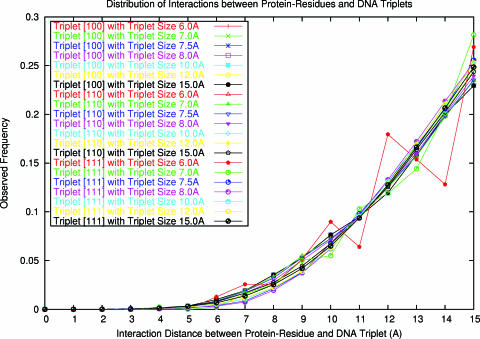
Here, a CRB is derived through the analysis of the distributions of the interactions between all protein–residues and DNA triplets in the structural data set. First, the distance bins of interactions are split evenly using a fixed bin-length. Then, the numbers of three types of interactions between three types of DNA triplets ([100], [110] and [111]) and protein residues in each distance-bin are counted, without distinguishing the types of protein residues and DNA nucleotides in triplets. All distributions of the three types of interactions converge to a same state, no matter what triplet size is used (Figure 1). Equation 2 shows that the observed frequency of interactions in the converged distribution has an exponential correlation with the distance, where F(r) is the observed frequency, r is the interaction distance, and A, B are correlation parameters. Further analysis revealed a rigorous correlation between the exponential parameter B and the bin-length X, which ranges from 1.0 to 5.0 Å, as shown in Equation 3. This provides a criterion for constructing a CRB for different distance bins. Note that function F(r) is an ideal reference background, as it does not take the specificity of protein residues and DNA nucleotides into account and it provides a general description of consensus interaction density in protein–DNA complexes. It can remove the distance effect and the impact of a finite complex system, as discussed above, through a background deduction. Actually our distance correction function F(r) has a similar distance exponential parameter to that in Zhou's work (26,27), though our parameter is more consistent and is derived directly from the background distribution of interactions. However, the function does not explicitly consider the correlation among interactions between protein–residues and DNA-triplets. This correlation will be handled by two other parameters, as discussed below. Finally, to obtain a more accurate potential, a small bin-length (X), 1.0 Å, is chosen, and the corresponding exponential parameter B is set at 3.345 obtained from Equation 3.
| 2 |
| 3 |
Figure 1.
The distribution of observed interactions between protein–residues and DNA triplets in the training set of 141 protein complexes. The interaction distributions of three kinds of triplet types [100], [110] and [111] are analyzed using different triplet size 6.0, 7.0, 7.5, 8.0, 10.0, 12.0 and 15.0 Å, respectively.
Development of a distance-dependent knowledge-based potential
We now derive a distance-dependent knowledge-based potential through a statistical analysis of the interactions between protein–residues and DNA triplets. The coordinate of Cβ atom of each residue is used to represent the residue position (a pseudo Cβ is used for Glycine based on its geometric shape). We found a total of 185 468 interactions between 20 types of amino acids and 34 types of DNA triplets within an interaction distance of 15.0 Å.
After counting the interactions between protein residues and DNA triplets in our structural data set, we correct the observed interaction frequency, translate the frequency to an energy term and build a transferable distance-dependent mean-force potential for the protein–DNA interactions. First, the initial number of interactions between protein residues and DNA triplets, , is corrected to satisfy the requirement of a ‘uniform density’ reference state. The idea is to eliminate the effect of triplet size using the function K(T,R), to remove the distance effect through distance-normalization using the function F(r), and to correct the correlations among residue–triplet interactions as outlined below.
Because of the inevitable incompleteness and redundancy of the training set, the observed fractions of interaction pairs are biased. It has also been pointed out by several groups that the correlations between observed interactions could affect the performance of the potential significantly (30,36). To correct for such effects, Lu and Skolnick (36) introduced the mole fractions of protein atoms into their threading potential function. Ishchenko and Shakhnovich (30) adopted the numbers of the interacting protein and ligand atoms, and optimized two new exponential parameters for the number of protein atoms and ligand atoms, respectively. Here, two exponential parameters α and β, similar to those of Ishchenko and Shakhnovich's but only for the fraction of the interacting protein residues and the fraction of DNA triplets, respectively, are introduced. As in Equation 4, Nij(r) is the corrected number of interactions, i and j represent a protein–residue and a DNA triplet, respectively.
| 4 |
Wresidue and Wtriplet represent the fraction of interacting protein residue i and the fraction of the interacting DNA triplet j, respectively; α and β are two exponential parameters, to be optimized using a Z-score optimization. For regions with no observed interactions or only a few interactions, an offset parameter σ is introduced, whose value will be determined through the Z-score optimization as well. Ncutoff is a cutoff on the number of interactions, which is set to 1. After these corrections, the observed numbers of interactions between protein–residue and DNA triplet Nij(r) should be normalized, which maximally follows the uniform density theory.
Just like in any knowledge-based potential, we define a distance cutoff to exclude the regions that do not have much contribution to the potential. On the basis of the normalized distribution of the interactions, it is found that there is no statistical significance for interactions beyond 15.0 Å. Hence, we used a distance cutoff 15.0 Å. Our method works for both regions with no observed interactions and regions beyond the cutoff distance. In such regions, specific interactions between protein–residue and DNA–triplet are practically the same as the reference state with uniform density, and have an average distribution. Therefore, their energy contribution is zero based on Equations 5–7. The relationship between the observed probability pij(r) and the statistical thermodynamic interaction energy Eij(r) of an interaction between residue i and triplet j with a distance r is given by Equation 5, where T is the temperature and Z is the partition function. The uniform density reference state is used for each distance bin (Equation 6), where is the mean probability of the interactions between residues and triplets with a distance r. The energy in the final potential is given as the corrected energy with respect to the reference mean energy (Equation 7).
| 5 |
| 6 |
| 7 |
Z-score optimization
The Z-score optimization is used to optimize the potential parameters α, β and σ. Based on the thermodynamic hypothesis that the native structure of a protein/complex has the lowest energy, Z-score optimization has been used successfully in optimizing potentials for protein folding, protein–peptide and protein–DNA interactions (20,43,44). For a protein–DNA complex conformation, the interaction energy E will be the sum of the energies of all possible residue–triplet interactions within 15.0 Å (Equation 8). The critical Z-score, Zt, measures how standout the native energy is when compared with the decoy energies, assuming that they are both random Gaussian variables from a continuous random energy model (REM). Zt measures the gap between the native energy ENative−t and the average energy <Et> of the decoy energies (Equation 9), where δ(Et) is the standard deviation of the decoy energies, and t refers to a protein–DNA complex t. For multiple complexes, an average Z-score is computed using Equation 10, where M is the number of complexes used. The average Z-score gives more weight to the larger Zt scores, representing those complexes whose natives are not easily distinguished from the decoys.
In this work, the Z-score optimization is carried out through optimally distinguishing native DNA sequences from the decoy sequences. The optimization is used to parameterize α, β and σ by a Monte Carlo annealing simulation. Five hundred decoy DNA sequences are generated for each complex through randomly shuffling the nucleotide sequence of each protein–DNA structure. If the number of non-redundant decoy sequences through reshuffling is fewer than 500 for a complex, random DNA sequences will be used to make it 500. Given the initial values of α, β and σ, the initial potential can be determined. Subsequently, the decoy sequences are threaded onto the corresponding native DNA structure without gaps (note that ‘threading’ is to simply put a decoy sequence onto the same DNA position where the native sequence is; no search is involved). The interaction energies between the decoy sequences and the protein are computed using the potential Equations 7 and 8, and the Z-score for each structure (Zt) and the average Z-score. By changing the values of α, β and σ to optimize the average Z-score using the Monte Carlo simulation, the final potential is obtained when a minimal Z-score is reached. In this work, the parameters α and β converge to 1.350 and 0.872, respectively, and σ converges to 0.033 which has less effect on the observed number of interactions. The three parameters are more conserved when optimized using structural sets of different sizes; hence the Z-score optimization does not affect the statistical distribution of interactions nor distort the final potential.
| 8 |
| 9 |
| 10 |
RESULTS
We have evaluated our optimized potential through predicting the binding affinities for the zinc-finger protein–DNA complexes and a large set of protein–DNA complexes. We have also used the potential for recognition of the DNA-binding sites from a pool of DNA sequences for each given structure of TFs. Particularly, we have applied the potential to identify the DNA-binding sites of the CRP family in Escherichia coli.
Evaluation of interactions between zinc-finger protein and binding sequences
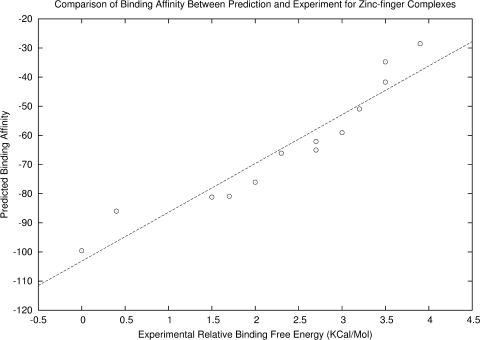
The zinc-finger binding motifs are prevalent in eukaryotic genomes as zinc-finger plays an important role in the regulation of gene expression in Drosophila, mice and human (39–42). The protein zif268 (PDB ID 1AAY) consists of three zinc-finger domains, each of which has three amino acids in contact with a DNA site consisting of 3 bp. The modular nature of the interactions between a single domain and its DNA-binding sites contributes to the binding affinity and binding specificity. Desjarlais and Berg (39) designed two consensus sequences ‘QNR-XXX-RHR’ and ‘GAG-NNN-GAT’ for the key interaction sites between the three zinc-finger domains and the three DNA-binding sites, respectively, where X, N represents an arbitrary amino acid and nucleotide, respectively. Experimentally, they have determined the relative binding free energies of 13 DNA-binding sequences (NNN = GCT, 0.0 kcal/mol; GCG, 0.4 kcal/mol; TCA, 1.5 kcal/mol; GCC, 1.7 kcal/mol; GAG, 2.0 kcal/mol; TTT, 2.3 kcal/mol; GAA, 2.7 kcal/mol; ACG, 2.7 kcal/mol; CCA, 3.0 kcal/mol; ACT, 3.2 kcal/mol; CGT, 3.5 kcal/mol; TTC, 3.5 kcal/mol; and CGA, 3.9 kcal/mol) to the protein domain QNR-QDR-RHR. We have used our potential function (Equations 7 and 8) to predict the binding affinities for these zinc-finger protein–DNA complexes, using 1AYY, which is not included in the training structural set. We found that the correlation coefficient between our predicted binding affinities and the experimental relative binding free energies is 0.950, which is significantly higher than that (0.79) in Mandel-Gutfreund and Margalit's work (Figure 2) (15), suggesting that our potential could rank the interaction free energy between TF–DNA more accurately.
Figure 2.
A comparison between predicted binding affinity and experimental relative binding free energy for zinc-finger protein–DNA complexes. A total of 13 DNA-binding sequences (GAG-NNN-GAT, where NNN = GCT, GCG, TCA, GCC, GAG, TTT, GAA, ACG, CCA, ACT, CGT, TTC, or CGA) were tested and the fitted linear correlation is r = 0.950.
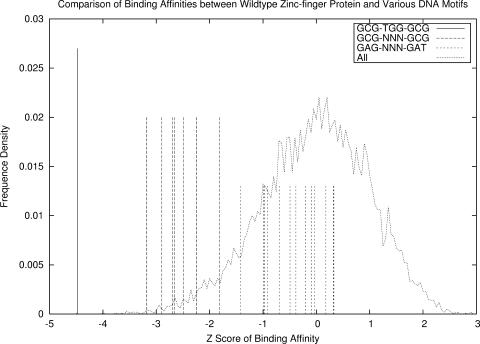
Subsequently, we have conducted a test of our potential function on 1AAY against 10 000 randomly generated sequences and the DNA motifs that are known to bind to various zinc-finger domains. The key residues involved in the interaction in the three zinc-finger domains are RER-RHT-RER. The binding affinities of all the DNA sequences to the native protein are computed and the Z-scores are shown in Figure 3. We can see that the native sequence GCG-TGG-GCG is ranked the highest with a Z-score of −4.48. In addition, seven other sequences GCG-GTG-GCG, GCG-GCG-GCG, GCG-TTG-GCG, GCG-GAT-GCG, GCG-GAC-GCG, GCG-GCA-GCG and GCG-GTA-GCG, known to bind to proteins with the same two zinc-finger domains but a different central domain, were also ranked high (top 4.4%). The high binding affinity of one of these sequences, GCG-TTG-GCG, has been validated by experiments with an equilibrium dissociation constants Kd = 3.0 ± 0.6 nM (41). We have also predicted the 13 aforementioned sequences with consensus pattern ‘GAG-NNN-GAT’, which are known to bind to zinc-finger domains with consensus pattern ‘QNR-XXX-RHR’. However, they are ranked relatively lower in binding presumably because their native bound proteins have different zinc-finger domains from Zif268 (1AAY). As for the 10 000 random sequences, their Z-score distribution follows a Gaussian distribution. The vast majority (95.6%) of them are ranked lower than the native sequence and the seven sequences that could potentially bind to the target protein. These results demonstrate that our potential could accurately distinguish the native DNA sequences from the decoys.
Figure 3.
Ranking of the binding affinities between DNA sequences and wild-type Zif268 protein. ‘GCG-TGG-GCG’ represents the rank of the native sequence GCG-TGG-GCG that binds to Zif268 mode RER-RHT-RER. ‘GCG-NNN-GCG’ corresponds to the ranks of seven sequences (GCG-GTG-GCG, GCG-GCG-GCG, GCG-TTG-GCG, GCG-GAT-GCG, GCG-GAC-GCG, GCG-GCA-GCG, GCG-GTA-GCG) binding to proteins with two zinc-finger domains of wild-type Zif268 but a different central domain. ‘GAG-NNN-GAT’ represents the 13 DNA sequences with motif GAG-NNN-GAT (NNN = GCT, GCG, TCA, GCC, GAG, TTT, GAA, ACG, CCA, ACT, CGT, TTC and CGA) that bind to another zinc-finger domain mode QNR-XXX-RHR. ‘All’ refers to the Z-score distribution of all sequences consisting of the aforementioned sequences and 10 000 random sequences.
General prediction of protein–DNA binding affinities
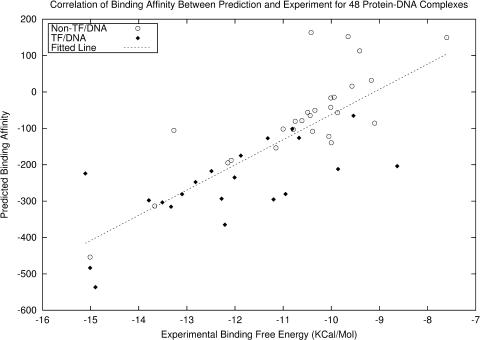
Our potential function is further tested on a set of 48 protein–DNA complex structures, which were extracted from the ProNIT website (http://www.rtc.riken.go.jp/jouhou/pronit/pronit_search.html), along with their binding free energies. As shown in Figure 4, the predicted results have a high correlation coefficient (0.800) with the experimental data (16). More importantly, our potential not only works well for TF/DNA complexes, but also achieves a high accuracy for the 27 non-TF/DNA complexes, suggesting that our potential function can possibly be used for the evaluation of protein–DNA binding beyond TFs.
Figure 4.
Correlation between predicted binding affinities and experimental binding free energies for 48 protein–DNA complexes. There is linear correlation of r = 0.800. Non-TF/DNA refers to 27 non-transcription factor/DNA complexes (including 1mse, 1tro, 1ca5, 2ezd, 1lcc, 1cjg, 1gcc, 1azp, 1az0, 1b69, 1tf3, 1bhm, 1ecr, 1cw0, 1hcr, 1yui, 1sx9, 7icr, 1qaa, 1jey, 1nk2, 1tau, 5gat, 1qrv, 1a73, 2gat and 1j1v), TF/DNA refers to remaining 21 transcription factor complexes (including 1lmb, 1cma, 1apl, 1par, 1run, 1glu, 1nfk, 1efa, 1mdy, 1tsr, 1ipp, 1ytf, 1vkx, 1oct, 1ihf, 1bc7, 1aay, 1cez, 1yrn, 1ysa and 1b3t).
Recognition of specific interactions between TFs and DNA-binding sites
We have also tested the recognition power of our potential in matching the DNA-binding motifs to their interacting TFs. For a set of protein–DNA complexes, we first extract all the binding DNA motifs and then append them into one long sequence of 2575 bp. We then scan the individual DNA motif along the long sequence using each complex structure. By doing this, we have generated more than 2500 sequence fragments for each complex structure. We then assess whether the potential function can distinguish the native binding DNA motif from a pool of decoy sequences for each TF. In PDB, there are at least 18 homeodomain protein–DNA complexes with distinct DNA sequences (PDB ID 1akh, 1apl, 1au7, 1b72, 1b8i, 1du0, 1e3o, 1fjl, 1ic8, 1ig7, 1jgg, 1lfu, 1nk2, 1oct, 1puf, 1yrn, 2hdd and 9ant, with SCOP classification as a.4.1.1). Because of the diversity of DNA sequences associated with the same protein structure, we consider that the recognition is correct if the native DNA sequence is ranked among top 25 (top 1%). In this test, 79.4% of the TFs correctly found their native binding DNA sequences based on this criterion. Particularly, 39.7% of the native DNA sequences have the highest ranking and 90.1% of TFs rank their native DNA sequences in the top 5% of all candidates (Table 2).
Table 2.
Recognition accuracy for specific interactions between TFs and native bound DNA sequences
| Accuracy | Top 1(%) | Top 10(%) | Top 20(%) | Top 1(%) | Top 5(%) |
|---|---|---|---|---|---|
| Whole structural set | 39.7 | 70.2 | 77.3 | 79.4 | 90.1 |
| α-Helixa | 26.3 | 54.4 | 63.2 | 66.7 | 82.5 |
| α-Helix + β-stranda | 49.4 | 79.2 | 85.7 | 87.0 | 94.8 |
aα-helix and β-strand refer to the secondary structures of DNA-binding sites. There are 57 proteins that bind DNA with α-helix only, 77 proteins bind to DNA with both α-helix and β-strand structures.
Identification of DNA-binding sites for TF CRP at genome scale
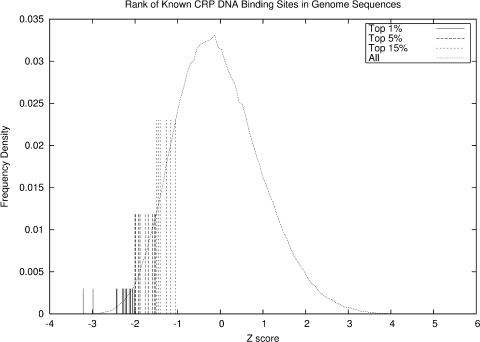
We have also tested our potential function in the identification of DNA-binding sites of TF CRP (CAMP regulatory protein) at the genome scale in E.coli K12. CRP is selected because its complex structure (PDB ID 1O3T, not included in the training structural set) with DNA has been solved, and the DNA sequence in 1O3T has high sequence-similarity (>60.0%) to at least one of the 32 known DNA-binding sites in E.coli, which proves that the 1O3T is the right structural template for CRP and its DNA-binding sites (Table 3). We extracted all 32 known binding sites and 5000 bp flanks at both ends of each binding site. We then scan these sequences on the DNA structure of 1O3T to find which sequence fragments can bind to the protein and whether they are known CRP-binding sites. Since the DNA sequence in the 1O3T protein–DNA complex contains additional bases than each of the 32 annotated binding motifs (i.e. the sequence is longer), we need to first determine which portion of the DNA corresponds to the annotated binding motifs. So we first align each of these 32 binding motifs with this DNA sequence in the 1O3T complex, and found that the best aligned positions go from the 7th to 25th nucleotides in DNA sequences of 1O3T, which is exactly located in the center of the sequence. So we will consider a correct prediction in our test only if a scanned motif contains a known binding motif and the corresponding part is aligned to the aforementioned positions. Our sequence scan generated 639 232 potential binding sequences. We use our potential function to rank these sequences based on the predicted binding affinity with CRP using the 1O3T complex structure (Figure 5 and Table 3). The computational results show that all 32 known binding sites were ranked among the top 15% of all candidates. Particularly, 1 of these 32 sequences (AAAATATAGATCTCCGTCA) is ranked No. 2; 16 of these sequences were ranked in top 1%; and 26 are ranked in the top 5%. The result demonstrates that our potential function can be used to effectively screen binding site candidates at the genome scale. In the test, some non-binding DNA sequences are also ranked high, which will be discussed below.
Table 3.
A genome-scale analysis of 32 known DNA-binding sites of CRP in E.coli.K12 predicted using 1O3T (PDB ID)
| Sequence sources | Sequence | Maximal similarity to sequence fragments of 1o3t with 19 nt (%) | Rank in total 639 232 sequences | ||
|---|---|---|---|---|---|
| Z-score | Position | Top (%) | |||
| DNA sequence in 1o3t | GCGAAAAATGCGATCTAGATCGCATTTTTCG | — | — | — | — |
| CRP-binding sites 1 | AGTAATCTGCTTTATGCCT | 47.37 | 1.5025 | 33 144 | 5.1850 |
| CRP-binding sites 2 | TTCTTCGTCAAATTTATCA | 57.89 | 1.5254 | 31 178 | 4.8774 |
| CRP-binding sites 3 | AGTGTGGAAGTATTGACCA | 36.84 | 1.9167 | 9199 | 1.4391 |
| CRP-binding sites 4 | GTTGTTACAAACATTACCA | 36.84 | 1.4114 | 41 799 | 6.5389 |
| CRP-binding sites 5 | TGCGTGACGAAGTTGCCAA | 42.11 | 1.1646 | 72 996 | 11.4193 |
| CRP-binding sites 6 | TTTAAATTAACTTATGTAA | 42.11 | 2.4214 | 1017 | 0.1591 |
| CRP-binding sites 7 | AATGTGACGGCAATCGATT | 57.89 | 2.0021 | 6759 | 1.0574 |
| CRP-binding sites 8 | TAGTTGAACCAGGTCACAA | 52.63 | 2.2871 | 2064 | 0.3229 |
| CRP-binding sites 9 | CGTAATGTGATTTATGCCT | 52.63 | 2.0207 | 6359 | 0.9948 |
| CRP-binding sites 10 | TGTGCGGGCGTGATCACAA | 57.89 | 1.9889 | 7114 | 1.1129 |
| CRP-binding sites 11 | AAATTGATCCCTTTTTAAC | 57.89 | 2.1399 | 3991 | 0.6243 |
| CRP-binding sites 12 | TGTGTGACGAAGTAACCAC | 42.11 | 1.4588 | 37 138 | 5.8098 |
| CRP-binding sites 13 | ATAGAGATCTACTTCACAA | 63.16 | 2.4383 | 928 | 0.1452 |
| CRP-binding sites 14 | GTTGTCACTCTAATGATAA | 42.11 | 2.0646 | 5,427 | 0.8490 |
| CRP-binding sites 15 | TATGTGACCTGGCAGCCAA | 52.63 | 2.2324 | 2685 | 0.4200 |
| CRP-binding sites 16 | AGATTAACTTATGTAACAG | 36.84 | 1.8809 | 10 403 | 1.6274 |
| CRP-binding sites 17 | GTTATCGTGACCTGGATCA | 52.63 | 1.5356 | 30 406 | 4.7566 |
| CRP-binding sites 18 | ACGATTGTGATTCGATTCA | 52.63 | 2.1064 | 4587 | 0.7176 |
| CRP-binding sites 19 | AACGTGATCAACCCCTCAA | 57.89 | 2.1973 | 3139 | 0.4911 |
| CRP-binding sites 20 | AAATTTGAGAGTTGAATCT | 57.89 | 2.1134 | 4427 | 0.6925 |
| CRP-binding sites 21 | AGAGTGATATGTATAACAT | 57.89 | 2.1929 | 3209 | 0.5020 |
| CRP-binding sites 22 | CGTTTCGTGACAGGAATCA | 36.84 | 1.0437 | 92 717 | 14.5044 |
| CRP-binding sites 23 | AGGAATGCGATTCCACTCA | 57.89 | 2.2785 | 2136 | 0.3342 |
| CRP-binding sites 24 | CTTCTCGTGATCAAGATCA | 52.63 | 1.7621 | 15480 | 2.4217 |
| CRP-binding sites 25 | TTTTTCTTGCTTACCGTCA | 36.84 | 2.1998 | 3112 | 0.4868 |
| CRP-binding sites 26 | TGCGATGAATGTCACATCC | 63.16 | 1.6900 | 19 371 | 3.0304 |
| CRP-binding sites 27 | ATCGTGCTCGCTTTCACGC | 52.63 | 1.2674 | 58398 | 9.1357 |
| CRP-binding sites 28 | AAAATATAGATCTCCGTCA | 57.89 | 3.2175 | 2 | 0.0003 |
| CRP-binding sites 29 | CAATTTGCGACGCGTCTCA | 47.37 | 1.5920 | 25 926 | 4.0558 |
| CRP-binding sites 30 | ACTGTAAAAGGAAACATCA | 42.11 | 1.5288 | 30 907 | 4.8350 |
| CRP-binding sites 31 | AATTCAATATTCATCACAC | 63.16 | 2.9837 | 29 | 0.0045 |
| CRP-binding sites 32 | TTTGTGAAGGCTATTAGCC | 42.11 | 2.0571 | 5590 | 0.8745 |
Figure 5.
A genome scale rank of binding affinities between CRP transcription factor and its 32 known DNA-binding sites. ‘Top 1%’ refers to 16 motifs ranked in top 1% of all 639 232 sequences. ‘Top 5%’ refers to 10 motifs ranked between top 1% and 5%, and ‘Top 15%’ refers to 6 motifs ranked between top 5% and 15%.
DISCUSSION
Determination of DNA triplet size
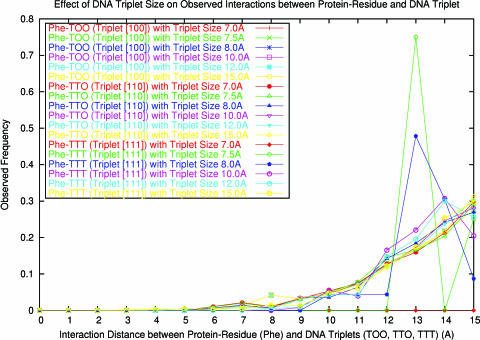
As defined in Methods, the DNA ‘triplet size’ is the cutoff distance between two real nucleotides in a triplet. The cutoff directly affects the observed number of DNA triplets, especially for types [110] and [111]. Hence, the triplet size is crucial to an accurate estimation of interactions between protein–residues and DNA triplets. First, a good triplet size should cover sufficient local environment information in DNA while keeping the stable distributions of all representative DNA triplets. We have to determine a minimal cutoff of the triplet size to satisfy this requirement. As in the case of determining the reference background, we have conducted a statistical analysis on the distance distribution of all interactions between protein residues and DNA triplets while neglecting the types of protein residues and the types of nucleotides in DNA–triplets (Figure 1). As the triplet size increases, the distributions tend to converge to the reference background. Note that such distributions fluctuate within the distance range between 6.0 and 7.0 Å. Therefore, to ensure a stable reference background, a minimal cutoff is set at 7.5 Å. On the other hand, the increase in the triplet size would dilute the contribution of each specific nucleotide, and hence reduce the statistical significance of interaction distribution. For example, we have analyzed the interactions between amino acid PHE and those DNA triplets involving only nucleotide T, which appears at the interface of protein–DNA complex with an average frequency (20,45). It was found that an increase in the triplet size would greatly smoothen the interaction distribution (Figure 6). The distribution has obvious statistical significance when the triplet size falls between 7.5 and 8.0 Å. When the triplet size is >10.0 Å, a lower statistical significance is observed, especially for the interaction between PHE and TTT (triplet type [111]). Therefore, the maximal cutoff is set at 10.0 Å. Based on the consideration of the aforementioned properties of the interaction distribution, we have set the triplet size at 8.0 Å. In this work, only DNA was described using triplet. If both protein and DNA are represented using triplet, more complete and more complicated multi-body effects could be modeled.
Figure 6.
The interaction distributions between protein residue PHE and DNA triplets containing nucleotide T (TOO, TTO and TTT) only in the training set. The TOO, TTO and TTT belong to triplet types [100], [110] and [111], respectively. The interaction distributions are analyzed using different triplet size of 7.0, 7.5, 8.0, 10.0, 12.0 and 15.0 Å, respectively.
Computational time
We have benchmarked the computing time of our method on the complex structure 1AAY against DNA sequences with different lengths, on a PC (Intel P4 2.8 GHz). We observed that the computing time is roughly linearly proportional to the length of DNA sequence. Typically, for a DNA fragment of 10 bp, the calculation would not be more than 0.002 s. Hence, for the binding-site screening at the genome scale for E.coli K12 (http://www.ncbi.nlm.nih.gov/genomes/MICROBES/Complete.html) with 4 639 675 bp, it takes about 154 min. Moreover, if we screen all possible TF-binding sites in the E.coli genome, and conduct a comprehensive threading study between all templates in the structural set and the E.coli genome, the computation will take ∼361 h. Because this is a data-parallel problem, running this program on a multi-cpu Linux cluster can practically achieve linear speed-up. So to run this computational job on our 128-cpu Linux cluster, it can finish the computation within 2 h.
Sensitivity and specificity of protein–DNA recognition
We computed the sensitivity and specificity of protein–DNA recognition using our
| 11 |
| 12 |
potential and compared the results with Ahmad et al.'s work (19). For consistency, we used the same definition of sensitivity and specificity as that in their work (Equations 11 and 12), where T is true, F is false, P is positive and N is negative. We should note that there is difference in the prediction methods, as they used DNA sequence to identify the exact binding TFs mainly using a sequence-based approach, while we used known TF to recognize the exact binding DNA motifs using structure-based technique. However, this difference should not affect the recognition ability between protein and DNA. We first compute the sensitivity and specificity of prediction for 141 TF/DNA complexes in the training set. For each complex, there are a total of ∼2500 tested sequences. The native binding sequence is treated as true sequence, and all other sequences are false because they specifically bind to different proteins and can be treated as random sequences for the given TF. In this computation, we consider a recognition as successful if the correct binding motif is placed among the top 5% of the predicted sequences. Using these criteria of top 5% as the positive, our method achieved an average sensitivity and specificity at 89.4 and 95.1%, respectively, on the training set, which compares favorably with 71.3 and 71.0%, respectively, by Ahmad et al.'s work. When computing these values on CRP set, which contains 32 true motifs, 639 200 false motifs, 31 961 positive predictions still using the same criterion of top 5% as the positive, and 26 found true positives, our method achieved an sensitivity and specificity at 81.3 and 95.0%, respectively, on the test set, compared with 68.6 and 63.4% by Ahmad et al.'s work. We should point out that the number of false sequences is much more than that of true motifs that in our prediction, which potentially increases the prediction specificity. In general, the discriminating power of our method is obvious.
Energy contribution of multi-body interactions
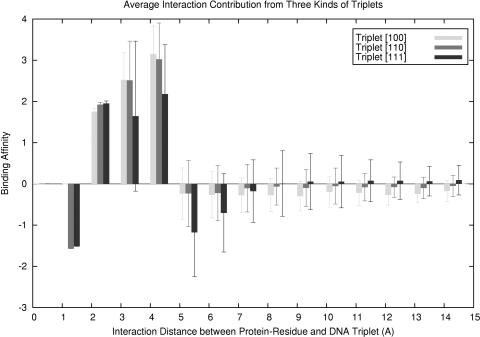
We have developed a triplet-based representation for DNA to represent the multi-body interactions and effects of DNA local environment. The energy contributions from the three kinds of triplet types were counted to evaluate the significance of the triplet representation. We systematically calculated all interactions between protein–residues and DNA triplet in the 141 training structures. The interactions were classified according to the triplet types, and the mean and standard deviation of the interaction energy for each triplet type in different distance bins were calculated (Figure 7). It was found that in each distance bin, the energy contributions from three-body interactions represented by triplet type [110] and from four-body interaction represented by triplet type [111] are comparable with that from two-body interactions represented by triplet type [100]. We found that the more complex the multi-body interactions are, the more fluctuations in energy are observed, which correspond to the exact variations in local DNA environments in different complexes. When the interaction distance between protein residue and DNA triplet is <7.0 Å, the interaction energies are significant as shown in Figure 7. If the interactions are in shortest distance, within 2.0 Å, the value of binding affinity is negative, suggesting that the attractive force draws protein and DNA close to each other. Note that the 2.0 Å is the distance between the center of triplet and protein residue, not the actual distance between residue and nucleotide. For interactions between 2.0 and 5.0 Å, the binding affinities are positive, which correspond to the repulsive forces and provide a balance effect in binding. The binding affinity will attenuate as the interaction distance increases further. Most of these interactions provide negative binding affinity and are related to remote attraction forces. For long-distance interactions, the energy deviations of four-body interactions are still obvious. Particularly, as the distance increases, more complicated multi-body interactions will dominate and will have more contributions to the total energy. Therefore, the multi-body interactions actually play important roles in protein–DNA interaction.
Figure 7.
The energy contribution of multi-body interactions represented by three different DNA triplet types in the training set. The solid black bar represents the energy contribution of four-body interaction between protein residue and DNA triplet type [111], the gray bar represents that of three-body interaction from DNA triplet type [110] and the light bar represents that of two-body pairwise interaction from triplet type [100].
The accuracy of knowledge-based potential
In our set of 141 complex structures, most TFs use α-helix (57) and α-helix+β-strand binding modes (77). Compared with the α-helix binding mode, α-helix+β-strand binding has higher prediction accuracy (Table 2). Our analysis of the known DNA–protein complexes showed that in most cases, an α-helix is the principal binding site that is inserted into the DNA groove and provides the binding specificity. Only when α-helix structure could not provide enough binding strength to stabilize the complex structure, a β-strand would be added to the binding interaction to provide an additional anchor in the complex structure.
As a knowledge-based potential, our method mainly calculates the static thermodynamic value between a protein and DNA, while neglecting the kinetic effect. Note that not all DNA sequences, which have strong thermodynamic interactions to a specific TF, are DNA-binding sites because of the kinetic constraint. This can explain why many DNA sequences that are not DNA-binding sites were ranked high in the binding affinity prediction using our method. In addition, the knowledge-based potential is at the level of residues and triplets, which lacks the detailed atomic level information. Such a model could lead to steric clash, which could not form the specific interaction between the amino acid side chains and the accessible functional groups of the base pairs. Apparently, one way to improve the accuracy is to combine the method with extra energy constraints derived from DNA sequence analysis and to consider the atomic structural examination to reduce the false-positive predictions.
CONCLUSION
‘DNA triplets’ is proposed to represent a DNA structure. A triplet could well describe the DNA local interaction environment and handle the multi-body interaction effect. Based on the assumption of ‘uniform density’ reference state, we have proposed a strategy of distance-normalization for distribution of interactions between residues and DNA triplets, and have constructed the CRB that could make a sensible statistical correction on the distance effect. Through a Z-score optimization, we developed a distance-dependent knowledge-based potential for validating predictions of TF–DNA binding, using sequence-based methods. The potential could accurately and quantitatively predict the binding affinities between TF and DNA, and even for a large class of protein–DNA interactions. It is possible to apply this capability to rank DNA-binding sites for a particular protein structure at the genome scale. With future improvement at the atomic level, the potential would be useful for the prediction of protein–DNA docking, protein–DNA complex study and drug design area.
Acknowledgments
This research was supported in part by National Science Foundation (#NSF/DBI-0354771, #NSF/ITR-IIS-0407204), and by the US Department of Energy's Genomes to Life program (http://doegenomestolife.org/) under project, ‘Carbon Sequestration in Synechococcus sp.: From Molecular Machines to Hierarchical Modeling’ (www.genomes2life.org). Funding to pay the Open Access publication charges for this article was provided by National Science Foundation (# NSF/DBI-0354771).
REFERENCES
- 1.Matthews B.W. Protein–DNA interaction. No code for recognition. Nature. 1988;335:294–295. doi: 10.1038/335294a0. [DOI] [PubMed] [Google Scholar]
- 2.Lawrence C.E., Altschul S.F., Boguski M.S., Liu J.S., Neuwald A.F., Wootton J.C. Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science. 1993;262:208–214. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- 3.Thompson W., Rouchka E.C., Lawrence C.E. Gibbs Recursive Sampler: finding transcription factor binding sites. Nucleic Acids Res. 2003;31:3580–3585. doi: 10.1093/nar/gkg608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benos P.V., Bulyk M.L., Stormo G.D. Additivity in protein–DNA interactions: how good an approximation is it? Nucleic Acids Res. 2002;30:4442–4451. doi: 10.1093/nar/gkf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benos P.V., Lapedes A.S., Stormo G.D. Is there a code for protein–DNA recognition? Probab(ilistical)ly. Bioessays. 2002;24:466–475. doi: 10.1002/bies.10073. [DOI] [PubMed] [Google Scholar]
- 6.Ellrott K., Yang C., Sladek F.M., Jiang T. Identifying transcription factor binding sites through Markov chain optimization. Bioinformatics. 2002;18((Suppl.2)):S100–S109. doi: 10.1093/bioinformatics/18.suppl_2.s100. [DOI] [PubMed] [Google Scholar]
- 7.Stormo G.D. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 8.Keich U., Pevzner P.A. Subtle motifs: defining the limits of motif finding algorithms. Bioinformatics. 2002;18:1382–1390. doi: 10.1093/bioinformatics/18.10.1382. [DOI] [PubMed] [Google Scholar]
- 9.Sze S.H., Gelfand M.S., Pevzner P.A. Finding weak motifs in DNA sequences. Pac. Symp. Biocomput. 2002:235–246. doi: 10.1142/9789812799623_0022. [DOI] [PubMed] [Google Scholar]
- 10.Keich U., Pevzner P.A. Finding motifs in the twilight zone. Bioinformatics. 2002;18:1374–1381. doi: 10.1093/bioinformatics/18.10.1374. [DOI] [PubMed] [Google Scholar]
- 11.Xu Y., Olman V., Xu D. Clustering gene expression data using a graph-theoretic approach: an application of minimum spanning trees. Bioinformatics. 2002;18:536–545. doi: 10.1093/bioinformatics/18.4.536. [DOI] [PubMed] [Google Scholar]
- 12.Olman V., Xu D., Xu Y. Cubic: identification of regulatory binding sites through data clustering. J. Bioinform. Comput. Biol. 2003;1:21–40. doi: 10.1142/s0219720003000162. [DOI] [PubMed] [Google Scholar]
- 13.Djordjevic M., Sengupta A.M., Shraiman B.I. A biophysical approach to transcription factor binding site discovery. Genome Res. 2003;13:2381–2390. doi: 10.1101/gr.1271603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mandel-Gutfreund Y., Baron A., Margalit H. A structure-based approach for prediction of protein binding sites in gene upstream regions. Pac. Symp. Biocomput. 2001:139–150. doi: 10.1142/9789814447362_0015. [DOI] [PubMed] [Google Scholar]
- 15.Mandel-Gutfreund Y., Margalit H. Quantitative parameters for amino acid–base interaction: implications for prediction of protein–DNA binding sites. Nucleic Acids Res. 1998;26:2306–2312. doi: 10.1093/nar/26.10.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prabakaran P., An J., Gromiha M.M., Selvaraj S., Uedaira H., Kono H., Sarai A. Thermodynamic database for protein–nucleic acid interactions (ProNIT) Bioinformatics. 2001;17:1027–1034. doi: 10.1093/bioinformatics/17.11.1027. [DOI] [PubMed] [Google Scholar]
- 17.Michael Gromiha M., Siebers J.G., Selvaraj S., Kono H., Sarai A. Intermolecular and intramolecular readout mechanisms in protein–DNA recognition. J. Mol. Biol. 2004;337:285–294. doi: 10.1016/j.jmb.2004.01.033. [DOI] [PubMed] [Google Scholar]
- 18.Higo J., Kono H., Nakamura H., Sarai A. Solvent density and long-range dipole field around a DNA-binding protein studied by molecular dynamics. Proteins. 2000;40:193–206. doi: 10.1002/(sici)1097-0134(20000801)40:2<193::aid-prot30>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 19.Ahmad S., Gromiha M.M., Sarai A. Analysis and prediction of DNA-binding proteins and their binding residues based on composition, sequence and structural information. Bioinformatics. 2004;20:477–486. doi: 10.1093/bioinformatics/btg432. [DOI] [PubMed] [Google Scholar]
- 20.Kono H., Sarai A. Structure-based prediction of DNA target sites by regulatory proteins. Proteins. 1999;35:114–131. [PubMed] [Google Scholar]
- 21.Kosikov K.M., Gorin A.A., Lu X.J., Olson W.K., Manning G.S. Bending of DNA by asymmetric charge neutralization: all-atom energy simulations. J. Am. Chem. Soc. 2002;124:4838–4847. doi: 10.1021/ja0125218. [DOI] [PubMed] [Google Scholar]
- 22.Kosikov K.M., Gorin A.A., Zhurkin V.B., Olson W.K. DNA stretching and compression: large-scale simulations of double helical structures. J. Mol. Biol. 1999;289:1301–1326. doi: 10.1006/jmbi.1999.2798. [DOI] [PubMed] [Google Scholar]
- 23.Olson W.K., Gorin A.A., Lu X.J., Hock L.M., Zhurkin V.B. DNA sequence-dependent deformability deduced from protein–DNA crystal complexes. Proc. Natl Acad. Sci. USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olson W.K., Babcock M.S., Gorin A., Liu G., Marky N.L., Martino J.A., Pedersen S.C., Srinivasan A.R., Tobias I., Westcott T.P. Flexing and folding double helical DNA. Biophys. Chem. 1995;55:7–29. doi: 10.1016/0301-4622(94)00139-b. [DOI] [PubMed] [Google Scholar]
- 25.Gorin A.A., Zhurkin V.B., Olson W.K. B-DNA twisting correlates with base-pair morphology. J. Mol. Biol. 1995;247:34–48. doi: 10.1006/jmbi.1994.0120. [DOI] [PubMed] [Google Scholar]
- 26.Zhang C., Liu S., Zhou H., Zhou Y. An accurate, residue-level, pair potential of mean force for folding and binding based on the distance-scaled, ideal-gas reference state. Protein Sci. 2004;13:400–411. doi: 10.1110/ps.03348304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou H., Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11:2714–2726. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muegge I., Martin Y.C. A general and fast scoring function for protein–ligand interactions: a simplified potential approach. J. Med. Chem. 1999;42:791–804. doi: 10.1021/jm980536j. [DOI] [PubMed] [Google Scholar]
- 29.Shimada J., Ishchenko A.V., Shakhnovich E.I. Analysis of knowledge-based protein–ligand potentials using a self-consistent method. Protein Sci. 2000;9:765–775. doi: 10.1110/ps.9.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishchenko A.V., Shakhnovich E.I. SMall Molecule Growth 2001 (SMoG2001): an improved knowledge-based scoring function for protein–ligand interactions. J. Med. Chem. 2002;45:2770–2780. doi: 10.1021/jm0105833. [DOI] [PubMed] [Google Scholar]
- 31.Xu Y., Xu D., Uberbacher E.C. An efficient computational method for globally optimal threading. J. Comput. Biol. 1998;5:597–614. doi: 10.1089/cmb.1998.5.597. [DOI] [PubMed] [Google Scholar]
- 32.Jiang L., Gao Y., Mao F., Liu Z., Lai L. Potential of mean force for protein–protein interaction studies. Proteins. 2002;46:190–196. doi: 10.1002/prot.10031. [DOI] [PubMed] [Google Scholar]
- 33.DeWitte R.S., Shakhnovich E.I. SMoG: de novo design method based on simple, fast, and accurate free energy estimates. 1. Methodology and supporting evidence. J. Am. Chem. Soc. 1996;118:11733–11744. [Google Scholar]
- 34.Sippl M.J. Calculation of conformational ensembles from potentials of mean force. An approach to the knowledge-based prediction of local structures in globular proteins. J. Mol. Biol. 1990;213:859–883. doi: 10.1016/s0022-2836(05)80269-4. [DOI] [PubMed] [Google Scholar]
- 35.Samudrala R., Moult J. An all-atom distance-dependent conditional probability discriminatory function for protein structure prediction. J. Mol. Biol. 1998;275:895–916. doi: 10.1006/jmbi.1997.1479. [DOI] [PubMed] [Google Scholar]
- 36.Lu H., Skolnick J. A distance-dependent atomic knowledge-based potential for improved protein structure selection. Proteins. 2001;44:223–232. doi: 10.1002/prot.1087. [DOI] [PubMed] [Google Scholar]
- 37.Westbrook J., Feng Z., Jain S., Bhat T.N., Thanki N., Ravichandran V., Gilliland G.L., Bluhm W., Weissig H., Greer D.S., et al. The Protein Data Bank: unifying the archive. Nucleic Acids Res. 2002;30:245–248. doi: 10.1093/nar/30.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murzin A.G., Brenner S.E., Hubbard T., Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 39.Desjarlais J.R., Berg J.M. Length-encoded multiplex binding site determination: application to zinc finger proteins. Proc. Natl Acad. Sci. USA. 1994;91:11099–11103. doi: 10.1073/pnas.91.23.11099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choo Y., Sanchez-Garcia I., Klug A. In vivo repression by a site-specific DNA-binding protein designed against an oncogenic sequence. Nature. 1994;372:642–645. doi: 10.1038/372642a0. [DOI] [PubMed] [Google Scholar]
- 41.Choo Y., Klug A. Selection of DNA binding sites for zinc fingers using rationally randomized DNA reveals coded interactions. Proc. Natl Acad. Sci. USA. 1994;91:11168–11172. doi: 10.1073/pnas.91.23.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choo Y., Klug A. Toward a code for the interactions of zinc fingers with DNA: selection of randomized fingers displayed on phage. Proc. Natl Acad. Sci. USA. 1994;91:11163–11167. doi: 10.1073/pnas.91.23.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Z., Dominy B.N., Shakhnovich E.I. Structural mining: self-consistent design on flexible protein–peptide docking and transferable binding affinity potential. J. Am. Chem. Soc. 2004;126:8515–8528. doi: 10.1021/ja032018q. [DOI] [PubMed] [Google Scholar]
- 44.Mirny L.A., Shakhnovich E.I. How to derive a protein folding potential? A new approach to an old problem. J. Mol. Biol. 1996;264:1164–1179. doi: 10.1006/jmbi.1996.0704. [DOI] [PubMed] [Google Scholar]
- 45.Luscombe N.M., Laskowski R.A., Thornton J.M. Amino acid–base interactions: a three-dimensional analysis of protein–DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]