Abstract
Cryo-electron tomography (cryo-ET) is a three-dimensional imaging technique that makes it possible to analyse the structure of complex and dynamic biological assemblies in their native conditions. The latest technological and image processing developments demonstrate that it is possible to obtain structural information at nanometre resolution. The sample preparation required for the cryo-ET technique does not require the isolation of a protein and other macromolecular complexes from its native environment. Therefore, cryo-ET is emerging as an important tool to study the structure of membrane-associated proteins including pores.
This article is part of the themed issue ‘Membrane pores: from structure and assembly, to medicine and technology’.
Keywords: cryo-electron tomography, cryo-electron microscopy, structural biology, membrane-associated proteins, sub-tomogram averaging
1. Introduction
Cells are complex environments populated with millions of proteins [1]. Every one of these proteins has multiple interaction partners, such as other proteins, DNA or RNA, and the duration and specificity of each macromolecular complex can be extremely variable [2]. For decades, structural biologists have been able to resolve the structure of isolated proteins or complexes and generate models representative of those isolated states. Currently, there are three major techniques commonly used for this purpose: X-ray crystallography, nuclear magnetic resonance (NMR) and single-particle cryo-electron microscopy (SP cryo-EM) [3]. All three of these approaches have demonstrated that the structure of a protein or complex can be resolved to atomic or near-atomic resolution; however, the overall picture of the macromolecular activity remains incomplete unless related to its native environment [4].
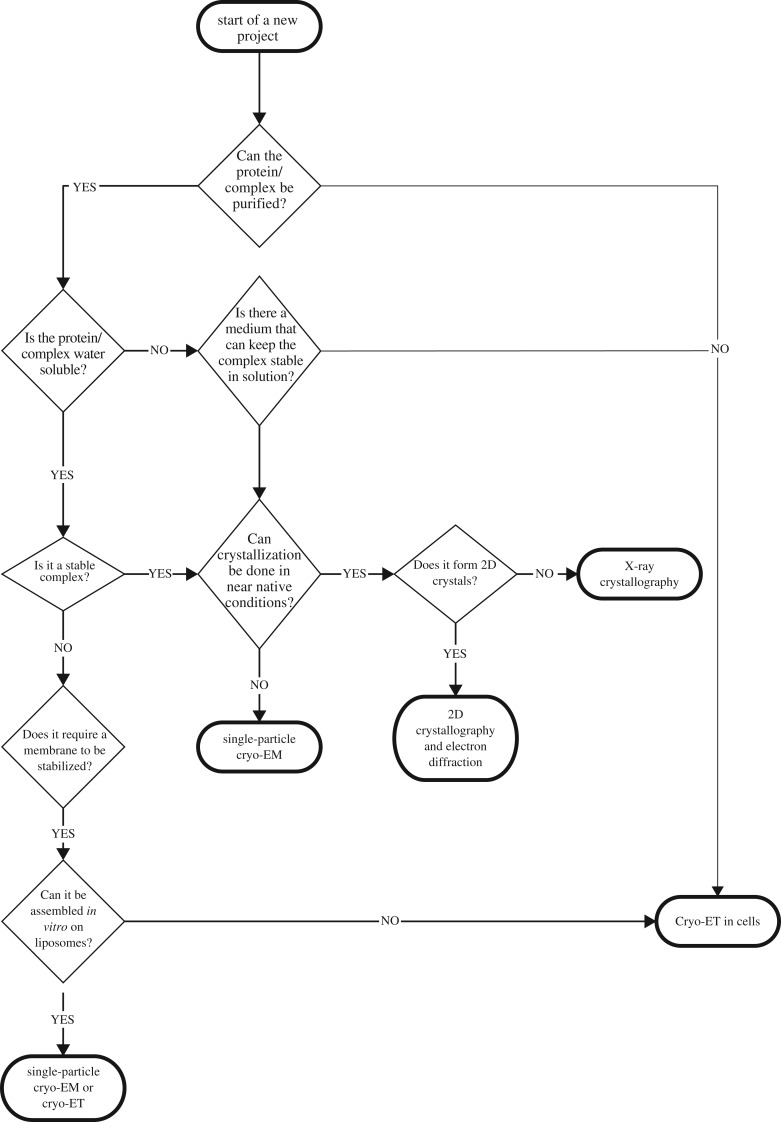
There have been a number of techniques developed to retrieve structural information directly from within the cell or organelle environment, and which ensures that the macromolecular complex is studied in its intact state. The most commonly used technique today is fluorescence microscopy, which can be used to visualize the dynamics of a protein directly in a living cell or organism [5]. Fluorescence microscopy can provide detailed information about the localization and the dynamics of a macromolecular complex, and over the past years there has been an increase in single-molecule-based techniques that can provide insights into the structural conformation of a protein [6,7]. However, the level of detail is limited by the fact that the signal comes from the fluorophore used for detection rather than from the target protein (figure 1).
Figure 1.
Schematic of the possible imaging modalities that can be applied to obtain structural information depending on the sample properties.
An alternative method to analyse the cell is electron tomography (ET), and although it provides a more static image, the details are at higher resolution compared to fluorescent microscopy. ET can be performed either on fixed and stained samples or under cryogenic conditions (cryo-ET). In cryo-ET, there is no need to fix or stain the sample, and therefore it is considered to reflect the native conditions of the macromolecular complex [8] (figure 1). When performing cryo-ET, the sample is immobilized by plunging it in liquid ethane, or by high-pressure freezing, and then stored in liquid nitrogen. This procedure ensures optimal structural preservation and transparency to electrons [9]. A target region is then imaged in a transmission electron microscope (TEM) from multiple angles, and a three-dimensional volume is computed from the projections. This technique has the capability to provide a very detailed map (3–8 nm resolution) of the molecules within the field of view; however, it is limited to image samples whose thickness is below 500 nm [10]. Given that cell thickness can range from 100 nm to 30 µm, depending on cell type and location, this limitation of the technique has meant that most of the cellular cryo-ET data collected to date have been performed on the periphery of the cell, where the thickness is compatible with data collection. In order to access regions in the centre of a cell, the techniques of cryo-sectioning of vitreous samples (CEMOVIS) [11] or cryo-focus ion-beam milling (cryo-FIB) have been developed [12].
Cryo-ET is the only technique of choice to analyse the ultrastructure of samples that are complex and multi-layered because of its three-dimensional information and its independence from labelling and staining. With single projections, it is impossible to assign the imaged densities to the correct molecular layer, resulting in difficult, if not impossible, structural analyses. Cryo-ET can be used for two main purposes: (i) the description of the landscape of a cellular region and (ii) the determination of structures of large macromolecular complexes. The data collection between these two applications changes significantly. If the goal is to obtain a detailed description of the cellular landscape, the magnification is typically lowered to allow the resolution of the structure of interest and to obtain a large overview of the landscape surrounding the target. The electron dose will be maximized (up to approx. 100 e− Å−2), and the defocus will be increased to provide the best possible contrast. Alternatively, if the goal is to obtain molecular structural information, the electron dose is typically minimized to prevent damage (approx. 50 e− Å−2) [13]. However, a recent report showed that it is possible to increase the dose applied to the sample, without incurring any resolution loss, by application of a dose-dependent filter throughout the tilt series [14]. The magnification needs to be increased depending on the information limit required, and defocus needs to be stable within a tilt series [15]. The recent introduction of direct electron detectors [16] has significantly improved the image quality and dramatically reduced the high noise levels that typically characterize cryo-electron tomograms. In addition, the introduction of Volta phase plates [17] makes it possible to collect data that are defocused by only a few tens of nanometres and yet have high contrast.
A typical problem arising upon the inspection of a tomogram collected on a cellular sample, as well as poorly purified samples, is that only a small fraction of the densities can be unequivocally recognized based on their shape or location. Over the past 5 years, the use of correlative light and electron microscopy (CLEM) has been applied to cryo-preserved samples, allowing the identification of rare events and associating a structure to a protein complex or event [18–20]. Current developments in cryo-CLEM showed that it is possible to predict the position of a molecule in three dimensions with a precision better than 200 nm, and it is therefore possible to target the cryo-FIB milling to prepare the sample for cryo-ET [21].
2. Cryo-electron tomography: an ideal tool to study membrane-associated complexes
For many years, the structure of membrane-associated proteins has been extremely difficult to resolve and study. About a quarter of all proteins are associated to a membrane but only 656 unique protein structures have been determined (reported in November 2016 on MemProtMD (http://blanco.biomol.uci.edu/mpstruc/)). In fact, the presence of exposed hydrophobic transmembrane regions makes the crystallization process difficult, and often the transmembrane domain is cleaved to increase chances of success [22]. Alternatively, the preparation of two-dimensional crystals, and their analysis through electron diffraction, allows resolution of the structure of membrane-associated proteins in a conformation that is the closest to the one displayed in the lipid bilayer [23,24]. Two-dimensional crystallization relies on the use of detergents that maintain the hydrophobic regions properly folded while also using conditions for crystallization that can be extremely difficult.
Over the past 5 years, the use of liposomes, lipid nanodiscs, detergents and amphipols has overcome the need for crystallization, allowing membrane-associated proteins to be studied in closer to native conditions through SP cryo-EM. The use of nanodiscs is particularly advantageous in the study of protein–lipid interactions [25]. Together with nanodiscs, the use of detergent micelles and amphipol showed reproducibly to allow sub-nanometre structural determination on a number of complexes, such as γ-secretase [26], the TRP channel [27] and ryanodine receptor C [28,29]. Another method for imaging isolated membrane-associated complexes uses liposomes [30,31]. Although the obtained resolution of liposome-embedded complexes has been lower than with the previously described detergent/amphipol solubilization methods, liposome–protein complexes have allowed the structure of lipid bilayer embedded protein complexes to be studied. For example, SP cryo-EM has been used to determine the structure of pore-forming toxins such as suilysin and pleurotolysin [32,33].
Altogether, the presented approaches have in common the need to either extract a protein from its native environment or assemble it within a non-native environment. This strategy removes complexity from the system; however, it does not allow the complete overview of the structure within the cellular context. Cryo-ET is not subject to the artefacts generated during purification because it can be performed directly within cells. This also ensures that the full spectrum of interactions of the target molecule is preserved for imaging [34,35].
3. Image processing techniques for cryo-electron tomography
The resolution in a cryo-tomogram is limited by two factors: Firstly, the limited electron dose applicable to the sample (ideally below 100 e−/Å2) [36,37], and its distribution across multiple images in the tilt series, results in a high level of noise and extremely poor contrast [38]. Secondly, the maximum achievable tilt range is approximately 140° due to the slab shape of the EM grid [38]. The limited tilt range results in an incomplete angular sampling with consequently a lower resolution along the beam axis; hence, the resolution is proportional to the maximum angle that can be imaged.
In order to overcome the high noise level, a range of filtering methods can be applied from simple frequency band pass and spatial blurring, to more targeted noise filtering methods based on anisotropic diffusion [39,40]. The user can choose the preferred method depending on the type of information required, the size of the dataset and the computing power available. Importantly, although filtering methods can highlight features that are already above the noise level, they cannot retrieve any information that is below the noise level.
The most effective way to overcome the low signal has been shown to be through averaging. If multiple copies of the same structure can be identified, aligned relative to each other and then averaged, then the conserved information will be boosted above noise level. Another advantage of averaging multiple copies of the same structure is to overcome the anisotropy of the tomogram data. If the individual copies of the structure analysed are randomly oriented relative to the beam axis, averaging leads to filling the missing angular information and therefore the resolution of the average will be isotropic [41–43]. This technique is called sub-tomogram averaging and has already been demonstrated to allow the resolution of macromolecular structures to sub-nanometre resolution for large viral assemblies [15,44], as well as near-atomic resolution on the HIV-1 immature capsid [14].
4. Analyses of glycoproteins in enveloped viruses
For a long time, viral proteins have been the most studied by structural biologists, not only for the high pathological relevance, but also because a significant proportion of the viruses studied over the years have a capsid that is organized following strict icosahedral symmetry [45]. Glycoproteins in icosahedral viruses are generally located in defined positions on the capsid. The structure in those cases can be obtained together with the capsid structure through cryo-EM and icosahedral reconstruction [46]. With the implementation of sub-tomogram averaging, it is now possible to resolve the structure of viral glycoproteins located in the membrane of enveloped viruses. Even without knowing the exact location of the target proteins, the typical workflow consists of the indiscriminate extraction of observed density located on the membrane surface followed by iterative alignment. This method works based on the idea that the most abundant feature will predominate in the final average, while all densities that do not match will be excluded based on correlation threshold when compared to the average. A few densities are manually picked based on the expected shape and size and are used to provide a reference model. The classification can be done by applying a threshold in the cross correlation value calculated during the alignment with the reference or with the sum of the densities after alignment.
Sub-tomogram averaging was used to successfully determine the structure of the gp120 trimer (figure 2a) on the surface of SIV and HIV-1 virions at a resolution close to 2 nm [47,48]. The analysis in this case allowed the binding dynamics and conformational changes induced by the neutralizing antibodies to be understood. In another example, the structure of the Lassa virus glycoprotein showed how conformational changes were induced by the pH variation directly in situ. This research supported the hypothesis that the viral entry mechanism depends on a lysosome-resident receptor (LAMP1) [49].
Figure 2.
Gallery of membrane-associated proteins, structures determined through the application of tomography and sub-tomogram averaging. Surfaces are coloured based on distance from the membrane with which the complex is associated. The red surface represents the membrane or the area where the membrane is associated, and the blue represents the component of the complex furthest away from the membrane. (a) The structure of the HIV-1 glycoprotein GP120 resolved on the virus. (b) The structure of the COPI coat resolved on the surface of in vitro assembled vesicles. In panel (c), we show the MAC, and in (d), there is a cumulative view of the NPC resolved on purified nuclear membranes. All panels have been generated from the density maps deposited on the EMDataBank (EMD: 5272, 2985, 3289, 3006–3009).
A study of Tula virus showed how, through in situ analyses, it is possible to map the exact position and relative orientation of each glycoprotein and identified the presence of ordered patches. The authors also inferred that those patches are driven by regulated interactions, which could be sufficient to drive the required membrane curvature for the viral budding [50]. Similarly, in herpesvirus, it was possible to identify the existence of lateral protein–protein interaction between multiple copies of glycoprotein B. The described interactions induced the formation of a locally continuous coat, suggesting the role of this protein as a potential driver for the fusion process during infection [51].
5. Large molecular motors
Tomography has demonstrated its value particularly when it comes to the analysis of large molecular machines that are difficult to purify or to assemble in vitro. For example, the study of type IVa pili, on the surface of the bacterium Myxococcus xanthus, provided an understanding of the assembly and functioning mechanisms of these large machineries [52]. Here, the authors combined topologic and structural analyses to be able to discriminate between different conformational states. The sub-tomogram averaging reconstructions further allowed structures at resolutions of 3–5 nm to be identified. The high resolution on such large protein complexes, together with the analyses of mutants, allowed the distinction of all subunits.
Other molecular motors have been studied using cryo-ET, including cilia and flagella, which are associated with cell motility. Owing to the stability of the structures, they could be studied both in situ and in vitro [53]. The first structure of a flagellar motor ever solved was done by averaging only 20 motors and led to a resolution of approximately 7 nm [54], and at this resolution it was possible to identify the rotor from the stator and resolve its 16-fold symmetry. Reconstructions of microtubules and microtubule-associated proteins (e.g. dynein), in the resolution range between 3 and 4 nm, showed how the sliding of microtubule doublets against each other induces bending of the flagella to control cell movement [55–57].
6. Analyses of cellular vesicles and organelles associated proteins
For years, clathrin-coated vesicles were the only cellular coated vesicle for which both the structure and the assembly mechanism were known [58,59]. The reason for this is linked to the highly ordered basic triskelion unit, which was first studied in an isolated state in 1983 [60]. The structure of the assembled cage could be resolved using standard SP cryo-EM [61].
COPII coats were first resolved by SP cryo-EM of in vitro assembled cages composed of the sub-complex (Sec13/31) but lacked both the membrane and cargo [60]. Using sub-tomogram averaging, it has recently been possible to describe the assembly of the complete COPII transport vesicle with a resolution of approximately 2 nm (figure 2b). In this case, the vesicles were also assembled in vitro but in the presence of membranes and the cargo-binding proteins (Sec23/24). Similarly, COPI-coated vesicles have recently been described through sub-tomogram averaging [62,63]. The difficulties linked to generate stable COPII complexes that were not associated with the membrane, together with the fact that every vesicle is different, required a method that could inspect a structure locally without expecting a rigidly ordered repetitive structure.
Over the past years, it has been possible to attempt structural biology directly within the cell. A sample preparation method that includes the use of a focused ion beam (FIB) to etch away a part of the sample that does not contain the structures of interest allows thick samples to be studied using cryo-ET [12]. This approach can be coupled to fluorescence light microscopy in order to specifically target the region containing the complex to be studied, using CLEM [21]. Using this method to prepare the sample, intracisternal protein arrays were observed in the Golgi apparatus [64]. Given the high level of repetitiveness of those arrays, it has been possible to obtain a structure with a resolution in the range of approximately 2 nm.
7. Transmembrane proteins and pores
Transmembrane proteins are, for structural biologists, among the most difficult samples. If they are part of a large complex, the stability of the complex itself is heavily dependent on the presence of the lipid bilayer [65]. An extremely powerful method to solve the structure of a transmembrane protein in a native-like condition has been through electron diffraction on two-dimensional crystals [65–67]. Despite the good results that this technique can bring, the process of two-dimensional crystallization can be difficult or can require conditions that are far from physiological. In cryo-ET, the sample preparation is minimized to the fast freezing of the sample that is coupled to a thinning method. The resolution is limited only by the flexibility of the complex and by the number of asymmetric units that can be collected. Cryo-ET has been applied only to a limited number of samples; however, the use of this technique is increasing with the recent advent of fully automated data collection, improved detectors and Volta phase plates.
An example of a transmembrane pore protein that was studied by cryo-ET has been presented by Sharp et al. [68]. The authors here acquired tomograms of partly and fully assembled membrane attack complex (MAC) pores on liposomes. Through sub-tomogram averaging, they could obtain a structure at a resolution between 2 and 3 nm for the fully assembled MAC (figure 2c). This study also represents the first attempt to image this machinery during its assembly phase. Similarly, a cryo-ET study of pneumolysin observed full-ring pores and incomplete rings, referred to as arcs, in both the prepore and pore state [69]. The structures analysed raise the question of whether arcs could be as effective as full pores in the biological function of pneumolysin and other CDCs.
As previously mentioned, the application of cryo-ET in situ often requires the sample to be thinned in order to be compatible with high-resolution imaging. One of the early examples of structural biology in situ is the analysis of the architecture of desmosomes [70]. Here, the authors performed 80 nm cryo-sections on a vitrified epithelial tissue and could get a complete view over the desmosomal region. They then applied sub-tomogram averaging to enhance the resolution on the extracellular region and obtained enough detail to allow fitting of pre-existing crystal structures.
Among the most studied large membrane-associated complexes is the nuclear pore complex (NPC). This machine is 124 MDa in size, highly dynamic and is composed of approximately 450 proteins [71]. The size of the NPC makes it impossible to purify or assemble the whole complex in vitro. The early structural studies were performed on Xenopus laevis oocytes [72] and provided a general understanding of the symmetry and architecture of the NPC to a resolution of approximately 12 nm. Subsequent studies performed on Dictyostelium discoideum as well as on human cells achieved a resolution down to 2 nm [73,74], providing a detailed understanding of the mechanisms used to allow the passage of molecules through the NPC (figure 2d).
8. Conclusion
As outlined in this review, cryo-ET is an emerging technique which has just started to show its potential. The hardware development that has occurred in the past 5 years, together with the recent advances in image processing techniques, has made cryo-ET available to a constantly growing community. Cryo-ET provides the possibility of performing structural biology studies of complexes within their native environment. This attribute makes cryo-ET the perfect tool to study membrane-associated proteins, particularly those complexes that are not stable when purified or that cannot crystallize.
It has been shown that it is possible to achieve near-atomic resolution with cryo-ET when used in conjunction with sub-tomogram averaging. A limiting factor at the moment is the high number of copies required to achieve such resolutions, and therefore it is unlikely that cryo-ET will be routinely used for high-resolution studies. However, it is also reasonable to predict that the use of cryo-ET will increase over the coming years and will become the technique of choice to study the structure of complexes that cannot be reconstituted in vitro or that can be reconstituted only in large liposomes. When used on cellular samples, it will allow an understanding of the distribution and the steps of assembly of macromolecular complexes inside their native environment.
Overall, the information retrieved will be complemented with data coming from classical structural biology techniques, such as X-ray crystallography, NMR or SP cryo-EM. In this scenario, each component of a macromolecular complex will be resolved at high resolution individually, while the assembled complex will be resolved at intermediate resolution (1–2 nm) through cryo-ET and individual components will be fitted into the low-resolution density. Given the current state of cryo-EM and cryo-ET techniques, the resolution at which a complex can be resolved is not limited by the technique in use but the flexibility of the target complex or by the number of copies that can be imaged. The observation of different states or flexibility of a macromolecular complex is in itself important information that can be used to inform crystallization approaches.
Although it has not been in the focus of this review, it is important to mention that there are multiple software packages that can be used to perform sub-tomogram averaging (Pytom, Dynamo, Jsubtomo, EMAN2, Relion2). All the available packages have proven to be functional although the workflows are not yet completely set, and procedure requires significant expertise to avoid the pitfalls of reference bias and noise alignment. The future perspectives in the field will likely see cryo-ET tightly bound to correlative microscopy, to allow any target to be recognized and imaged. The exciting developments in the fields of cryo-FIB milling, when mature, will allow reproducible imaging in any region of the cell.
Authors' contributions
A.d.M. co-wrote the text and designed and made the figures. M.A.D. co-wrote the text.
Competing interests
The authors have no competing interests.
Funding
M.A.D. is supported by an NHMRC Career Development Fellowship and an ARC Future Fellowship.
References
- 1.Milo R. 2013. What is the total number of protein molecules per cell volume? A call to rethink some published values. Bioessays 35, 1050–1055. ( 10.1002/bies.201300066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berendsen HJ, Hayward S. 2000. Collective protein dynamics in relation to function. Curr. Opin. Struct. Biol. 10, 165–169. ( 10.1016/S0959-440X(00)00061-0) [DOI] [PubMed] [Google Scholar]
- 3.Campbell ID. 2002. Timeline: the march of structural biology. Nat. Rev. Mol. Cell Biol. 3, 377–381. ( 10.1038/nrm800) [DOI] [PubMed] [Google Scholar]
- 4.Asano S, Engel BD, Baumeister W. 2016. In situ cryo-electron tomography: a post-reductionist approach to structural biology. J. Mol. Biol. 428, 332–343. ( 10.1016/j.jmb.2015.09.030) [DOI] [PubMed] [Google Scholar]
- 5.Ettinger A, Wittmann T. 2014. Fluorescence live cell imaging. Methods Cell Biol. 123, 77–94. ( 10.1016/B978-0-12-420138-5.00005-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forkey JN, Quinlan ME, Goldman YE. 2000. Protein structural dynamics by single-molecule fluorescence polarization. Prog. Biophys. Mol. Biol. 74, 1–35. ( 10.1016/S0079-6107(00)00015-8) [DOI] [PubMed] [Google Scholar]
- 7.Schuler B. 2013. Single-molecule FRET of protein structure and dynamics—a primer. J. Nanobiotechnol. 11(Suppl. 1), S2 ( 10.1186/1477-3155-11-S1-S2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai XC, McMullan G, Scheres SH. 2015. How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 40, 49–57. ( 10.1016/j.tibs.2014.10.005) [DOI] [PubMed] [Google Scholar]
- 9.Adrian M, Dubochet J, Lepault J, McDowall AW. 1984. Cryo-electron microscopy of viruses. Nature 308, 32–36. ( 10.1038/308032a0) [DOI] [PubMed] [Google Scholar]
- 10.Feja B, Aebi U. 1999. Determination of the inelastic mean free path of electrons in vitrified ice layers for on-line thickness measurements by zero-loss imaging. J. Microsc. 193, 15–19. ( 10.1046/j.1365-2818.1999.00436.x) [DOI] [PubMed] [Google Scholar]
- 11.Al-Amoudi A, et al. 2004. Cryo-electron microscopy of vitreous sections. EMBO J. 23, 3583–3588. ( 10.1038/sj.emboj.7600366) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rigort A, et al. 2010. Micromachining tools and correlative approaches for cellular cryo-electron tomography. J. Struct. Biol. 172, 169–179. ( 10.1016/j.jsb.2010.02.011) [DOI] [PubMed] [Google Scholar]
- 13.Karuppasamy M, Karimi Nejadasl F, Vulovic M, Koster AJ, Ravelli RBG. 2011. Radiation damage in single-particle cryo-electron microscopy: effects of dose and dose rate. J. Synchrotron Radiat. 18, 398–412. ( 10.1107/S090904951100820X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schur FK, Obr M, Hagen WJ, Wan W, Jakobi AJ, Kirkpatrick JM, Sachse C, Kräusslich HG, Briggs JA. 2016. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science 353, 506–508. ( 10.1126/science.aaf9620) [DOI] [PubMed] [Google Scholar]
- 15.Koestler SA, et al. 2013. Arp2/3 complex is essential for actin network treadmilling as well as for targeting of capping protein and cofilin. Mol. Biol. Cell 24, 2861–2875. ( 10.1091/mbc.E12-12-0857) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McMullan G, McMullan Clark AT, Turchetta R, Faruqi AR. 2009. Enhanced imaging in low dose electron microscopy using electron counting. Ultramicroscopy 109, 1411–1416. ( 10.1016/j.ultramic.2009.07.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danev R, Buijsse B, Khoshouei M, Plitzko JM, Baumeister W. 2014. Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc. Natl Acad. Sci. USA 111, 15 635–15 640. ( 10.1073/pnas.1418377111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sartori A, Gatz R, Beck F, Rigort A, Baumeister W, Plitzko JM. 2007. Correlative microscopy: bridging the gap between fluorescence light microscopy and cryo-electron tomography. J. Struct. Biol. 160, 135–145. ( 10.1016/j.jsb.2007.07.011) [DOI] [PubMed] [Google Scholar]
- 19.Schorb M, Briggs JAG. 2014. Correlated cryo-fluorescence and cryo-electron microscopy with high spatial precision and improved sensitivity. Ultramicroscopy 143, 24–32. ( 10.1016/j.ultramic.2013.10.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jasnin M, Ecke M, Baumeister W, Gerisch G. 2016. Actin organization in cells responding to a perforated surface, revealed by live imaging and cryo-electron tomography. Structure 24, 1031–1043. ( 10.1016/j.str.2016.05.004) [DOI] [PubMed] [Google Scholar]
- 21.Arnold J, et al. 2016. Site-specific cryo-focused ion beam sample preparation guided by 3D correlative microscopy. Biophys. J. 110, 860–869. ( 10.1016/j.bpj.2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carpenter EP, Beis K, Cameron AD, Iwata S. 2008. Overcoming the challenges of membrane protein crystallography. Curr. Opin. Struct. Biol. 18, 581–586. ( 10.1016/j.sbi.2008.07.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glaeser RM, Downing KH. 1993. High-resolution electron crystallography of protein molecules. Ultramicroscopy 52, 478–486. ( 10.1016/0304-3991(93)90064-5) [DOI] [PubMed] [Google Scholar]
- 24.Renault L, et al. 2006. Milestones in electron crystallography. J. Comput. Aided Mol. Des. 20, 519–527. ( 10.1007/s10822-006-9075-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuler MA, Denisov IG, Sligar SG. 2013. Nanodiscs as a new tool to examine lipid-protein interactions. Methods Mol. Biol. 974, 415–433. ( 10.1007/978-1-62703-275-9_18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu P, et al. 2014. Three-dimensional structure of human gamma-secretase. Nature 512, 166–170. ( 10.1038/nature13567) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moiseenkova-Bell VY, Wensel TG. 2009. Hot on the trail of TRP channel structure. J. Gen. Physiol. 133, 239–244. ( 10.1085/jgp.200810123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samso M, Wagenknecht T, Allen PD. 2005. Internal structure and visualization of transmembrane domains of the RyR1 calcium release channel by cryo-EM. Nat. Struct. Mol. Biol. 12, 539–544. ( 10.1038/nsmb938) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludtke SJ, Baker ML, Cong Y, Topf M, Eramian D, Sali A, Hamilton SL, Chiu W. 2008. Subnanometer-resolution electron cryomicroscopy-based domain models for the cytoplasmic region of skeletal muscle RyR channel. Proc. Natl Acad. Sci. USA 105, 9610–9615. ( 10.1073/pnas.0803189105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rigaud JL, Levy D. 2003. Reconstitution of membrane proteins into liposomes. Methods Enzymol. 372, 65–86. ( 10.1016/S0076-6879(03)72004-7) [DOI] [PubMed] [Google Scholar]
- 31.Simeonov P, Werner S, Haupt C, Tanabe M, Bacia K. 2013. Membrane protein reconstitution into liposomes guided by dual-color fluorescence cross-correlation spectroscopy. Biophys. Chem. 184, 37–43. ( 10.1016/j.bpc.2013.08.003) [DOI] [PubMed] [Google Scholar]
- 32.Leung C, et al. 2014. Stepwise visualization of membrane pore formation by suilysin, a bacterial cholesterol-dependent cytolysin. eLife 3, e04247 ( 10.7554/eLife.04247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukoyanova N, et al. 2015. Conformational changes during pore formation by the perforin-related protein pleurotolysin. PLoS Biol. 13, e1002049 ( 10.1371/journal.pbio.1002049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medalia O, Weber I, Frangakis AS, Nicastro D, Gerisch G, Baumeister W. 2002. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science 298, 1209–1213. ( 10.1126/science.1076184) [DOI] [PubMed] [Google Scholar]
- 35.Lucic V, Rigort A, Baumeister W. 2013. Cryo-electron tomography: the challenge of doing structural biology in situ. J. Cell Biol. 202, 407–419. ( 10.1083/jcb.201304193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conway JF, Trus BL, Booy FP, Newcomb WW, Brown JC, Steven AC. 1993. The effects of radiation damage on the structure of frozen hydrated HSV-1 capsids. J. Struct. Biol. 111, 222–233. ( 10.1006/jsbi.1993.1052) [DOI] [PubMed] [Google Scholar]
- 37.Glaeser RM. 1971. Limitations to significant information in biological electron microscopy as a result of radiation damage. J. Ultrastruct. Res. 36, 466–482. ( 10.1016/S0022-5320(71)80118-1) [DOI] [PubMed] [Google Scholar]
- 38.Koster AJ, Grimm R, Typke D, Hegerl R, Stoschek A, Walz J, Baumeister W. 1997. Perspectives of molecular and cellular electron tomography. J. Struct. Biol. 120, 276–308. ( 10.1006/jsbi.1997.3933) [DOI] [PubMed] [Google Scholar]
- 39.Narasimha R, Aganj I, Bennett AE, Borgnia MJ, Zabransky D, Sapiro G, McLaughlin SW, Milne JLS, Subramaniam S. 2008. Evaluation of denoising algorithms for biological electron tomography. J. Struct. Biol. 164, 7–17. ( 10.1016/j.jsb.2008.04.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frangakis AS, Hegerl R. 2001. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J. Struct. Biol. 135, 239–250. ( 10.1006/jsbi.2001.4406) [DOI] [PubMed] [Google Scholar]
- 41.Forster F, Medalia O, Zauberman N, Baumeister W, Fass D. 2005. Retrovirus envelope protein complex structure in situ studied by cryo-electron tomography. Proc. Natl Acad. Sci. USA 102, 4729–4734. ( 10.1073/pnas.0409178102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. 2007. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 26, 2218–2226. ( 10.1038/sj.emboj.7601664) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zanetti G, Briggs JAG, Grünewald K, Sattentau QJ, Fuller SD. 2006. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2, e83 ( 10.1371/journal.ppat.0020083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bharat TA, Russo CJ, Löwe J, Passmore LA, Scheres SHW. 2015. Advances in single-particle electron cryomicroscopy structure determination applied to sub-tomogram averaging. Structure 23, 1743–1753. ( 10.1016/j.str.2015.06.026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prasad BV, Schmid MF. 2012. Principles of virus structural organization. Adv. Exp. Med. Biol. 726, 17–47. ( 10.1007/978-1-4614-0980-9_3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mancini EJ, de Haas F, Fuller S. 1997. High-resolution icosahedral reconstruction: fulfilling the promise of cryo-electron microscopy. Structure 5, 741–750. ( 10.1016/S0969-2126(97)00229-3) [DOI] [PubMed] [Google Scholar]
- 47.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455, 109–113. ( 10.1038/nature07159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White TA, et al. 2010. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog. 6, e1001249 ( 10.1371/journal.ppat.1001249) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li S, et al. 2016. Acidic pH-induced conformations and LAMP1 binding of the Lassa virus glycoprotein spike. PLoS Pathog. 12, e1005418 ( 10.1371/journal.ppat.1005418) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huiskonen JT, Hepojoki J, Laurinmaki P, Vaheri A, Lankinen H, Butcher SJ, Grunewald K. 2010. Electron cryotomography of Tula hantavirus suggests a unique assembly paradigm for enveloped viruses. J. Virol. 84, 4889–4897. ( 10.1128/JVI.00057-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maurer UE, et al. 2013. The structure of herpesvirus fusion glycoprotein B-bilayer complex reveals the protein-membrane and lateral protein-protein interaction. Structure 21, 1396–1405. ( 10.1016/j.str.2013.05.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang YW, Rettberg LA, Treuner-Lange A, Iwasa J, Sogaard-Andersen L, Jensen GJ. 2016. Architecture of the type IVa pilus machine. Science 351, aad2001 ( 10.1126/science.aad2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicastro D. 2009. Cryo-electron microscope tomography to study axonemal organization. Methods Cell Biol. 91, 1–39. ( 10.1016/S0091-679X(08)91001-3) [DOI] [PubMed] [Google Scholar]
- 54.Murphy GE, Leadbetter JR, Jensen GJ. 2006. In situ structure of the complete Treponema primitia flagellar motor. Nature 442, 1062–1064. ( 10.1038/nature05015) [DOI] [PubMed] [Google Scholar]
- 55.Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T. 2009. Asymmetry of inner dynein arms and inter-doublet links in Chlamydomonas flagella. J. Cell Biol. 186, 437–446. ( 10.1083/jcb.200903082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoog JL, Bouchet-Marquis C, McIntosh JT, Hoenger A, Gull K. 2012. Cryo-electron tomography and 3-D analysis of the intact flagellum in Trypanosoma brucei. J. Struct. Biol. 178, 189–198. ( 10.1016/j.jsb.2012.01.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koyfman AY, Schmid MF, Gheiratmand L, Fu CJ, Khant HA, Huang D, He CY, Chiu W. 2011. Structure of Trypanosoma brucei flagellum accounts for its bihelical motion. Proc. Natl Acad. Sci. USA 108, 11 105–11 108. ( 10.1073/pnas.1103634108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmid SL. 1997. Clathrin-coated vesicle formation and protein sorting: an integrated process. Annu. Rev. Biochem. 66, 511–548. ( 10.1146/annurev.biochem.66.1.511) [DOI] [PubMed] [Google Scholar]
- 59.McMahon HT, Boucrot E. 2011. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12, 517–533. ( 10.1038/nrm3151) [DOI] [PubMed] [Google Scholar]
- 60.Ungewickell E. 1983. Biochemical and immunological studies on clathrin light chains and their binding sites on clathrin triskelions. EMBO J. 2, 1401–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fotin A, Cheng Y, Grigorieff N, Walz T, Harrison SC, Kirchhausen T. 2004. Structure of an auxilin-bound clathrin coat and its implications for the mechanism of uncoating. Nature 432, 649–653. ( 10.1038/nature03078) [DOI] [PubMed] [Google Scholar]
- 62.Faini M, Prinz S, Beck R, Schorb M, Riches JD, Bacia K, Brugger B, Wieland FT, Briggs JAG. 2012. The structures of COPI-coated vesicles reveal alternate coatomer conformations and interactions. Science 336, 1451–1454. ( 10.1126/science.1221443) [DOI] [PubMed] [Google Scholar]
- 63.Dodonova SO, Diestelkoetter-Bachert P, von Appen A, Hagen WJ, Beck R, Beck M, Wieland F, Briggs JA. 2015. Vesicular transport. A structure of the COPI coat and the role of coat proteins in membrane vesicle assembly. Science 349, 195–198. ( 10.1126/science.aab1121) [DOI] [PubMed] [Google Scholar]
- 64.Engel BD, Schaffer M, Albert S, Asano S, Plitzko JM, Baumeister W. 2015. In situ structural analysis of Golgi intracisternal protein arrays. Proc. Natl Acad. Sci. USA 112, 11 264–11 269. ( 10.1073/pnas.1515337112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abeyrathne PD, Chami M, Pantelic RS, Goldie KN, Stahlberg H. 2010. Preparation of 2D crystals of membrane proteins for high-resolution electron crystallography data collection. Methods Enzymol. 481, 25–43. ( 10.1016/S0076-6879(10)81001-8) [DOI] [PubMed] [Google Scholar]
- 66.Chou HT, Evans JE, Stahlberg H. 2007. Electron crystallography of membrane proteins. Methods Mol. Biol. 369, 331–343. ( 10.1007/978-1-59745-294-6_16) [DOI] [PubMed] [Google Scholar]
- 67.Goldie KN, Abeyrathne P, Kebbel F, Chami M, Ringler P, Stahlberg H. 2014. Cryo-electron microscopy of membrane proteins. Methods Mol. Biol. 1117, 325–341. ( 10.1007/978-1-62703-776-1_15) [DOI] [PubMed] [Google Scholar]
- 68.Sharp TH, Koster AJ, Gros P. 2016. Heterogeneous MAC initiator and pore structures in a lipid bilayer by phase-plate cryo-electron tomography. Cell Rep. 15, 1–8. ( 10.1016/j.celrep.2016.03.002) [DOI] [PubMed] [Google Scholar]
- 69.Sonnen AF, Plitzko JM, Gilbert RJ.. 2014. Incomplete pneumolysin oligomers form membrane pores. Open Biol. 4, 140044 ( 10.1098/rsob.140044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Al-Amoudi A, Díez DC, Betts MJ, Frangakis AS. 2007. The molecular architecture of cadherins in native epidermal desmosomes. Nature 450, 832–837. ( 10.1038/nature05994) [DOI] [PubMed] [Google Scholar]
- 71.Alber F, et al. 2007. Determining the architectures of macromolecular assemblies. Nature 450, 683–694. ( 10.1038/nature06404) [DOI] [PubMed] [Google Scholar]
- 72.Stoffler D, Feja B, Fahrenkrog B, Walz J, Typke D, Aebi U. 2003. Cryo-electron tomography provides novel insights into nuclear pore architecture: implications for nucleocytoplasmic transport. J. Mol. Biol. 328, 119–130. ( 10.1016/S0022-2836(03)00266-3) [DOI] [PubMed] [Google Scholar]
- 73.Beck M, Förster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeiaster W, Medalia O. 2004. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science 306, 1387–1390. ( 10.1126/science.1104808) [DOI] [PubMed] [Google Scholar]
- 74.Eibauer M, Pellanda M, Turgay Y, Dubrovsky A, Wild A, Medalia O. 2015. Structure and gating of the nuclear pore complex. Nat. Commun. 6, 7532 ( 10.1038/ncomms8532) [DOI] [PMC free article] [PubMed] [Google Scholar]