Abstract
Ion transporters and channels are able to identify and act on specific substrates among myriads of ions and molecules critical to cellular processes, such as homeostasis, cell signalling, nutrient influx and drug efflux. Recently, we designed Rocker, a minimalist model for Zn2+/H+ co-transport. The success of this effort suggests that de novo membrane protein design has now come of age so as to serve a key approach towards probing the determinants of membrane protein folding, assembly and function. Here, we review general principles that can be used to design membrane proteins, with particular reference to helical assemblies with transport function. We also provide new functional and NMR data that probe the dynamic mechanism of conduction through Rocker.
This article is part of the themed issue ‘Membrane pores: from structure and assembly, to medicine and technology’.
Keywords: computational design, membrane protein folding, metallotransporter
1. Introduction
De novo design is an attractive approach to test and extend principles derived from studying natural systems [1]. Water-soluble protein design has advanced towards expanding the undiscovered fold space [2], catalytic function [3–5], vaccines [6,7] and materials [8–12]. Although transmembrane (TM) protein design has advanced less rapidly due to technical difficulties in experimental verifications of designs [13], significant progress has been made [14,15].
Much work has focused on the design of helical peptides that self-associate to form membrane-spanning helical bundles [14–16]. These systems have been useful for testing the principles of helix–helix association in membranes as well as functions, including TM electron [17] and ion transport. One recent accomplishment from our group was the de novo design of Rocker (Transmembrane Translocation Kinetics Enhancer), a 25-residue TM peptide that assembles into a four-helix bundle capable of antiporting Zn2+ and protons [18]. Natural proton-coupled transporters are believed to use a TM proton gradient to drive active ligand transport by rocking between two states, the substrate-binding site being alternately accessible to either side of the bilayer [18–22]. The binding and release of substrates and protons induce conformational changes required for active transport. In designing Rocker, we thus aimed to test this so-called alternating-access model of proton-linked transport (figure 1). At a high level, this goal required: (i) design and structure determination of a membrane protein; (ii) positioning polar ionizable ligands, ordinarily excluded from membranes, into the bilayer; (iii) linking proton binding to changes in metal ion affinity; and (iv) control of dynamics to facilitate ion transport. X-ray crystallography, solution NMR and SSNMR showed the bundle was comprised of two tightly interacting pairs of helices that coalesce along a more dynamic interface to form a tetramer. The tight interface is important for positioning the essential ligands, while the dynamic interface enabled conformational transitions required for Zn2+ translocation. As Zn2+ ions diffuse down their concentration gradients, protons are antiported.
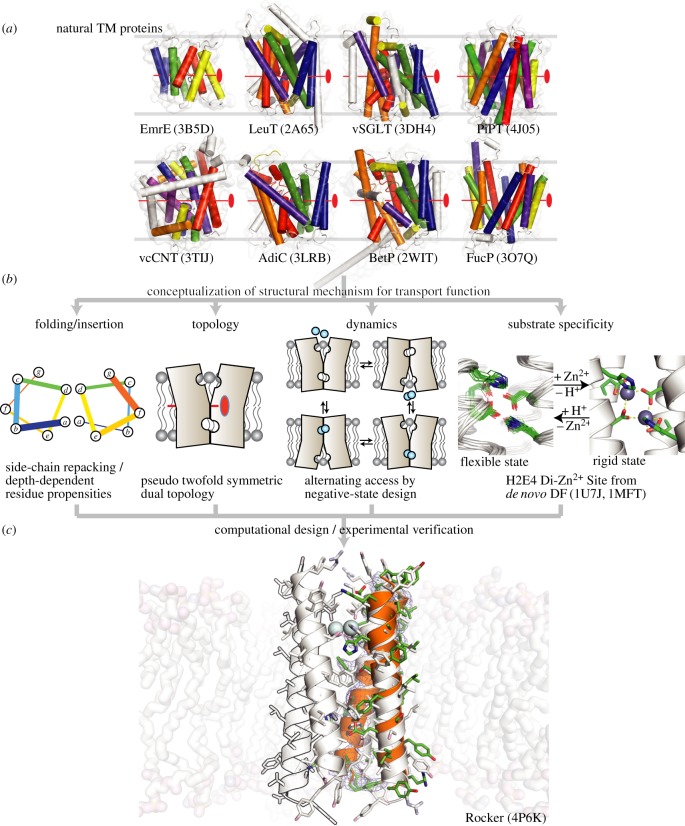
Figure 1.
Concepts used in the design of Rocker. Natural TM proteins shown in (a) [23–31] show pseudo twofold symmetry in their structures. (b) Principles used in the design of Rocker, including a di-metal binding site similar to that found in DF proteins [32] ((b), far right). (c) The designed structure of tetrameric Rocker ((c), white cartoon) superposed on the X-ray structure of the corresponding dimer in orange cartoon. PDB codes are in parentheses.
In this paper, we first review fundamental principles that can be used to design membrane proteins, with particular emphasis on the principles used in the design of Rocker. We then summarize recent experimental data on Rocker and present previously unpublished data relating to its proton flux and conformational dynamics.
2. Principles of membrane protein design
α-Helical TM proteins comprise 25–30% of all proteins in several genomes [33]. Unlike water, the chemical environment within the lipid bilayer, where these proteins reside, is highly heterogeneous [34,35], confounding both experiment [13] and design [14]. The residues projecting towards the membrane-inaccessible core of the protein must compete with abundant lipid hydrocarbons, so there is little hydrophobic driving force to stabilize the regions of proteins that span the most apolar region of the bilayer. The lipid-facing residues must adapt to the depth-dependent hydrophobicity of the surroundings [36]. To deconvolute this complexity in TM protein stability, Engelman and co-workers conceptualized folding as a processes in which individual TM helices are inserted [37–40] and assembled [41,42] in the two-stage folding paradigm [41]. We first consider features required for appropriate TM insertion, and then assembly into folded structures.
(a). Transmembrane helix sequence and topology
Identifying the location and membrane-insertion orientation of TM helices is the first challenge in the TM protein-folding problem. For large natural proteins, the TM helix could generally be identified by finding a stretch of approximately 20 mostly hydrophobic residues, which form a helix with an adequate length that could span across approximately 30 Å deep hydrocarbon core of the bilayer [43]. Polar group–containing large sidechains are overrepresented at either end of the helix, barring the flipping of the helix by anchoring the polar side-chain atoms at the termini on to the lipid–water interface, known as polar-atom snorkeling [44]. The overall topology could be predicted using the limited sequence-structure information found early on, such as the Positive-Inside rule [45] that describes the propensity for positively charged Lys or Arg residues to reside in the cytosolic domains.
Predicting the insertion orientations for individual TM helices has also become possible. Our analysis of growing numbers of high-resolution membrane protein structures revealed depth-dependent propensities for residues along the TM helix [36,40]. Distribution of many residues between the cytosolic- and extracellular-leaflet sides of helices is asymmetric. Lys and Arg maintain their strong propensity to reside at the cytosolic termini. Acidic residues were found slightly enriched also in the cytosolic termini. Cysteine is overrepresented at the extracellular termini where the environment is non-reducing to allow disulphides. By only using such sequence information, the correct topology could be determined for 80% of TM helices with known structure. The knowledge-based potential, Ez-3D, derived from these distributions [40], accurately recapitulated the protein orientations in OPM database [46].
(b). Transmembrane helix-interaction motifs
Analysis of the structural database has been extended to discover frequently occurring TM helix-interaction motifs [42,47]. Pairs of interacting helices from the structures of primarily multi-span helical proteins were identified and clustered based on geometric similarity. A total of 16 highly recurrent pairs were identified, and sequence profiles could be derived for the more frequently occurring helix interaction geometries. Interestingly, the clusters display a fold space similar to the most predominant water-soluble counterparts, with a slight bias towards tighter helical distance, owing to the higher tendency for small hydrophobic residues to be selected for inter-helical packing in TM compared with water-soluble proteins.
Feng & Barth [48] recently found that TM proteins could be conceptualized as assemblies of helical trimers, most of which could be clustered into six discrete motifs. The motif-associated residues are predictors of the trimer unit geometry. According to their computation, these residues make up rigid regions within proteins, and provide a relatively large contribution to protein stability when compared with the non-motif residues. These results are consistent with the experimental finding that TM proteins can adopt conformational dynamics for function while maintaining architectural stability consolidated via a strategically localized folding core with well-packed residues, often including polar interactions [49].
Helical packing motifs and their corresponding sequence profiles can be used both in membrane protein structure prediction as well as design. For example, we have developed a method dubbed CHAMP (Computed Helical Anti Membrane Protein) to design peptides that bind TM helical targets. The computational procedure starts with the target TM helix sequence, using position-dependent amino acid propensities of common TM interaction motifs to predict a preferred geometry for interaction with a second helix. Next, the sequence of the CHAMP peptide is designed using side-chain repacking algorithms to interact with the target in the predicted structural motif. Using this method, we have designed CHAMP peptides that bind the integrin αIIb TM domain [50,51].
(c). Experimental evaluation of helix–helix interaction in transmembrane
A motif first found in glycophorin A (GpA) has been the subject of numerous early and ongoing investigations [35,39,52–55]. The TM domain of this protein forms a tightly associating parallel dimer featuring a GxxxG motif. The small Gly residues form a surface that allows close interaction of the backbone of the helices within a dimer when they pack with a right-handed crossing angle. In related motifs, the Gly residues can be substituted for other small Ala or Ser residues. GxxxG is overrepresented in interacting TM helices, and β-branched residues sometimes reside in the neighbouring positions [56]. The presence of flanking β-branched residues is reminiscent of the packing motif originally found in GpA [53] and, more recently, in integrin αIIbβ3 TM domain complex [57]. Small residues, namely, Gly, Ala or Ser (GAS), occupy the corresponding positions for Gly in GxxxG or Gly-Zipper in the structures for TM helix dimers of several members of receptor tyrosine kinase (RTK) family [58–64]. The tight packing of GAS in TM results in a higher level of side-chain and main-chain burial compared with water-soluble domains, highlighting the importance of tight packing and weakly polar interactions in the absence of a strong hydrophobic driving force for folding [65,66]. Recent calculation shows that parallel dimerization involving GxxxG-like motifs optimizes the network and geometry of interaction between Cα-hydrogen and carbonyl (Cα-H···O=C, or Cα H-bond) [59], suggesting Cα H-bond as a driving force for TM helix interaction. GxxxG alone is not a sufficient predictor for helix interaction, however, as the sequence context and membrane properties strongly modulate the interaction propensity [67].
Small residues are also important for antiparallel interactions between TM helices. In one particularly frequently occurring class of tightly interacting antiparallel TM helices, the small amino acids are spaced every seven residues in a repeating pattern of the sequence. When the small residue is a Ser, the motif is designated a serine zipper [68], and synthetic peptides incorporating this motif have a high propensity to form antiparallel dimers [69]. In other work, a series of TM peptides were synthesized with small residues at each ‘a’ and Leu at each ‘d’ position of the TM domain [70] (illustration on the left panel in figure 1b). As the ‘a’ position changed from Val to Ala to Gly, the helices had an increased propensity to form antiparallel heterodimers. Natural proteins also show a high propensity for small GAS residues at position ‘a’ in antiparallel TM helix interaction [42].
Hydrogen bonds between polar sidechains can help stabilize the folds of membrane proteins [35,71]. The energetic contributions to the protein stability by polar side-chain interactions vary widely [72,73]. Ser is the most frequently occurring polar residue in TM regions [56], yet their contribution in Ser–zippers of designed TM helix dimerization was no larger than the VDW packing by Ala substituted at their positions [69]. Contrastingly, single Asn at the hydrocarbon core drove dimeric and trimeric self-assemblies of GCN4-based model peptides, including MS1 [74–76].
3. Design of proton channel peptides
De novo protein design has been extensively used to examine the features required for efficient protein conduction. For example, in early studies [77–85], we designed a series of minimalist peptides intended to assemble into TM bundles that mimic the conducting paths of ion channel proteins, for which structures were not yet available. One peptide with the sequence (LSLLLSL)3 self-assembled in bilayers to form very proton-selective tetramers. Although high-resolution structures were not solved at the time, spectroscopic and electrophysiological experiments strongly supported a tetrameric arrangement with the Ser residues lining a pore that was impermeable to substances other than water or protons. A more polar peptide with one additional Ser per heptad, (LSSLLSL)3, also formed highly proton-selective channels that were assigned to a tetrameric association state. However, a second higher-conductance state was also observed. A variety of studies indicated that it corresponded to a hexameric channel that was mildly selective for cations including Li+, Na+, K+, Cs+ and guanidinium, with conductance and selectivity very similar to that seen for the acetylcholine receptor channel. In the recent molecular dynamics (MD) simulations of (LSLLLSL)3 and (LSLLLSL)3 in phospholipid bilayers, these peptides remained stable in hexameric conformations, although the strict rotational symmetry of the bundles was distorted in individual snapshots along the trajectory [86].
4. Design of Rocker, a Zn2+ proton antiporter
More recently, we addressed the more challenging goal of designing a Zn2+ proton antiporter that transports transition metal ions using a dynamic mechanism, inspired by natural transporters and facilitators (figure 1). Various complex mechanisms for proton-dependent transport [87] share a simple underlying principle: global structural change interconverts the substrate affinity and accessibility across the bilayer during transport, alternating between binding of the substrate at one side of the membrane and releasing to the other side. Rigid/inflexible symmetric structures would disallow rocking between states, whereas the frustrated symmetry found in natural transporters appears better fit to facilitate the necessary structural changes. Rocker is an abstract version of complex natural transporters that use frustrated symmetry [88] to enable global rocking.
(a). Design of the backbone conformation and metal-binding site of Rocker
To test the importance of frustrated symmetry, we chose a very simple antiparallel four-helix bundle topology formed by tetramerization of a single TM helix. The highest symmetry achievable for such an antiparallel tetramer is D2, a dihedral symmetry group with a twofold rotational axis running down the axis of the bundle as well as two additional twofold rotational axes orthogonal to the central twofold axis (figure 2a). The axial twofold rotational symmetry axis relates each of the diagonally disposed helices to one another, while the other two orthogonal axes relate neighbouring helices. Thus, each helix is geometrically equivalent in a four-helix bundle with D2 symmetry (figure 2a). The next lower-symmetry group would be C2, which has only a single twofold symmetry axis (figure 2b). A C2 four-helix bundle has two geometrically non-equivalent helices; if the sequence of each helix is identical, the overall bundle will have two energetically equivalent conformations that may facilitate ion translocation (figure 2b).
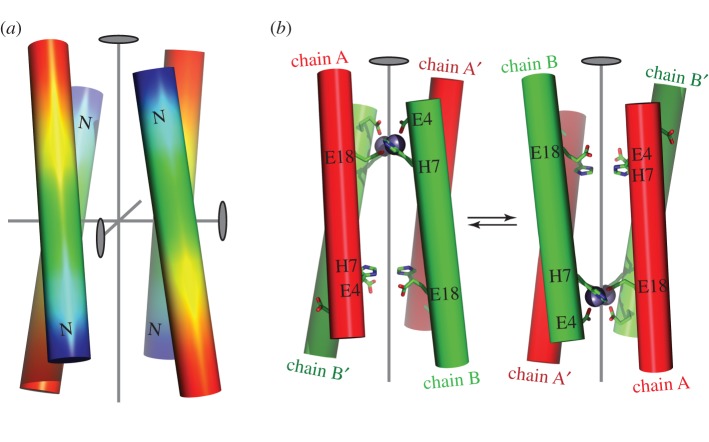
Figure 2.
Design of principles used in the construction of the fold for Rocker. The highest symmetry for an antiparallel four-helix bundle is D2, as shown in (a). The positions of the three mutually orthogonal twofold symmetry axes are shown as ovals, and the N-termini are designated by the letter N. Glu and His residues were positioned at three locations of the helix (E4, H7 and E18) such that they would form two di-metal–binding sites in an antiparallel four-helix bundle. (b) Binding of metal ion to only one of the sites decreases the symmetry to C2, making two of the helices non-equivalent (red versus green shading). The resulting structure has two energetically degenerate conformations, which should be in dynamic equilibrium. A major goal in designing the sequence of Rocker was to introduce negative cooperativity, such that they would prefer the C2 conformation.
We chose to design a helical bundle with two metal-binding sites per four-helix bundle. In earlier work, we had designed series of water-soluble proteins (designated DF for Due Ferro) that bind di-Fe(II/III) and di-Zn2+ ions in a site comprised of 2 His and 4 Glu sidechains (H2E4 site) [89]. An attractive feature of the DF proteins is that the Glu sidechains on the interior of the protein have very high pKa values, such that they release protons when metal ions bind to the site. Thus, protonation and metal-binding are thermodynamically linked, providing a basis for driving transport of protons using a Zn2+ gradient, or vice versa.
Rocker contains two dimetal-binding sites per a four-helix bundle. As in DF, the site is comprised of two His and four Glu residues, although the arrangement of these sidechains was redesigned in Rocker to support its different symmetry (see below). To avoid falling into a deep energy well with metal ions bound in both H2E4 sites in a fully symmetrical protein, we aimed to stabilize the C2-symmetric state relative to the D2-symmetric state. In this way, binding of di-Zn2+ to one site would substantially decrease the affinity of the second site (i.e. the sites would exhibit negative cooperativity). Thus, we were presented with the challenge of designing a single TM helix that would prefer to form a C2 rather than D2-symmetrical tetramer.
As discussed below, we used the principle of computational negative design to stabilize the C2 over the D2 structure [90]. This requires the generation of backbone coordinates for the C2 and D2 bundles, which was accomplished using simple parametric models for four-helix bundles [91,92]. The natural topology for modelling the D2-symmetric state was as the antiparallel four-helix coiled coil—a motif in which helices gently wind around a central axis in a super-helical manner, creating D2 symmetry [91,92]. In this undesired state, two equivalent H2E4 sites would be formed by the coalescence of Glu4 and His7 in one pair of opposing chains with Glu18 from the remaining pair oriented anti-parallel to the first (figure 2). Each H2E4 site was designed to bind two Zn2+ ions, with two bridging Glu sidechains and two chelating sidechains. The two His sidechains are oriented trans to one another, whereas they are oriented in a cis geometry in the DF protein.
The C2-symmetric structure was designed by straightening the curved helices at one of the two metal sites into idealized straight helices (figure 2). While the H2E4 site where the helices were straightened retained the metal-binding geometry, the second H2E4 at the other end of the bundle was distorted and incapable of metal binding. Because the two ends of the structure are different, the bundle is no longer fully symmetric. If the sequence preferentially stabilizes this C2-symmetric state, the protein should be able to rock between the two off-symmetric conformations.
(b). Design of the sequence of Rocker
The specific backbone configurations to represent the above D2- and C2-symmetric topologies were chosen by meeting two constraints: (i) favourable metal coordination geometry at H2E4 sites, and (ii) high designability of the two different helix–helix interfaces in the bundle. The latter was aimed at assuring, prior to the beginning of sequence design, that the defined backbone configurations can, in fact, be stabilized with natural amino acids and thus represent relevant states. The constraint was met by ensuring that interface fragments from respective structures represented geometries richly populated in the structural database [18,93,94].
Having defined the representative structural templates, the overall design procedure was split into two stages. In the first stage, we sought to find a set of sequences predicted to insert into the membrane and associate into an anti-parallel four-helix bundle with one H2E4 site organized at each end. The calculations in this stage used the EZ membrane potential to model membrane solubility [36,40] and standard molecular mechanics with the rotamer approximation to ensure favourable packing in the bundle [18]. Thus, the goal of this stage was to provide general stability to the membrane-inserted four-helix–bundle topology.
The second stage aimed to encode specificity towards the C2-symmetric state relative to the D2 alternative, using negative design. Towards this end, metal-binding constraints were used to define thermodynamic ensembles around the representative structure of each state—i.e. the C2-symmetric ensemble was defined by constraining for metal coordination at one end of the bundle, but no such possibility at the other end. Similarly, the D2-symmetric ensemble was defined by requiring that both ends support metal coordination. These ensembles were then used in conjunction with MD-based free-energy calculations [95] to screen sequences emergent form the first stage for those with a preference towards C2 symmetry [18]. We found that due to the homo-tetrameric nature of the bundle, the higher D2 symmetry was generally preferred with C2-preferring sequences rare. However, a recurring theme among a handful of sequences that did exhibit C2 preference was two very different helix–helix interfaces: a tight/narrow interface lined with Ala residues that remained stable in MD, and a looser/wider interface rich in Phe residues that was much more dynamic. Rocker was chosen as the sequence with the strongest C2 preference among those that exhibited sufficient room in the core to support the passage of an ion.
(c). Comparison of the design of Rocker with natural transporters
Our strategy borrows from the alternating-access theory in the adoption of frustrated symmetry as a design principle. However, there are a number of differences between the expected mechanism for Rocker and that of native alternating-access transporters. The great majority of natural proteins that conduct via alternating access have a single ligand-binding site near the centre of the bilayer, while we used two sites in our design. Also, most transporters have two tightly packed ‘gaskets’ that alternately open and close to maintain a tightly sealed structure. The hydrophobic packing near the ends of the bundle of Rocker was less extensive than in natural proteins, which may lead to weaker coupling between the transport of protons and metal ions than in native counterparts.
5. Characterization of Rocker
(a). Structural studies
As expected from design, Rocker forms a four-helix bundle in micelles and bilayers [18]. Sedimentation equilibrium of the peptide in density-matched detergent micelles indicated that Rocker formed a tight dimer, which was in equilibrium with a tetramer. The tetrameric species was strongly stabilized in the presence of Zn2+, and NMR studies confirmed that Zn2+ bound directly to the protein ligands. NMR titrations of Zn2+ into micelle-bound Rocker showed metal-binding stoichiometry of two Zn2+ per tetramer, indicating the saturation of only one of the two potential Zn2+ sites. Only at very high Zn2+ was the second site bound, confirming that the designed negative cooperativity between the sites had been achieved. We have been unable to crystallize the protein in the tetrameric state, possibly because the tetramer is excessively dynamic (by design). Nevertheless, the structure of the dimeric form, determined by X-ray crystallography, showed the two helices interacting in a tight Ala-coil interaction, as anticipated by design.
Using solid-state NMR, we also probed the structure in phospholipid membranes, which are a more favourable environment for assembly when compared with micelles [69,96,97]. These experiments confirmed that Rocker formed tetramers in bilayers, and that spectral shifts upon addition of Zn2+ were consistent with binding. Distance measurements were also consistent with the designed structure. In particular, inter-residue distances within the DMPC-bound apo Rocker monitored by 19F CODEX of para 19F-Phe at position 19 using solid-state NMR verified the wide interface formed by Phenylalanine residues at positions 10 and 14 (F10 and F14).
(b). Functional measurements
When reconstituted in large unilamellar lipid vesicles, Rocker demonstrated a substrate-specific antiporter-like function [18]. Rocker moved first-row transition metal ions, Zn2+ and Co2+, across the bilayer, but not the hard divalent-metal cation, Ca2+. These findings are consistent with the ligand environment of designed H2E4 sites [89,98–101], which approximately follows the Irving Williams series [102] for metal-binding affinity. Substituting Glu4 and Glu18 in H2E4 with Gln eliminated the Zn2+ transport activity, corroborating the intended mode of binding and transport. Rocker harnessed the concentration gradient of Zn2+ to antiport H+ against the H+-concentration gradient, and vice versa, displaying active co-transport function.
Rocker is not as efficient as natural transporters in terms of the rate of Zn2+ transport or efficient use of a proton gradient to pump Zn2+. While the KM value of 280 ± 90 µM in Michaelis–Menten kinetics for Zn2+ transport by Rocker was comparable to the KM of 105 µM Zn2+ for the natural zinc-transporter ZitB [103,104], the maximum velocity (Vmax) was 1 Zn2+ min−1 per tetramer in the designed transporter, approximately 100 times slower compared with the Vmax of 142 Zn2+ min−1 per protein for ZitB. The H+ : Zn2+ antiport molar ratio of 4 : 1 when Zn2+ drives Rocker to antiport H+ changes to 240 : 1 when H+ drives Rocker to antiport Zn2+ into the vesicle. In our previous work, we hypothesized that the relatively inefficient coupling between protons and Zn2+ resulted from Rocker's lack of a hydrophobic gasket that might prevent non-coupled leaky flux of protons.
To assess this possibility, here we describe the pH-dependence of proton conduction. Rocker was reconstituted in phospholipid vesicles, as in our earlier work. The initial pH of the vesicle interior was held at 6.8, while the outer pH was varied from 4.5 to 8. In the absence of Zn2+, Rocker did not conduct H+ (figure 3). Thus, Rocker must be primed with Zn2+ for H+ conduction to occur. In the presence of 1 mM Zn2+, the proton conduction curve is well described by saturable Michaelis–Menten kinetics, with a maximum flux of 141 ± 8 H+ s−1 per tetramer. The Michaelis–Menten kinetics are consistent with a transporter-like mechanism in which protons are specifically bound in the channel, and the levelling of the rate at high proton concentration (low pH) indicates that the protein does not act as a simple non-selective pore. For a simple pore, one would expect the rate to rise linearly over this range of proton concentration [77,80–82,105,106]. For transporters in which protons are bound to ionizable sidechains and then released during a conductance cycle, the rate saturates due to rate-determining proton dissociation and/or a slow conformational change. Indeed, the rate of shuttling for Rocker is close to the values of 50–200 proton s−1 per tetramer, estimated for the TM four-helix bundle of a bona fide H+-channel, M2, from influenza A virus [107].
Figure 3.
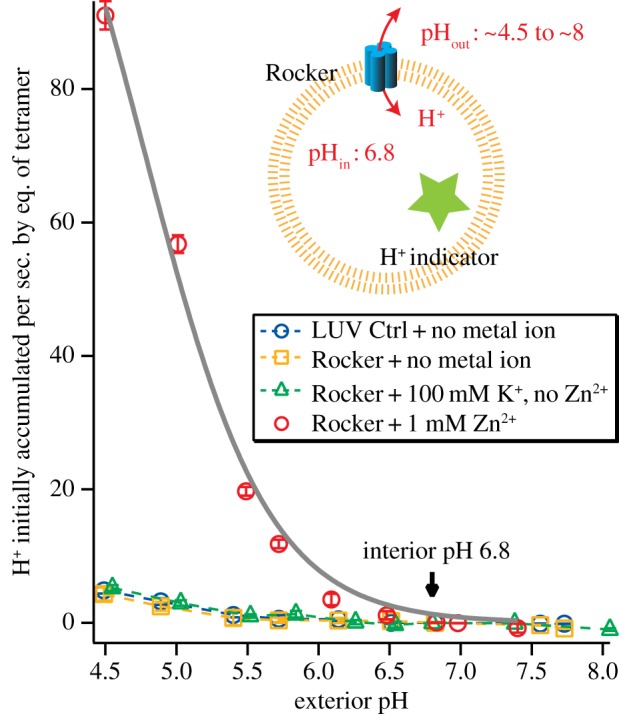
Zn2+ is required for proton conduction mediated by Rocker. The initial rates of H+-flux into large unilamellar vesicles at an interior pH of 6.8 are plotted as a function of the exterior pH. LUV reconstituted with Rocker and preloaded with HPTS H+-indicator was monitored via stop-flow fluorescence flux assay. Fitted curve and traces are shown as solid and dashed lines, respectively.
In summary, we find protons diffuse through Rocker at a rate that is consistent with a binding–unbinding mechanism that involves protonation of His or Glu residues along the conductance path. Significantly, proton flux through Rocker requires Zn2+, which has been shown to stabilize the tetramer. Because Zn2+ is not conducted at as high a rate as protons, we speculate that the protons can diffuse past the Zn2+ ions, probably by transiently protonating a Glu or His ligand in the pore.
(c). Conformational dynamics of Rocker
The conformational dynamics of Rocker in phospholipid bilayers have been probed using solid-state NMR [18], using peptide labelled with 13C at four positions (A8, I15, I19 and A22). In DMPC bilayers, the one-dimensional 13C peak positions were found to be invariant over a wide range of temperature from −40 to 40°C in CP-MAS of both apo and Zn2+-bound Rocker in DMPC, indicating a stable bundle. The dynamics in both the absence and presence of Zn2+ increased with temperature, indicated by peak broadening. Zn2+ binding decreased the rigidity of the structure across the A8 and L19 at the interface, indicated by weaker 2D A8–L19 13C–13C correlation cross peak upon adding Zn2+. Dephasing of 13C–1H dipolar coupling in both apo and Zn2+-bound Rocker was also indicative of dynamics on a microsecond time-scale.
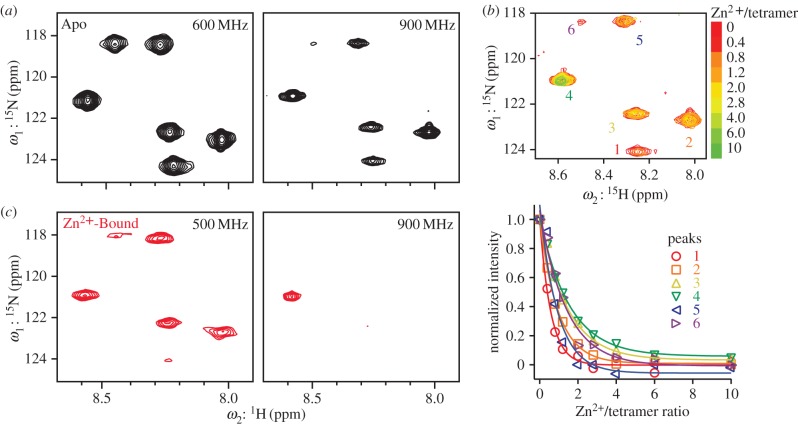
Here, we describe previously unpublished results of solution NMR experiments on Rocker labelled with 15N at six positions (A8, A12, L13, F14, A15 and L16) in isotropic bicelles. Tetramerization detected in DPC detergent by AUC is expected to be more predominant in a bicelle that better mimics the bilayer than micelles do. Access to multiple different NMR field strengths allows interrogation of dynamics on the microsecond-to-millisecond time-scale. The intensities of the peaks in the 15N HSQC spectra of apo Rocker decrease when the magnetic field is changed from 600 to 900 MHz (figure 4a). One peak, in particular, is almost completely absent at 900 MHz. The near-complete loss of this signal without the appearance of a new resonance or additional broadening is indicative of conformational exchange with a state that is not visible by NMR.
Figure 4.
Backbone conformational dynamics of Rocker in bicelles probed by NMR. (a) 15N HSQC spectra for apo Rocker in bicelles at magnetic-field strength of 600 and 900 MHz. (b) Superposition of HSQC spectra from 15N-labelled Rocker in bicelles in increasing Zn2+ (upper panel) at 900 MHz. The lower panel shows the peak intensities plotted as a function of increasing Zn2+ for the six protons. (c) In the presence of four equivalences of Zn2+ per a tetramer, peaks labelled 1 and 6 in (b) decrease in intensity to almost below the level of detection at 500 MHz, and all but one peak is lost from the spectrum at 900 MHz.
Significant field-dependent changes in intensity are also seen in response to the addition of Zn2+. Titration of Zn2+ led to pronounced decreases in intensity of the peaks without affecting the peak width (figure 4b). Surprisingly, different titration curves are seen for each labelled amide. Some peaks decrease in intensity by more than a factor of two before the sample is half-saturated, suggesting that Zn2+ can exchange between binding sites of the same or different bundles, giving rise to exchange processes that specifically affect some of the proton's resonances. Other peaks show behaviour similar to that previously reported in micelles, in which a saturable binding curve is observed with a limiting stoichiometry of two Zn2+ per tetramer. Field effects are again observed, with much a greater loss in peak intensity seen at 900 MHz when compared with 500 MHz.
A full understanding of this complex behaviour would require deeper investigation. Clearly, there are numerous phenomena leading to peak intensity decrease, ranging from conformational dynamics that can be seen in the apo and Zn2+-bound forms of the protein to metal binding/unbinding kinetics within or between tetramers that are seen at intermediate metal ion concentrations. However, the observation of field-dependent effects strongly suggests that the protein exists in an equilibrium between a state with well-resolved spectra and one with dynamic properties that preclude direct detection. The field dependence most probably indicates that the chemical shifts of specific amides differ between the two states. Assuming a typical proton chemical shift of 1 ppm, rates on the order of high microsecond to low millisecond are calculated. This is similar to the rate of proton exchange seen in the proton channel measurement. It is also interesting to note that the dynamics on this time-scale increase in the presence of zinc ions at both intermediate as well as saturating Zn2+ concentrations. These findings provide support for a dynamic rocking mechanism of proton conduction.
6. Discussion
Structural biophysics of TM proteins has steadily grown since the first TM protein crystal structure was published in 1985 [108], 3 decades after the first X-ray structure of a water-soluble protein was determined at atomic resolution [109]. Similarly, there has been a large temporal gap in our understanding of the principles of membrane protein structure, and in testing this understanding by designing membrane proteins entirely from scratch. The design of Rocker demonstrated a significant milestone in de novo design. It represents the first example of the successful design of a membrane protein with a predetermined structure, and, for the first time, demonstrated the design of an energetic landscape to support a dynamic functional mechanism. Clearly, the de novo design of complex architectures with predefined functional dynamics is in the realm of possibility. The future of membrane protein design is limited only by the creativity and audacity of future young scientists entering the field—and their ability to fire the imagination of funding agencies that support their work.
7. Experimental methods
(a). Materials
Peptides were synthesized as C-terminal amides using Fmoc solid-phase chemistry and purified as trifluoroacetate salts by HPLC using a C4 reverse-phase prep column. Lyophilized peptides dissolved in ethanol or isopropanol stock were kept at less than −60°C for reconstitution in experiments.
Reagents used in experiments include POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine), POPG (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)), DMPC (1,2-dimyritoyl-sn-glycero-3-phosphocholine, cholesterol, DPC (n-dodecylphosphocholine), deuterated DPC, DHPC (1,2-dihexanoyl-sn-glycero-3-phosphocholine) (Avanti Polar Lipids); OG (n-Octyl-β-d-Glucopyranoside (Affymetrix); K+ ionophore valinomycin, protonophore CCCP (carbonyl cyanide m-chlorophenyl hydrazone); HPTS fluorescent dye; vesicle-impermeable control cation N-methyl d-glucamonium and fluorescent quencher DPX (Invitrogen).
(b). Liposome flux assay of Rocker
Ion flux was tested in lipid vesicles reconstituted with pre-determined number of Rocker bundles (see electronic supplementary material, Methods). The vesicles, reconstituted in interior buffers, and washed in exterior buffers by size exclusion, are mixed with assay buffers in stopped-flow to initiate ion flux. Upon initiating the flux, the change in the intravesicular ion concentration is monitored by detecting the fluorescence of preloaded membrane-impermeable dye. Initial rates of ion flux were evaluated prior to build-up of an electrical potential. The pH of all buffers (interior, exterior and assay buffers) was adjusted using 5 mM MOPSO; and the ionic strength and osmolarity, using 100 mM K2SO4, or 80 mM vesicle-impermeable non-metal control salt, N-methyl d-glucamonium sulphate. Zinc sulphate at a concentration of 0 or 1 mM was included to test the effect of Zn2+ binding on proton conduction by Rocker.
Each interior buffer consisted 500 µM HPTS to be encapsulated in vesicles. The exterior buffer was identical to the respective interior buffer, except the HPTS was not included in the exterior buffer. The respective assay buffer is identical to the corresponding exterior buffer, except the assay buffer varied the pH from 4.5 to 8 to allow the generation of proton concentration gradient across the bilayer. To generate a standard curve for encapsulated HPTS, LUV standards were prepared. Aliquots of LUVs preloaded with HPTS and potassium were incubated with equal volumes of assay buffers containing 800 nM CCCP and 800 nM valinomycin with varying pHs for 30 min to allow the interior and exterior pHs to equilibrate. The emission spectra of each sample were determined at above two excitation wavelengths by using the TEACAN infinite M1000 plate reader; and the pH values, by the Thermo Orion pH meter equipped with a Pinnacle semi-micro pH electrode. To generate HPTS/pH calibration, the ratio of integrals of emission spectra at the two excitations (F455/F415) was plotted against the corresponding pH values. The best-fitting curve is described by 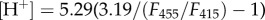 obtained by nonlinear curve fitting to describe the plots obtained for the HPTS/pH calibration (electronic supplementary material, figure S1).
obtained by nonlinear curve fitting to describe the plots obtained for the HPTS/pH calibration (electronic supplementary material, figure S1).
The KinetAsyst Stopped-Flow system was used to monitor H+ flux at 18°C. Equal volumes of the proteoliposome sample and assay buffer are loaded into two separate mixing syringes, and ion flux is initiated by instantaneously injecting 75 µl of each reactant through the mixing cell simultaneously. For monitoring proton, vesicles preloaded with HPTS were excited at 455 and 415 nm, and emissions at the wavelength greater than 500 nm were recorded using a 500 nm long-wave pass filter. The ratio of emission intensities at two excitation wavelengths (F455/F416) decreases with decreasing pH. The initial interior pH calculated from the unprocessed stopped-flow data deviated from the known interior pH and approached the exterior pH, suggesting the presence of HPTS on the vesicle exterior probably due to incomplete washing of the vesicles by spin size exclusion. To gauge the proton flux, the measured F455/F416 data were corrected to account for any unwashed HPTS (see electronic supplementary material, Methods).
(c). Solutions nuclear magnetic resonance
The Rocker peptide that contained 15N-labelled amino acids at the positions Ala8, Ala12, Leu13, Phe14, Ala15 and Leu16 was synthesized by Fmoc solid-phase chemistry, and purified in a trifluoroacetate salt form by reverse-phase HPLC.
For two-dimensional 15N-1H HSQC spectra of 15N-labelled Rocker, the peptide at a final concentration of 1 mM in isotropic bicelle solution comprising 50 mM DMPC, 150 mM DHPC, 50 mM KCl, 10 mM HEPES pH 7, 10% D2O with varying concentration of ZnSO4 was monitored in 500, 600 or 900 MHz NMR spectrometer at 45°C. The spectra were recorded with the standard bruker pulse sequence trosyetf3gpsi with t2,max = 64–95 ms, t1,max = 27–69 ms, 16–32 scans. 1H chemical shift was referenced with respect to residual water peak at 4.60 ppm and 15N chemical shift was referenced indirectly via gyromagnetic ratios. The two-dimensional spectra were processed and analysed using the program nmrPipe [110]. Time domain data of the 15N indirect dimension were doubled by linear prediction. Time domain data were multiplied by sine square bell window functions shifted by 90o and zero-filled once before Fourier transformation.
Supplementary Material
Authors' contributions
N.H.J., G.G., Y.W. and W.F.D. designed the experiments, interpreted the data and wrote the manuscript. G.G. conducted computational design. N.H.J. and Y.W. performed the experiments.
Competing interests
We declare we have no competing interests.
Funding
This work was funded by NIH F32 GM096727 to N.H.J. W.F.D. acknowledges support from NIH R01 GM054616 and NSF grants nos CHE-141329 and DMR1120901.
References
- 1.Lupas AN. 2014. Protein design. What I cannot create, I do not understand. Science 346, 1455–1456. ( 10.1126/science.aaa2721) [DOI] [PubMed] [Google Scholar]
- 2.Kuhlman B, Dantas G, Ireton G, Varani G, Stoddard B, Baker D. 2003. Design of a novel globular protein fold with atomic-level accuracy. Science 302, 1364–1368. ( 10.1126/science.1089427) [DOI] [PubMed] [Google Scholar]
- 3.Jiang L, et al. 2008. De novo computational design of retro-aldol enzymes. Science 319, 1387–1391. ( 10.1126/science.1152692) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Röthlisberger D, et al. 2008. Kemp elimination catalysts by computational enzyme design. Nature 453, 190–195. ( 10.1038/nature06879) [DOI] [PubMed] [Google Scholar]
- 5.Robertson DE, et al. 1994. Design and synthesis of multi-haem proteins. Nature 368, 425–432. ( 10.1038/368425a0) [DOI] [PubMed] [Google Scholar]
- 6.Jardine J, et al. 2013. Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716. ( 10.1126/science.1234150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Correia BE, et al. 2014. Proof of principle for epitope-focused vaccine design. Nature 507, 201–206. ( 10.1038/nature12966) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grigoryan G, Kim YH, Acharya R, Axelrod K, Jain RM, Willis L, Drndic M, Kikkawa JM, DeGrado WF. 2011. Computational design of virus-like protein assemblies on carbon nanotube surfaces. Science 332, 1071–1076. ( 10.1126/science.1198841) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King NP, Bale JB, Sheffler W, McNamara DE, Gonen S, Gonen T, Yeates TO, Baker D. 2014. Accurate design of co-assembling multi-component protein nanomaterials. Nature 510, 103–108. ( 10.1038/nature13404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hsia Y, et al. 2016. Design of a hyperstable 60-subunit protein icosahedron. Nature 535, 136–139. ( 10.1038/nature18010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim KH, et al. 2016. Protein-directed self-assembly of a fullerene crystal. Nat. Commun. 7, 11429 ( 10.1038/ncomms11429) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mustata GM, Kim YH, Zhang J, DeGrado WF, Grigoryan G, Wanunu M. 2016. Graphene symmetry amplified by designed peptide self-assembly. Biophys. J. 110, 2507–2516. ( 10.1016/j.bpj.2016.04.037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong H, Joh NH, Bowie JU, Tamm LK. 2009. Methods for measuring the thermodynamic stability of membrane proteins. Methods Enzymol. 455, 213–236. ( 10.1016/S0076-6879(08)04208-0) [DOI] [PubMed] [Google Scholar]
- 14.Ghirlanda G. 2009. Design of membrane proteins: toward functional systems. Curr. Opin. Chem. Biol. 13, 643–651. ( 10.1016/j.cbpa.2009.09.017) [DOI] [PubMed] [Google Scholar]
- 15.Barth P, Senes A. 2016. Toward high-resolution computational design of the structure and function of helical membrane proteins. Nat. Struct. Mol. Biol. 23, 475–480. ( 10.1038/nsmb.3231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghirlanda G, Senes A. 2013. Membrane proteins: folding, association, and design, 246 p. New York, NY: Humana Press. [Google Scholar]
- 17.Korendovych IV, et al. 2010. De novo design and molecular assembly of a transmembrane diporphyrin-binding protein complex. J. Am. Chem. Soc. 132, 15 516–15 518. ( 10.1021/ja107487b) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joh NH, Wang T, Bhate MP, Acharya R, Wu Y, Grabe M, Hong M, Grigoryan G, DeGrado WF. 2014. De novo design of a transmembrane Zn2+-transporting four-helix bundle. Science 346, 1520–1524. ( 10.1126/science.1261172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell P. 1957. A general theory of membrane transport from studies of bacteria. Nature 180, 134–136. ( 10.1038/180134a0) [DOI] [PubMed] [Google Scholar]
- 20.Law CJ, Maloney PC, Wang DN. 2008. Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 62, 289–305. ( 10.1146/annurev.micro.61.080706.093329) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forrest LR, Krämer R, Ziegler C. 2011. The structural basis of secondary active transport mechanisms. Biochim. Biophys. Acta 1807, 167–188. ( 10.1016/j.bbabio.2010.10.014) [DOI] [PubMed] [Google Scholar]
- 22.Forrest LR. 2015. Structural symmetry in membrane proteins. Annu. Rev. Biophys. 44, 311–337. ( 10.1146/annurev-biophys-051013-023008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrison EA, DeKoster GT, Dutta S, Vafabakhsh R, Clarkson MW, Bahl A, Kern D, Ha T, Henzler-Wildman KA. 2012. Antiparallel EmrE exports drugs by exchanging between asymmetric structures. Nature 481, 45–50. ( 10.1038/nature10703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen YJ, Pornillos O, Lieu S, Ma C, Chen AP, Chang G. 2007. X-ray structure of EmrE supports dual topology model. Proc. Natl Acad. Sci. USA 104, 18 999–19 004. ( 10.1073/pnas.0709387104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. 2005. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature 437, 215–223. ( 10.1038/nature03978) [DOI] [PubMed] [Google Scholar]
- 26.Faham S, Watanabe A, Besserer GM, Cascio D, Specht A, Hirayama BA, Wright EM, Abramson J. 2008. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science 321, 810–814. ( 10.1126/science.1160406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedersen BP, et al. 2013. Crystal structure of a eukaryotic phosphate transporter. Nature 496, 533–536. ( 10.1038/nature12042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson ZL, Cheong CG, Lee SY. 2012. Crystal structure of a concentrative nucleoside transporter from Vibrio cholerae at 2.4 A. Nature 483, 489–493. ( 10.1038/nature10882) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao X, Lu F, Zhou L, Dang S, Sun L, Li X. 2009. Structure and mechanism of an amino acid antiporter. Science 324, 1565–1568. ( 10.1126/science.1173654) [DOI] [PubMed] [Google Scholar]
- 30.Ressl S, Terwisscha van Scheltinga AC, Vonrhein C, Ott V, Ziegler C. 2009. Molecular basis of transport and regulation in the Na+/betaine symporter BetP. Nature 458, 47–52. ( 10.1038/nature07819) [DOI] [PubMed] [Google Scholar]
- 31.Dang S, Sun L, Huang Y, Lu F, Liu Y, Gong H, Wang J, Yan N. 2010. Structure of a fucose transporter in an outward-open conformation. Nature 467, 734–738. ( 10.1038/nature09406) [DOI] [PubMed] [Google Scholar]
- 32.Pasternak A, Kaplan J, Lear JD, Degrado WF. 2001. Proton and metal ion-dependent assembly of a model diiron protein. Protein Sci. 10, 958–969. ( 10.1110/ps.52101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wallin E, von Heijne G. 1998. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 7, 1029–1038. ( 10.1002/pro.5560070420) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White S, Wimley W. 1999. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 28, 319–365. ( 10.1146/annurev.biophys.28.1.319) [DOI] [PubMed] [Google Scholar]
- 35.Mackenzie K, Fleming K. 2008. Association energetics of membrane spanning alpha-helices. Curr. Opin. Struct. Biol. 18, 412–419. ( 10.1016/j.sbi.2008.04.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Senes A, Chadi DC, Law PB, Walters RF, Nanda V, Degrado WF. 2007. E(z), a depth-dependent potential for assessing the energies of insertion of amino acid side-chains into membranes: derivation and applications to determining the orientation of transmembrane and interfacial helices. J. Mol. Biol. 366, 436–448. ( 10.1016/j.jmb.2006.09.020) [DOI] [PubMed] [Google Scholar]
- 37.Hessa T, White S, von Heijne G. 2005. Membrane insertion of a potassium-channel voltage sensor. Science 307, 1427 ( 10.1126/science.1109176) [DOI] [PubMed] [Google Scholar]
- 38.Hessa T, Meindl-Beinker N, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G. 2007. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450, 1026–1030. ( 10.1038/nature06387) [DOI] [PubMed] [Google Scholar]
- 39.Senes A, Engel D, DeGrado W. 2004. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr. Opin. Struct. Biol. 14, 465–479. ( 10.1016/j.sbi.2004.07.007) [DOI] [PubMed] [Google Scholar]
- 40.Schramm CA, Hannigan BT, Donald JE, Keasar C, Saven JG, Degrado WF. 2012. Knowledge-based potential for positioning membrane-associated structures and assessing residue-specific energetic contributions. Structure 20, 924–935. ( 10.1016/j.str.2012.03.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Popot J, Engelman D. 1990. Membrane protein folding and oligomerization: the two-stage model. Biochemistry 29, 4031–4037. ( 10.1021/bi00469a001) [DOI] [PubMed] [Google Scholar]
- 42.Zhang SQ, Kulp DW, Schramm CA, Mravic M, Samish I, DeGrado WF. 2015. The membrane- and soluble-protein helix–helix interactome: similar geometry via different interactions. Structure 23, 527–541. ( 10.1016/j.str.2015.01.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engelman DM, Steitz TA, Goldman A. 1986. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu. Rev. Biophys. Biophys. Chem. 15, 321–353. ( 10.1146/annurev.bb.15.060186.001541) [DOI] [PubMed] [Google Scholar]
- 44.Chamberlain AK, Lee Y, Kim S, Bowie JU. 2004. Snorkeling preferences foster an amino acid composition bias in transmembrane helices. J. Mol. Biol. 339, 471–479. ( 10.1016/j.jmb.2004.03.072) [DOI] [PubMed] [Google Scholar]
- 45.von Heijne G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225, 487–494. ( 10.1016/0022-2836(92)90934-C) [DOI] [PubMed] [Google Scholar]
- 46.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. 2012. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 40(Database issue), D370–D376. ( 10.1093/nar/gkr703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walters R, DeGrado W. 2006. Helix-packing motifs in membrane proteins. Proc. Natl Acad. Sci. USA 103, 13 658–13 663. ( 10.1073/pnas.0605878103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng X, Barth P. 2016. A topological and conformational stability alphabet for multipass membrane proteins. Nat. Chem. Biol. 12, 167–173. ( 10.1038/nchembio.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baker RP, Urban S. 2012. Architectural and thermodynamic principles underlying intramembrane protease function. Nat. Chem. Biol. 8, 759–768. ( 10.1038/nchembio.1021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yin H, et al. 2007. Computational design of peptides that target transmembrane helices. Science 315, 1817–1822. ( 10.1126/science.1136782) [DOI] [PubMed] [Google Scholar]
- 51.Fong KP, Zhu H, Span LM, Moore DT, Yoon K, Tamura R, Yin H, DeGrado WF, Bennett JS. 2016. Directly activating αllbβ3 initiates outside-in signaling by causing αllbβ3 clustering. J. Biol. Chem. 291, 11 706–11 716. ( 10.1074/jbc.M116.716613) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lemmon MA, Treutlein HR, Adams PD, Brunger AT, Engelman DM. 1994. A dimerization motif for transmembrane alpha-helices. Nat. Struct. Biol. 1, 157–163. ( 10.1038/nsb0394-157) [DOI] [PubMed] [Google Scholar]
- 53.MacKenzie K, Prestegard J, Engelman D. 1997. A transmembrane helix dimer: structure and implications. Science 276, 131–133. ( 10.1126/science.276.5309.131) [DOI] [PubMed] [Google Scholar]
- 54.Russ WP, Engelman DM. 2000. The GxxxG motif: a framework for transmembrane helix–helix association. J. Mol. Biol. 296, 911–919. ( 10.1006/jmbi.1999.3489) [DOI] [PubMed] [Google Scholar]
- 55.Trenker R, Call ME, Call MJ. 2015. Crystal structure of the glycophorin A transmembrane dimer in lipidic cubic phase. J. Am. Chem. Soc. 137, 15 676–15 679. ( 10.1021/jacs.5b11354) [DOI] [PubMed] [Google Scholar]
- 56.Senes A, Gerstein M, Engelman D. 2000. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J. Mol. Biol. 296, 921–936. ( 10.1006/jmbi.1999.3488) [DOI] [PubMed] [Google Scholar]
- 57.Anthis NJ, et al. 2009. The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J. 28, 3623–3632. ( 10.1038/emboj.2009.287) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sarabipour S, Hristova K. 2016. Mechanism of FGF receptor dimerization and activation. Nat. Commun. 7, 10262 ( 10.1038/ncomms10262) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mueller BK, Subramaniam S, Senes A. 2014. A frequent, GxxxG-mediated, transmembrane association motif is optimized for the formation of interhelical Cα-H hydrogen bonds. Proc. Natl Acad. Sci. USA 111, E888–E895. ( 10.1073/pnas.1319944111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arkhipov A, Shan Y, Das R, Endres NF, Eastwood MP, Wemmer DE., Wemmer DE, Kuriyan J, Shaw DE. 2013. Architecture and membrane interactions of the EGF receptor. Cell 152, 557–569. ( 10.1016/j.cell.2012.12.030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mineev KS, Bocharov EV, Pustovalova YE, Bocharova OV, Chupin VV, Arseniev AS. 2010. Spatial structure of the transmembrane domain heterodimer of ErbB1 and ErbB2 receptor tyrosine kinases. J. Mol. Biol. 400, 231–243. ( 10.1016/j.jmb.2010.05.016) [DOI] [PubMed] [Google Scholar]
- 62.Escher C, Cymer F, Schneider D. 2009. Two GxxxG-like motifs facilitate promiscuous interactions of the human ErbB transmembrane domains. J. Mol. Biol. 389, 10–16. ( 10.1016/j.jmb.2009.04.002) [DOI] [PubMed] [Google Scholar]
- 63.Bocharov EV, et al. 2008. Spatial structure of the dimeric transmembrane domain of the growth factor receptor ErbB2 presumably corresponding to the receptor active state. J. Biol. Chem. 283, 6950–6956. ( 10.1074/jbc.M709202200) [DOI] [PubMed] [Google Scholar]
- 64.Bocharov EV, Mayzel ML, Volynsky PE, Goncharuk MV, Ermolyuk YS, Schulga AA. 2008. Spatial structure and pH-dependent conformational diversity of dimeric transmembrane domain of the receptor tyrosine kinase EphA1. J. Biol. Chem. 283, 29 385–29 395. ( 10.1074/jbc.M803089200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Joh NH, Oberai A, Yang D, Whitelegge JP, Bowie JU. 2009. Similar energetic contributions of packing in the core of membrane and water-soluble proteins. J. Am. Chem. Soc. 131, 10 846–10 847. ( 10.1021/ja904711k) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oberai A, Joh NH, Pettit FK, Bowie JU. 2009. Structural imperatives impose diverse evolutionary constraints on helical membrane proteins. Proc. Natl Acad. Sci. USA 106, 17 747–17 750. ( 10.1073/pnas.0906390106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Teese MG, Langosch D. 2015. Role of GxxxG motifs in transmembrane domain interactions. Biochemistry 54, 5125–5135. ( 10.1021/acs.biochem.5b00495) [DOI] [PubMed] [Google Scholar]
- 68.Adamian L, Liang J. 2002. Interhelical hydrogen bonds and spatial motifs in membrane proteins: polar clamps and serine zippers. Proteins 47, 209–218. ( 10.1002/prot.10071) [DOI] [PubMed] [Google Scholar]
- 69.North B, Cristian L, Fu Stowell X, Lear J, Saven J, Degrado W. 2006. Characterization of a membrane protein folding motif, the Ser zipper, using designed peptides. J. Mol. Biol. 359, 930–939. ( 10.1016/j.jmb.2006.04.001) [DOI] [PubMed] [Google Scholar]
- 70.Zhang Y, Kulp DW, Lear JD, DeGrado WF. 2009. Experimental and computational evaluation of forces directing the association of transmembrane helices. J. Am. Chem. Soc. 131, 11 341–11 343. ( 10.1021/ja904625b) [DOI] [PubMed] [Google Scholar]
- 71.Grigoryan G, Degrado WF. 2008. Modest membrane hydrogen bonds deliver rich results. Nat. Chem. Biol. 4, 393–394. ( 10.1038/nchembio0708-393) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Joh NH, Min A, Faham S, Whitelegge JP, Yang D, Woods VL. 2008. Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins. Nature 453, 1266–1270. ( 10.1038/nature06977) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gao J, Bosco DA, Powers ET, Kelly JW. 2009. Localized thermodynamic coupling between hydrogen bonding and microenvironment polarity substantially stabilizes proteins. Nat. Struct. Mol. Biol. 16, 684–690. ( 10.1038/nsmb.1610) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choma C, Gratkowski H, Lear JD, DeGrado WF. 2000. Asparagine-mediated self-association of a model transmembrane helix. Nat. Struct. Biol. 7, 161–166. ( 10.1038/72440) [DOI] [PubMed] [Google Scholar]
- 75.Zhou FX, Merianos HJ, Brunger AT, Engelman DM. 2001. Polar residues drive association of polyleucine transmembrane helices. Proc. Natl Acad. Sci. USA 98, 2250–2255. ( 10.1073/pnas.041593698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou FX, Cocco MJ, Russ WP, Brunger AT, Engelman DM. 2000. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat. Struct. Biol. 7, 154–160. ( 10.1038/72430) [DOI] [PubMed] [Google Scholar]
- 77.Lear JD, Wasserman ZR, DeGrado WF. 1988. Synthetic amphiphilic peptide models for protein ion channels. Science 240, 1177–1181. ( 10.1126/science.2453923) [DOI] [PubMed] [Google Scholar]
- 78.DeGrado WF, Wasserman ZR, Lear JD. 1989. Protein design, a minimalist approach. Science 243, 622–628. ( 10.1126/science.2464850) [DOI] [PubMed] [Google Scholar]
- 79.Åkerfeldt K, Kim RM, Camac D, Groves JT, Lear JD, DeGrado WF. 1992. Tetraphilin: a four-helix proton channel built on a tetraphenylporphyrin framework. J. Am. Chem. Soc. 114, 9656–9657. ( 10.1021/ja00050a054) [DOI] [Google Scholar]
- 80.Åkerfeldt KS, Lear JD, Waserman ZR, Chung LA, DeGrado WF. 1993. Synthetic peptides as models for ion channel proteins. Acc. Chem. Res. 26, 191–197. ( 10.1021/ar00028a009) [DOI] [Google Scholar]
- 81.Kienker PK, DeGrado WF, Lear JD. 1994. A helical-dipole model describes the single-channel rectification of an uncharged peptide ion channel. Proc. Natl Acad. Sci. USA 91, 4859–4863. ( 10.1073/pnas.91.11.4859) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lear JD, Wasserman ZR, DeGrado WF. 1994. The use of synthetic peptides for the study of membrane protein structure. In Membrane protein structure (ed. White SH.), pp. 335–354. New York, NY: Oxford University Press. [Google Scholar]
- 83.Kienker PK, Lear JD. 1995. Charge selectivity of the designed uncharged peptide ion channel Ac-(LSSLLSL)3-CONH2. Biophys. J. 68, 1347–1358. ( 10.1016/S0006-3495(95)80307-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Åkerfeldt KS, Kienker PK, Lear JD, DeGrado WF. 1996. Structure and conduction mechanisms of minimalist ion channels. In Comprehensive supramolecular chemistry. Supramolecular technology, vol. 10 (ed. Reinhoudt DN.), pp. 659–686. London, UK: Pergamon. [Google Scholar]
- 85.Lear JD, Schneider JP, Kienker PK, DeGrado WF. 1997. Electrostatic effects on ion selectivity and rectification in designed ion channel peptides. J. Am. Chem. Soc. 119, 3212–3217. ( 10.1021/ja9629672) [DOI] [Google Scholar]
- 86.Randa HS, Forrest LR, Voth GA, Sansom MS. 1999. Molecular dynamics of synthetic leucine-serine ion channels in a phospholipid membrane. Biophys. J. 77, 2400–2410. ( 10.1016/S0006-3495(99)77077-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Slotboom DJ. 2014. Structural and mechanistic insights into prokaryotic energy-coupling factor transporters. Nat. Rev. Microbiol. 12, 79–87. ( 10.1038/nrmicro3175) [DOI] [PubMed] [Google Scholar]
- 88.Theobald DL, Miller C. 2010. Membrane transport proteins: surprises in structural sameness. Nat. Struct. Mol. Biol. 17, 2–3. ( 10.1038/nsmb0110-2) [DOI] [PubMed] [Google Scholar]
- 89.Lombardi A, Summa CM, Geremia S, Randaccio L, Pavone V, DeGrado WF. 2000. Retrostructural analysis of metalloproteins: application to the design of a minimal model for diiron proteins. Proc. Natl Acad. Sci. USA 97, 6298–6305. ( 10.1073/pnas.97.12.6298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grigoryan G, Reinke AW, Keating AE. 2009. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature 458, 859–864. ( 10.1038/nature07885) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.North B, Summa CM, Ghirlanda G, DeGrado WF. 2001. D(n)-symmetrical tertiary templates for the design of tubular proteins. J. Mol. Biol. 311, 1081–1090. ( 10.1006/jmbi.2001.4900) [DOI] [PubMed] [Google Scholar]
- 92.Grigoryan G, Degrado WF. 2011. Probing designability via a generalized model of helical bundle geometry. J. Mol. Biol. 405, 1079–1100. ( 10.1016/j.jmb.2010.08.058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang J, Grigoryan G. 2013. Mining tertiary structural motifs for assessment of designability. Methods Enzymol. 523, 21–40. ( 10.1016/B978-0-12-394292-0.00002-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou J, Grigoryan G. 2015. Rapid search for tertiary fragments reveals protein sequence-structure relationships. Protein Sci. 24, 508–524. ( 10.1002/pro.2610) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Grigoryan G. 2013. Absolute free energies of biomolecules from unperturbed ensembles. J. Comput. Chem. 34, 2726–2741. ( 10.1002/jcc.23448) [DOI] [PubMed] [Google Scholar]
- 96.Cristian L, Lear JD, DeGrado WF. 2003. Use of thiol-disulfide equilibria to measure the energetics of assembly of transmembrane helices in phospholipid bilayers. Proc. Natl Acad. Sci. USA 100, 14 772–14 777. ( 10.1073/pnas.2536751100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.DeGrado WF, Gratkowski H, Lear JD. 2003. How do helix–helix interactions help determine the folds of membrane proteins? Perspectives from the study of homo-oligomeric helical bundles. Protein Sci. 12, 647–665. ( 10.1110/ps.0236503) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Summa CM, Rosenblatt MM, Hong JK, Lear JD, DeGrado WF. 2002. Computational de novo design, and characterization of an A(2)B(2) diiron protein. J. Mol. Biol. 321, 923–938. ( 10.1016/S0022-2836(02)00589-2) [DOI] [PubMed] [Google Scholar]
- 99.Kaplan J, DeGrado WF. 2004. De novo design of catalytic proteins. Proc. Natl Acad. Sci. USA 101, 11 566–11 570. ( 10.1073/pnas.0404387101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Geremia S, Di Costanzo L, Randaccio L, Engel DE, Lombardi A, Nastri F, DeGrado WF. 2005. Response of a designed metalloprotein to changes in metal ion coordination, exogenous ligands, and active site volume determined by X-ray crystallography. J. Am. Chem. Soc. 127, 17 266–17 276. ( 10.1021/ja054199x) [DOI] [PubMed] [Google Scholar]
- 101.Calhoun JR, Kono H, Lahr S, Wang W, DeGrado WF, Saven JG. 2003. Computational design and characterization of a monomeric helical dinuclear metalloprotein. J. Mol. Biol. 334, 1101–1115. ( 10.1016/j.jmb.2003.10.004) [DOI] [PubMed] [Google Scholar]
- 102.Irving HMNH, Williams RJP. 1953. The stability of transition-metal complexes. J. Chem. Soc. 8, 3192–3210. ( 10.1039/jr9530003192) [DOI] [Google Scholar]
- 103.Chao Y, Fu D. 2004. Kinetic study of the antiport mechanism of an Escherichia coli zinc transporter, ZitB. J. Biol. Chem. 279, 12 043–12 050. ( 10.1074/jbc.M313510200) [DOI] [PubMed] [Google Scholar]
- 104.Chao Y, Fu D. 2004. Thermodynamic studies of the mechanism of metal binding to the Escherichia coli zinc transporter YiiP. J. Biol. Chem. 279, 17 173–17 180. ( 10.1074/jbc.M400208200) [DOI] [PubMed] [Google Scholar]
- 105.DeCoursey TE. 2008. Voltage-gated proton channels: what's next? J. Physiol. 586, 5305–5324. ( 10.1113/jphysiol.2008.161703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Decoursey TE. 2003. Voltage-gated proton channels and other proton transfer pathways. Physiol. Rev. 83, 475–579. ( 10.1152/physrev.00028.2002) [DOI] [PubMed] [Google Scholar]
- 107.Leiding T, Wang J, Martinsson J, DeGrado WF, Arskold SP. 2010. Proton and cation transport activity of the M2 proton channel from influenza A virus. Proc. Natl Acad. Sci. USA 107, 15 409–15 414. ( 10.1073/pnas.1009997107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Deisenhofer J, Epp O, Miki K, Huber R, Michel H. 1985. Structure of the protein subunits in the photosynthetic reaction centre of Rhodopseudomonas viridis at 3A resolution. Nature 318, 618–624. ( 10.1038/318618a0) [DOI] [PubMed] [Google Scholar]
- 109.Kendrew JC, Bodo G, Dintzis HM, Parrish RG, Wyckoff H, Phillips DC. 1958. A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature 181, 662–666. ( 10.1038/181662a0) [DOI] [PubMed] [Google Scholar]
- 110.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293. ( 10.1007/BF00197809) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.