Abstract
Efficient construction of BAC-based human artificial chromosomes (HACs) requires optimization of each key functional unit as well as development of techniques for the rapid and reliable manipulation of high-molecular weight BAC vectors. Here, we have created synthetic chromosome 17-derived alpha-satellite arrays, based on the 16-monomer repeat length typical of natural D17Z1 arrays, in which the consensus CENP-B box elements are either completely absent (0/16 monomers) or increased in density (16/16 monomers) compared to D17Z1 alpha-satellite (5/16 monomers). Using these vectors, we show that the presence of CENP-B box elements is a requirement for efficient de novo centromere formation and that increasing the density of CENP-B box elements may enhance the efficiency of de novo centromere formation. Furthermore, we have developed a novel, high-throughput methodology that permits the rapid conversion of any genomic BAC target into a HAC vector by transposon-mediated modification with synthetic alpha-satellite arrays and other key functional units. Taken together, these approaches offer the potential to significantly advance the utility of BAC-based HACs for functional annotation of the genome and for applications in gene transfer.
INTRODUCTION
Alpha-satellite DNA is the major species of repetitive element found at the centromere of all normal primate chromosomes. It is organized in a hierarchical structure based on a ∼171 bp monomeric unit that is tandemly multimerized into a higher-order repeat (HOR), which is itself repeated in tandem over hundreds to thousands of kilobases in the centromeric region of all normal human chromosomes [reviewed in (1–3)]. Centromeric alpha-satellite acts to organize the recruitment of key centromeric proteins (CENPs) to form a trilaminar protein–DNA complex, the kinetochore, which mediates the interactions between the chromosome and the spindle apparatus that are responsible for coordinated chromosome movements during cell division (4–6). While functional kinetochores have been observed at chromosomal locations not containing any alpha-satellite [so-called ‘neocentromeres’; reviewed in (7)], only cloned alpha-satellite DNA has thus far been shown to form centromeres de novo when introduced into the cell nucleus by transfection or microinjection in artificial chromosome assays (8–11).
The ability to create human artificial chromosomes (HACs) was pioneered through the development of techniques to synthesize large alpha-satellite arrays in vitro (12). These artificial chromosome vectors may have eventual applications in human gene transfer (8,13); e.g. HACs containing the HPRT genomic locus have been shown to complement HPRT-deficient cell lines (14,15), and we have observed sustained expression of the β-globin gene from HACs carrying the entire 150 kb β-globin genomic region (J. Basu, unpublished data). In addition, HACs incorporating the GCH1 locus have been shown to reproduce the expression pattern of GCH1 in vivo (16). Finally, artificial chromosome vectors provide a methodological platform for the identification and functional analysis of genomic elements in alpha-satellite that are critical for centromere function (9,12,17–20).
Two fundamentally different approaches have been developed for the creation of HACs: top down, whereby an existing chromosome is systematically shortened by telomere-mediated truncation (21); bottom up, whereby cloned chromosomal elements including alpha-satellite DNA, with or without telomeric DNA and genomic DNA, are preassembled into a defined YAC- or BAC-based artificial chromosome vector or are assembled by the host cell through a combination of nonhomologous recombination and DNA repair mechanisms [reviewed in (22)]. BAC-based artificial chromosomes may be either linear or circular and minimally require only a mammalian selectable marker and a cloned alpha-satellite array, which may be of natural or synthetic origin (9,23).
Variation in the efficiency of de novo centromere formation between alpha-satellite templates derived from different human chromosomes (9,19,24) have suggested a causal link between the density of sequence elements, such as CENP-B boxes, and de novo centromere seeding efficiency (25). The CENP-B box is the biochemically defined motif ‘PyTTCGTTGGAAPuCGGGA’ minimally responsible for mediating binding of the constitutive centromeric protein CENP-B to human alpha-satellite DNA (26,27).
In order to address the functional significance of the CENP-B box in human alpha-satellite and in HAC formation, we have developed methodologies to directly vary the density and distribution of CENP-B boxes in the D17Z1 chromosome 17-derived HOR unit, which in its natural configuration contains a CENP-B box in 5 of its 16 constituent monomers (28). We have constructed entirely synthetic D17Z1-based derivatives in which each of the 16 tandem monomeric repeats contains either a consensus CENP-B box or a mutated sequence element that does not bind CENP-B (29,30). Here, we report that the efficiency of formation of HACs is proportional to the density of CENP-B boxes in the HAC vector, thus demonstrating a dependence on CENP-B boxes in centromeric chromatin assembly.
In addition, we describe the development of a novel, single-step, transposition-based technique for retrofitting genomic BACs with synthetic alpha-satellite arrays and other key chromosomal components using a modified Tn5 transposon vector (31). We have established the conditions required to mediate successful transfer of a functionally competent 86 kb alpha-satellite array with head-to-head oriented telomeres and selectable markers into a 100–200 kb genomic BAC. These BAC–HAC constructs can be introduced into cultured mammalian cells and are shown to be capable of forming synthetic artificial chromosomes de novo. Data on the composition, stability and function of these HACs demonstrate that transposon-mediated retrofitting of genomic BACs is an effective and universally applicable methodology for converting any genomic BAC into a HAC vector. Taken together, these approaches have significant implications for the design and further development of HACs for potential applications in gene transfer and functional genomics.
MATERIALS AND METHODS
Synthesis of modified 2.7 kb D17Z1 repeats
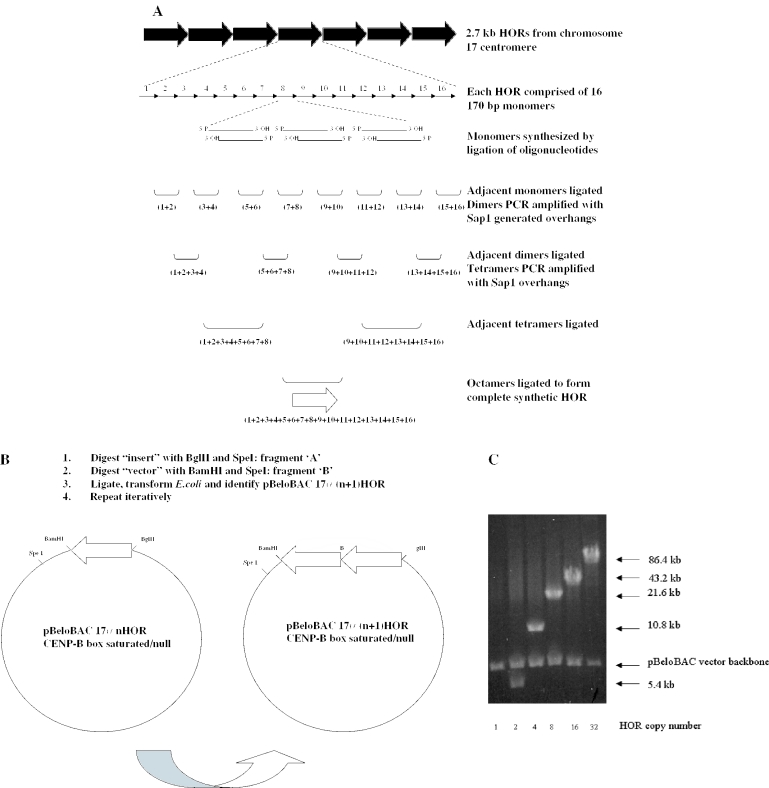
The sequence of the 2.7 kb D17Z1 HOR (28) was modified such that each of the 16 ∼171 bp monomer units contained the consensus CENP-B box element 5′-TTT CGT TGG AAA CGG GA-3′ (26) or the Y alpha-satellite derived null element AGA TGG TGG AAA AGG AA. Each of the 16 modified monomer units was then synthesized by ligation of two to three pairs of overlapping oligonucleotides (Operon Technologies, CA). Pairs of engineered monomer units were then ligated together appropriately to form dimers, such that a given monomer n would be ligated to monomer (n + 1) or (n − 1). In addition, the EcoRI sites of monomers 1 and 16 were altered to create a BamHI site at the 5′ end of monomer 1 and a BglII site at the 3′ end of monomer 16 (12). Each gel-purified dimer was then PCR amplified with a BsaI or SapI restriction site such that upon digestion each dimer would produce a defined overhang exactly complementary to an overhang in the adjacent dimer. The resultant tetramers (containing no extraneous sequence) were then T/A subcloned into pGem-Teasy (Promega) and sequence verified. These tetrameric subunits were then ligated together in the appropriate orientation, using SapI (or NotI and SapI for monomers 1 and 16), to generate the required overhang. Finally, the resultant octamers were further gel purified and ligated together to produce the completed synthetic 16mer, representing a single synthetic D17Z1 repeat unit, with NotI overhangs. This repeat unit was then subcloned as a NotI fragment into the BAC cloning vector pBeloBAC11 (32). The overall strategy is outlined in Figure 1A.
Figure 1.
(A) Outline of iterative scheme for synthesis of mutated versions of D17Z1 alpha-satellite arrays. Each of the 16 individual monomers comprising a single 2.7 kb higher-order repeat (HOR) was synthesized as 2–3 oligonucleotide pairs (60–100 bp each), which were directly ligated together and gel purified. Adjacent HORs were subsequently ligated to form dimers as shown, and PCR modified to introduce SapI recognition sites at both ends as appropriate. Digestion with SapI allows seamless ligation of adjacent dimers to create tetramers without introduction of extraneous non-alpha-satellite sequences. Two additional rounds of serial ligation resulted in formation of a complete synthetic HOR, which was subcloned into the BAC vector pBeloBAC. (B) Outline of scheme for directional multimerization of mutated HORs. A synthetic, 86 kb alpha-satellite array consisting of 32 tandem copies of the HOR was created as follows. pBAC17α1 was digested with BglII and SpeI and the alpha-satellite containing fragment (fragment ‘A’) was isolated and gel purified. The same construct was separately digested with BamHI and SpeI, and the larger fragment (fragment ‘B’) was isolated and gel purified. Ligation of fragment ‘A’ to fragment ‘B’ is directional, resulting in head-to-tail multimerization of adjacent repeats. The resulting pBAC17α(n + 1) construct was then isolated following transformation of the ligation reaction into E.coli. This process was repeated iteratively to create the final pBAC17α32 arrays. (C) Pulsed-field gel electrophoresis (PFGE) analysis of intermediates in the construction of 17α32 HOR/BeloBAC constructs. Each intermediate was digested with NotI, which drops out the entire subcloned alpha-satellite array from the pBeloBAC vector backbone. Each lane shows the completed intermediate en route to 32 copies of the mutated HOR. The number beneath each lane corresponds to the number of copies of the HOR in each construct, and the arrows to the right indicate the size of the alpha-satellite insert and the vector backbone. The insert in lane labeled ‘1’ is 2.7 kb and therefore too small to be resolved by PFGE.
Directional multimerization of the CENP-B box enriched/null repeat units
The 2.7 kb CENP-B box enriched or CENP-B box null synthetic D17Z1 repeat was multimerized directionally as follows. The cloned synthetic repeat (in pBeloBAC11, as outlined above) was digested with BglII and SpeI, and this band (fragment ‘A’) was gel purified by standard procedures (Qiagen). A second fragment (fragment ‘B’) was generated by digesting the same cloned repeat with BamHI and SpeI. The appropriate fragment ‘B’ was subsequently gel purified and ligated to fragment ‘A’. This ligation reaction was transformed into Escherichia coli (GibcoBRL), and recombinant clones were identified by NotI digestion and pulsed-field gel electrophoresis (Figure 1B). This process was repeated iteratively to create clones containing 4, 8, 16 and 32 copies of the CENP-B box enriched/CENP-B box null repeat unit in pBeloBAC (Figure 1C). Finally, for use as a selectable marker in mammalian cells, a cDNA cassette conferring resistance to puromycin was introduced into each clone by transposition into the pBeloBAC vector backbone.
A ∼86 kb synthetically assembled alpha-satellite array, derived from directional multimerization of the naturally occurring D17Z1 repeat unit (12), was subcloned as a BamHI/BglII fragment into the BamHI site of pBeloBAC11. This construct was further modified by transposition with the puromycin resistance selectable marker, as above. The structural integrity of all modified arrays and of the original D17Z1-based array was confirmed by sequencing, restriction digestion and fluorescence in situ hybridization (FISH) [as described in (12)], using the array as probe.
Mobility shift analysis
The effect of the mutations described above on CENP-B binding to the synthetic repeat units was evaluated by a gel mobility shift assay. Cloned tetramer units assembled from CENP-B box enriched and CENP-B box null monomers were digested with NotI and the inserts were gel purified. After incubation with purified recombinant CENP-B protein (Diarect, Germany) for 25 min at room temperature in CENP-B binding buffer (26), protein–DNA complexes were electrophoresed through a 2% agarose gel in 0.5× TBE buffer. Following electrophoresis, a SybrGold (Molecular Probes) stain was used to visualize DNA bands.
Transposon engineering
BAC-based transposon vectors containing synthetic centromeric and telomeric arrays were constructed as follows. Selectable markers encoding resistance to neomycin/kanamycin and puromycin were subcloned as PCR products into the PstI and SmaI sites, respectively, of the pMod transposon vector (Epicentre). This modified transposon cassette was PCR-amplified to introduce BglII overhangs on either side and subcloned into the BamHI site of the BAC vector pBeloBAC11. Synthetic telomeric DNA was then subcloned into the BAC transposon vector such that two 800 bp telomere seeds were arranged in a head-to-head orientation separated by an adapter containing the recognition site for the ultrarare restriction enzyme I-CeuI (NEB). A synthetically assembled D17Z1-based array (12) was subcloned into the BamHI site of the BAC-based transposon vector. The structure of the final transposon vector construct pBAC17α32 HTH Tel TN was confirmed by extensive restriction analysis, sequencing and Southern blotting (data not shown). Independent versions of this transposon vector were created for the CENP-B box enriched and CENP-B box null synthetic arrays described above, in addition to the natural D17Z1 array.
Preparation of the transposon
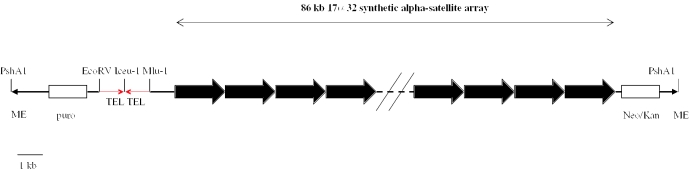
Digestion of the transposon vector with PshAI (NEB) releases a linear transposon containing an 86 kb alpha-satellite array, neo/kan and puroR selectable markers, and synthetic telomeres in a head-to-head orientation (Figure 3). The transposon fragment was separated by agarose gel electrophoresis, isolated in a gel slice and purified by standard techniques.
Figure 3.
Digestion of pBAC 17α32 HTH Tel TN with PshA1 releases the ∼100 kb linear transposon vector, shown with an 86 kb chromosome 17-derived alpha-satellite array, as well as ∼800 bp synthetic telomeres separated by a recognition site for the ultrarare endonuclease I-CeuI. Mammalian cell line selection is based on a pgk-puro cassette conferring resistance to puromycin and a neo/kan cassette conferring resistance to geneticin. The transposon vector is flanked by 19 bp transposase recognition elements (ME).
Transposition reaction
A collection of genomic BAC clones of average insert size 100–150 kb was created by shotgun subcloning a NotI digest of whole genomic DNA from human HT1080 cells into the BAC vector VJ104, a BAC108L derivative (12). The chromosomal origin of each genomic fragment was established by direct sequencing of each genomic insert and comparison to public sequence databases. In addition, BAC 2202F23 was identified by a BLAST search of the HTGS database through NCBI with the human growth hormone (HGH) cDNA sequence. These genomic BACs were used as targets for transposon-mediated modification to establish the feasibility of transposition as a simple, one-step methodology for converting genomic BACs into HAC vectors.
Transposition reactions were extensively optimized to determine the ideal reaction volume, total amount of DNA, molar ratio of transposon to target, reaction conditions and transformation conditions. The entire transposition reaction was electroporated into DH10B E.coli and plated onto Cm/kan plates to select for successful transposition events. Individual colonies were miniprepped and screened by NotI digestion to establish if the transposon had integrated into the genomic insert or the BAC vector backbone. Both classes of integrants could be readily obtained without extensive initial screening. Clones were further screened by digestion with EcoRI, PstI, PvuII and HindIII to confirm the presence of D17Z1 alpha-satellite DNA in the genomic BAC. For clones with integration events in the genomic insert, sequencing with transposon-based primers was used to directly establish the site of integration of the transposon.
Cell transfection
Human HT1080 cells were transfected using Fugene 6 (Roche) reagent according to the manufacturer's instructions and stable clones were identified on the basis of resistance to puromycin (Kayla) at 3 μg/ml. Clones appeared after 7–10 days and were subsequently expanded to generate clonal lines for further analysis.
Cytogenetic analysis and validation of HACs
Clonal populations of cells containing putative HACs were analyzed as described (9,12,17,19). HACs were considered validated if they showed a positive hybridization signal with a FISH probe derived from the synthetic array as well as positive immunoreactivity for CENP-C, a centromere protein localized to the outer kinetochore and specific for active centromeres (12,33), and if they were mitotically stable.
Briefly, cells were arrested at metaphase using colchicine (Gibco) at 40 μg/ml for 45 min at 37°C, then treated with hypotonic solution (0.075 M KCl, 12 min, 37°C) and applied to slides using a Shandon Cytospin 3. Slides were subsequently fixed in 2% formaldehyde and immunoreacted with rabbit anti-CENP-C antibody (12) at a dilution of 1/2000 in PBS and detected with goat anti-rabbit IgG (Molecular Probes). DNA probes were labeled by nick-translation using the Vysis system according to the manufacturer's instructions. Immunoreacted slides were fixed (3:1, methanol:acetic acid), subjected to denaturation (70% formamide, 72°C, 8 min) and hybridized to denatured probes as described (9,12). Mitotic stability was evaluated by cytogenetic analysis after growth in the presence or absence of drug selection for up to 6 weeks.
RESULTS
Previous studies have established that vectors containing multiple copies of certain alpha-satellite arrays can seed formation of de novo centromeres in human HT1080 cells (12,20). However, the overall frequency of generation of HACs has been reported to be variable and often quite low (9,17,18,24,34). Therefore, we have undertaken to develop a general approach to increase the efficiency of HAC formation and to evaluate the sequence dependency of de novo centromere seeding.
Construction of engineered, D17Z1-based HAC vectors
The BAC–HAC system provides a platform to systematically evaluate the functional significance of sequence elements within human alpha-satellite DNA. We developed methodologies to construct modified, synthetic D17Z1 repeat units that are either enriched for or depleted in the density of CENP-B box DNA binding elements. In order to generate engineered repeat units, each of the 16 monomers was synthesized by the serial, stepwise assembly of oligonucleotide pairs, each between 60 and 100 bp in length, as shown in Figure 1A. Adjacent monomer units could then be gel purified and ligated to form dimers. This process of PCR and ligation assembly was serially repeated until the complete 16-monomer HOR was constructed (see Materials and Methods). The resulting synthetic repeat was then subcloned and directionally concatamerized to 32 copies (Figure 1B and C), using methods previously developed in our laboratory (12).
CENP-B boxes are required for efficient centromere formation de novo
We used the techniques described above to create a modified variant of D17Z1 alpha-satellite in which all of the consensus CENP-B boxes or elements resembling the consensus in each of the 16 monomer units were replaced with a sequence derived from Y chromosome alpha-satellite. As Y alpha-satellite does not bind CENP-B (30), this approach allowed us to knock out any interaction between CENP-B and its biochemically defined consensus element, as well as any interactions between CENP-B and elements resembling the consensus that might potentially occur in vivo. Confirmation of abolishment of CENP-B binding to the synthetic repeat was shown by loss of mobility shift in a gel shift assay (data not shown).
Constructs based on the naturally occurring, unmodified D17Z1 repeat have been used previously to generate mitotically stable HACs in 10–50% of drug-resistant HT1080 clones after transfection (9,12,17,24). Here, cytogenetically detectable HACs were identified in 4 of 38 colonies (Table 1, Figure 2), consistent with previous studies. However, when using the CENP-B null construct in which all CENP-B boxes had been modified, only a single clone was identified to have a putative de novo centromere out of 40 clones screened (Table 1). The fact that the observed rate of de novo HAC formation is low but is not zero is consistent with other reports that some alpha-satellite arrays that do not contain CENP-B boxes may form apparent HACs at very low frequencies (24,34); however, this single clone (as well as those in the literature) may represent a false positive that has acquired HT1080 alpha-satellite from an endogenous chromosome, as documented by us previously (9). This point notwithstanding, our data indicating a dependence on CENP-B boxes are in agreement with Masumoto and colleagues, who used an alternative approach to modify CENP-B boxes in a repeat unit derived from chromosome 21 (18). Combined, the two studies provide strong evidence that CENP-B boxes are required for efficient formation of de novo centromeres in HAC systems.
Table 1.
Effect of CENP-B box density on rate of HAC formation
| Vector | No. of CENP-B boxes | No. of experiments | No. of clones analyzed | No. of clones with HACsa | HAC formation (%) |
|---|---|---|---|---|---|
| pBAC17α32-all | 16 | 15 | 45 | 10 | 22 |
| pBAC17α32-natural | 5 | 6 | 38 | 4 | 10.5 |
| pBAC17α32-null | 0 | 10 | 40 | 1 | 2.5 |
aCytogenetically detectable. Validated by FISH and mitotic stability (see Materials and Methods).
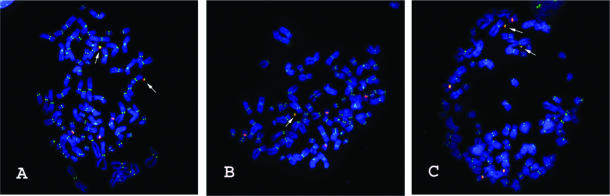
Figure 2.
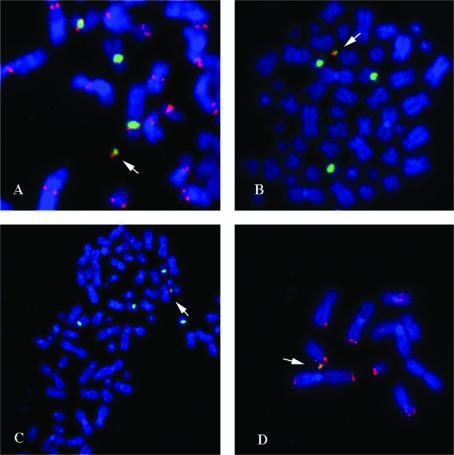
Cytogenetic analysis of HACs from synthetic chromosome 17-derived alpha-satellite arrays. Arrows designate HACs. Immunostaining with an anti-CENP-C antibody (green) identifies functional centromeres. FISH with the synthetic alpha-satellite as probe (red) hybridizes with the HAC as well as to the centromeres of the endogenous HT1080 chromosome 17s. (HT1080 is quasitetraploid, so there are four copies of the endogenous chromosome 17s.) DAPI-stained chromosomes are shown in blue. (A) HT1080 clone generated by transfection with pBAC17α32-all construct (Table 1) showing the presence of two HACs. (B) HT1080 clone generated by transfection with pBAC17α32-natural. A single HAC is visible. (C) HT1080 clone generated by transfection with pBAC17α32-null. Two HACs are present.
As a corollary to the data presented above and by Ohzeki et al. (18), we reasoned that if the density of CENP-B boxes was indeed critical for de novo centromere formation, it should be possible to create synthetic alpha-satellite arrays with a CENP-B box density even higher than their naturally occurring counterparts. These novel synthetic arrays might form a more efficient template for centromere formation de novo than arrays with the natural density of CENP-B box elements. To test this hypothesis, we used the strategy described above to construct a synthetic D17Z1-derived alpha-satellite array saturated with CENP-B boxes such that each of the 16 monomers in the D17Z1 repeat contained a consensus CENP-B box. Upon introduction into HT1080 cells, these saturated synthetic arrays formed HACs de novo more than twice as efficiently as arrays containing the natural density of CENP-B boxes (Table 1) (P < 0.09, Fisher's exact test) and almost nine times as efficiently as arrays without CENP-B boxes (P < 0.01, Fisher's exact test). Although the HAC formation frequency of the latter in this particular study does not differ significantly from that of arrays containing the natural density of CENP-B boxes (P = 0.14, Fisher's exact test), the overall trends nonetheless demonstrate that the rates of de novo centromere formation are dependant on the density of the CENP-B box and that arrays saturated for the CENP-B box clearly form de novo centromeres more effectively than arrays lacking the CENP-B box, confirming and extending previous studies based on chromosome 21 (18). The frequency of HACs within any one clone was observed to vary from 10 to 100%, similar to the ranges observed in cell lines derived from transfection with control natural arrays (9,20,23).
Consistent with other studies, cytogenetic estimates suggested that the detected HACs (from all versions of the array) are several megabases in size (Figure 2). In all cases, HACs were shown to be mitotically stable in the absence of selection for 6 weeks and to bind the centromere-specific protein CENP-C (Figure 2 and data not shown). Thus, the effect of changing the density of CENP-B boxes appears to be limited to the efficiency of formation of HACs and not to their subsequent behavior in mitotic segregation. More subtle effects on nondisjunction or chromosome lag (17), however, could not be assessed with the techniques used in this study.
Retrofitting of genomic BACs by transposition with synthetic alpha-satellite arrays
The utility of HAC vectors for gene transfer would be improved by enhancing the ability to combine elements capable of efficient de novo centromere formation with genomic fragments containing genes or other elements of interest. Thus, we next proceeded to develop strategies for the rapid introduction of the various CENP-B box optimized alpha-satellite arrays into genomic BACs. Figure 3 shows the critical functional elements in pBAC17α32 HTH Tel TN. Digestion of this construct with the restriction enzyme PshAI results in the release of a transposon containing the 86 kb synthetic alpha-satellite array, as well as 800 bp telomere seeds that are capable (upon linearization with I-CeuI) of seeding de novo telomeres (35). Transposition of this linear fragment into a target genomic BAC retrofits the latter into a BAC–HAC vector. We have constructed analogous transposon vectors containing the original D17Z1 array, as well as those containing the CENP-B box enriched or null versions of the array.
The fidelity of the transposition reaction was high, as >80% of BAC clones examined showed no detectable loss or rearrangement of genomic insert or alpha-satellite sequence, as evidenced by extensive restriction analysis on pulsed field and standard gels (Figure 4). Partial loss of telomeric sequence was occasionally observed, but recovery of retrofitted BAC clones containing nearly full-length telomeres was typically possible. We mapped the site of integration by direct sequencing outward from the transposon integration site and documented successful transpositions into both genomic insert and vector backbone (Figure 4) at the predicted frequencies.
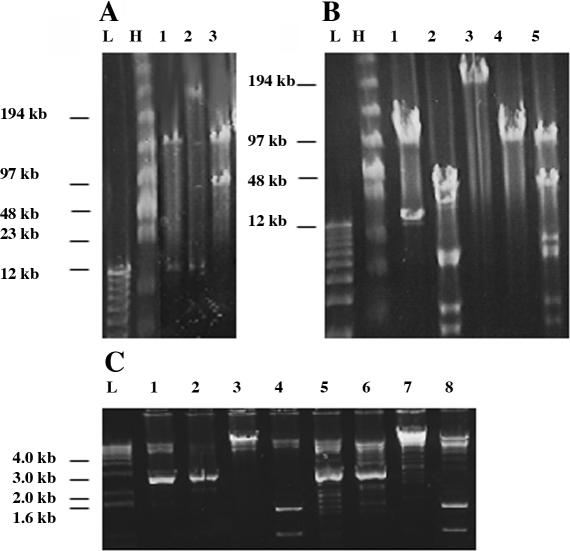
Figure 4.
Detection and validation of transposition events in genomic BACs. (A) Identification of transposition events by NotI digest. Lane 1, target genomic BAC. Lane 2, transposon insertion into genomic region of target BAC. Note that insert fragment excised by NotI is much larger than the starting fragment from lane 1. Lane 3, transposon insertion into vector backbone of target BAC. Note that vector fragment is much larger than the starting vector fragment from lane 1, while genomic insert fragment is unchanged. (B) Confirmation of successful transposition of complete 86 kb alpha-satellite array. Lane 1, target BAC cut with NotI. Lane 2, target BAC cut with BamHI. Lanes 3–5, clone with putative transposition event cut with I-CeuI (lane 3), Not I (lane 4) or BamHI (lane 5). The 86 kb band in lane 5 represents the size of the alpha-satellite array present in the clone. (C) Analysis of alpha-satellite array integrity in BAC modified by transposition. pBAC17α32 HTH Tel TN control cut with PstI (lane 1), PvuII (lane 2), EcoRI (lane 3) and HindIII (lane 4). Clone with putative transposition event cut with PstI (lane 5), PvuII (lane 6), EcoRI (lane 7) and HindIII (lane 8). Both PstI and PvuII digests generate a multicopy 2.7 kb HOR fragment from the alpha-satellite array. HindIII is predicted to produce bands at 1.4 and 0.9 kb. In A, B and C, L and H represent low-molecular weight and high-molecular weight markers, respectively.
In order to assess the generality of this approach as a method for BAC retrofitting, we assembled a collection of genomic BACs containing inserts of about 100 kb derived either by shotgun subcloning of whole genomic DNA or obtained through the public databases. We modified each BAC with the pBAC17α32 HTH Tel TN-derived transposon vector and confirmed the size and structural integrity of the retrofitted BAC by restriction analysis and Southern blotting (Figure 4 and data not shown). We then evaluated the ability of the resultant BAC–HAC vectors to generate HACs de novo.
Several representative BAC–HAC vectors (Table 2) were linearized by digestion with the ultrarare endonuclease I-CeuI and introduced into HT1080 cells by transfection. Potential HAC formation was confirmed by FISH analysis with probes against the chromosome 17 alpha-satellite, BAC vector backbone, genomic insert and telomeric DNA, and was validated as described above. Representative examples of such HACs are shown in Figure 5. While the current experiments have not rigorously addressed whether the HACs are linear or circular, they do contain telomere repeats (Figure 5D), as predicted from the structure of the BAC–HAC vectors used. The efficiency of de novo HAC formation using different BAC–HAC vectors is summarized in Table 2 and is within the range shown with other vectors (9,18,24).
Table 2.
Examples of genomic BACs engineered by transposition
| Target genomic BAC | Chromosomal origin | Insert size (kb) | Transposition site | HAC frequency |
|---|---|---|---|---|
| G1 | Chr 2 | 106 | Vector | 2/24 (8%) |
| Genomic insert | 1/12 (8%) | |||
| G3 | Chr 17 | 100 | Genomic insert | 1/25 (4%) |
| G7 | Chr 15 | 93 | Genomic insert | 2/14 (14%) |
| 2202F23 (HGH) | Chr 17 | 156 | Genomic insert | 2/13 (15%) |
Figure 5.
Cytogenetic analysis of HACs created from unimolecular BAC–HAC vectors. (A) Dual FISH/immunostaining with anti-CENP-C antibodies (red) and D17Z1 probe (green). (B) Two-color FISH analysis: D17Z1 probe (green) and 150 kb genomic fragment probe from BAC–HAC vector (red). (C) Two-color FISH analysis: D17Z1 probe (green) and BAC vector backbone probe (red). (D) Two-color FISH analysis: D17Z1 probe (green) and telomeric DNA (red). In all panels, DAPI-stained DNA in blue.
DISCUSSION
Since the original reports of de novo centromere and HAC formation (12,20), a number of groups have described related approaches to further develop and optimize artificial chromosome systems [reviewed in (8,22)]. The creation of HACs has now been established as a tractable (if laborious) approach to systematically identify and dissect elements that are critical for chromosome function (17–19). In this report, we describe the further refinement of the BAC–HAC system as a methodological platform to undertake a functional analysis of the role of the density of CENP-B box elements in human alpha-satellite DNA, as well as a novel approach for the rapid and reliable manipulation of genomic BACs with these synthetic arrays.
CENP-B is a constitutively present DNA-binding protein found in the underlying centric heterochromatin of all human chromosomes except for the Y chromosome (30). The corresponding DNA sequence element that defines the cognate binding site, the CENP-B box, has been identified as PyTTCGTTGGAAPuCGGGA (26,29) and is found distributed within some, but not all, of the monomer units of alpha-satellite DNA from human centromeres (3,34,36). However, the role of CENP-B, if any, in specifying centromeric identity in endogenous chromosomes remains unsettled (37). Y chromosome centromeres do not associate with CENP-B (30), and African green monkey centromeres lack CENP-B boxes even though the CENP-B protein itself is present (38). Furthermore, Cenp-B knockout mice show only modest phenotypic effects and appear to have fully functional centromeres, as evidenced by the lack of chromosome missegregation phenotypes (39–41).
Nevertheless, studies of de novo centromere formation with cloned alpha-satellite arrays appear to support a direct correlation between the density of CENP-B boxes and the frequency of de novo centromere formation. For example, comparison of cloned alpha-satellite arrays from chromosomes Y, X, 17 and 21 show that 17- and 21-derived arrays form de novo centromeres more efficiently than X- and Y-derived arrays (9,17,24). In addition, alpha-satellite from a CENP-B box-rich region of the chromosome 21 centromere forms de novo centromeres in a HAC system, while alpha-satellite from a neighboring CENP-B box-depleted region is inefficient (25). Further, the de novo centromere nucleation ability of the chromosome 21-derived alpha-satellite array can be disrupted by mutation of its constituent CENP-B boxes (18), an outcome that parallels our observations reported here on mutation of CENP-B boxes in chromosome 17-derived alpha-satellite. Finally, it has also been established that CENP-B boxes outside the context of alpha-satellite DNA are not competent to nucleate de novo centromere assembly (18), showing that elements other than the CENP-B box are required for centromere function. Taken together, our data and earlier observations unambiguously establish the presence of CENP-B and its cognate binding element as a critical, but not sole, element for de novo centromere formation in artificial chromosome assays.
Notwithstanding the clear role of the CENP-B box in assembly of HACs, the role of CENP-B in its endogenous chromosomal context remains open to debate. At least three CENP-B-like proteins have been identified in fission yeast, and double mutants exhibit severe chromosome segregation defects (42). Such functional redundancy may explain the lack of a major phenotype in mouse knockouts of Cenp-B (39–41), and also explain why CENP-B appears dispensable for function of the Y chromosome in both mice and humans as well as for function of neocentromeres (43). In addition, it remains to be established whether the distribution of CENP-B boxes within an array of monomers or even within a single monomer is also of importance, as might be expected if CENP-B participates in nucleosome positioning (44,45).
In addition to the effect of manipulating CENP-B boxes demonstrated here and by Ohzeki et al. (18), it is apparent that other sequences within alpha-satellite may influence the efficiency of HAC formation, as even arrays with a similar number of CENP-B boxes can differ quite substantially in their ability to seed HACs (17,34). This possibility may now be investigated systematically using synthetic alpha-satellite arrays where the distribution of CENP-B boxes and/or other sequences in each monomer has been manipulated, using the approach outlined here.
Determination of the ideal density and distribution of such sequences in alpha-satellite will maximize the efficiency with which BAC-based HAC vectors carrying potentially therapeutic genes might eventually be assembled in human cells (8,10,14). The ability to create HACs de novo from defined, synthetically assembled chromosomal components cloned into BAC vectors may provide significant advantages for practical development as therapeutic agents. Unlike engineered microchromosomes created by top-down methodologies relying on the truncation and manipulation of existing human chromosomes [reviewed in (46)], such BAC–HAC vectors can be propagated in a massive scale in E.coli and purified using well-established methodologies under GMP conditions, making them suitable for immediate use as human therapeutics (47). Once optimized, BAC-based HAC vectors may offer an alternative approach towards analysis of gene and genome function, gene transfer and potentially gene therapy that may circumvent many of the problems associated with conventional, retroviral-based gene delivery systems (48). Transgenes in viral vectors are also susceptible to position effects and silencing (49), and the possibility of activation or knockout of host oncogenes or tumor suppressors during viral integration is now an empirically observed phenomenon (50).
In order for BAC-based HACs to become practical vector systems for biotechnology or other high-throughput applications, methods for the manipulation of large (>100 kb) genomic fragments and large repetitive arrays have to be developed and optimized. Traditional subcloning techniques (51) are notoriously inefficient for this application, with significant levels of BAC deletion and rearrangement being routinely observed. A number of recombination-based BAC engineering systems have been developed in an attempt to address these concerns. For example, the Cre recombinase has been applied to the modification of BACs with mammalian selectable markers (52,53), as well as to the creation of BAC-based HACs (54). In addition, homologous recombination in E.coli has been used to engineer BACs with selectable markers (55,56). However, these approaches are probably not sufficiently efficient or flexible to make the high-throughput conversion of BACs into BAC–HAC vectors practical.
We have therefore developed and implemented a novel methodology for the rapid creation of unimolecular HAC vectors, based on Tn5 transposons (31) and containing synthetic alpha-satellite arrays and other key functional units. We show in this report that this technique allows for the rapid and reliable manipulation of target BACs and that the resultant species are capable of forming functionally validated HACs. Such transposon vectors may be used to introduce future iterations of optimized synthetic alpha-satellite, as well as other key functional elements impacting artificial chromosome formation and stability, into genomic BAC targets of interest. Taken together, we believe that the strategies described here will significantly facilitate the use of BAC-based HACs for gene transfer and functional studies of the genome.
Acknowledgments
We thank Linda Woods, Mark Frey, Jessica Dolhonde and Gwen Hau for technical assistance, and members of the artificial chromosome group for helpful discussions. This research was funded in part by a Franklin Delano Roosevelt Award to H.F.W. from the March of Dimes Birth Defects Foundation. Funding to pay the Open Access publication charges for this article was provided by Duke University Institute for Genome Sciences and Policy.
REFERENCES
- 1.Lee C., Wevrick R., Fisher R.B., Ferguson-Smith M.A., Lin C.C. Human centromeric DNAs. Hum. Genet. 1997;100:291–304. doi: 10.1007/s004390050508. [DOI] [PubMed] [Google Scholar]
- 2.Rudd M.K., Schueler M.G., Willard H.F. Characterization and functional annotation of human centromeres. Cold Spring Harb. Symp. Quant. Biol. 2004;68:141–150. doi: 10.1101/sqb.2003.68.141. [DOI] [PubMed] [Google Scholar]
- 3.Rudd M.K., Willard H.F. Analysis of the centromeric regions of the human genome assembly. Trends Genet. 2004;20:529–533. doi: 10.1016/j.tig.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Pluta A.F., Mackay A.M., Ainsztein A.M., Goldberg I.G., Earnshaw W.C. The centromere: hub of chromosomal activities. Science. 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- 5.Sullivan B.A., Blower M.D., Karpen G.H. Determining centromere identity: cyclical stories and forking paths. Nature Rev. Genet. 2001;2:584–596. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- 6.Cleveland D.W., Mao Y., Sullivan K.F. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- 7.Amor D.J., Choo K.H. Neocentromeres: role in human disease, evolution, and centromere study. Am. J. Hum. Genet. 2002;71:695–714. doi: 10.1086/342730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saffery R., Choo K.H. Strategies for engineering human chromosomes with therapeutic potential. J. Gene Med. 2002;4:5–13. doi: 10.1002/jgm.236. [DOI] [PubMed] [Google Scholar]
- 9.Grimes B.R., Rhoades A.A., Willard H.F. Alpha-satellite DNA and vector composition influence rates of human artificial chromosome formation. Mol. Ther. 2002;5:798–805. doi: 10.1006/mthe.2002.0612. [DOI] [PubMed] [Google Scholar]
- 10.Grimes B.R., Warburton P.E., Farr C.J. Chromosome engineering: prospects for gene therapy. Gene Ther. 2002;9:713–718. doi: 10.1038/sj.gt.3301763. [DOI] [PubMed] [Google Scholar]
- 11.Willard H.F. Neocentromeres and human artificial chromosomes: an unnatural act. Proc. Natl Acad. Sci. USA. 2001;98:5374–5376. doi: 10.1073/pnas.111167398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrington J.J., Van Bokkelen G., Mays R.W., Gustashaw K., Willard H.F. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nature Genet. 1997;15:345–355. doi: 10.1038/ng0497-345. [DOI] [PubMed] [Google Scholar]
- 13.Willard H.F. Genomics and gene therapy. Artificial chromosomes coming to life. Science. 2000;290:1308–1309. doi: 10.1126/science.290.5495.1308. [DOI] [PubMed] [Google Scholar]
- 14.Mejia J.E., Willmott A., Levy E., Earnshaw W.C., Larin Z. Functional complementation of a genetic deficiency with human artificial chromosomes. Am. J. Hum. Genet. 2001;69:315–326. doi: 10.1086/321977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimes B.R., Schindelhauer D., McGill N.I., Ross A., Ebersole T.A., Cooke H.J. Stable gene expression from a mammalian artificial chromosome. EMBO Rep. 2001;2:910–914. doi: 10.1093/embo-reports/kve187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ikeno M., Inagaki H., Nagata K., Morita M., Ichinose H., Okazaki T. Generation of human artificial chromosomes expressing naturally controlled guanosine triphosphate cyclohydrolase I gene. Genes Cells. 2002;7:1021–1032. doi: 10.1046/j.1365-2443.2002.00580.x. [DOI] [PubMed] [Google Scholar]
- 17.Rudd M.K., Mays R.W., Schwartz S., Willard H.F. Human artificial chromosomes with alpha-satellite-based de novo centromeres show increased frequency of nondisjunction and anaphase lag. Mol. Cell. Biol. 2003;23:7689–7697. doi: 10.1128/MCB.23.21.7689-7697.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohzeki J., Nakano M., Okada T., Masumoto H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 2002;159:765–775. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schueler M.G., Higgins A.W., Rudd M.K., Gustashaw K., Willard H.F. Genomic and genetic definition of a functional human centromere. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- 20.Ikeno M., Grimes B., Okazaki T., Nakano M., Saitoh K., Hoshino H., McGill N.I., Cooke H., Masumoto H. Construction of YAC-based mammalian artificial chromosomes. Nat. Biotechnol. 1998;16:431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- 21.Barnett M.A., Buckle V.J., Evans E.P., Porter A.C., Rout D., Smith A.G., Brown W.R. Telomere directed fragmentation of mammalian chromosomes. Nucleic Acids Res. 1993;21:27–36. doi: 10.1093/nar/21.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larin Z., Mejia J.E. Advances in human artificial chromosome technology. Trends Genet. 2002;18:313–319. doi: 10.1016/S0168-9525(02)02679-3. [DOI] [PubMed] [Google Scholar]
- 23.Ebersole T.A., Ross A., Clark E., McGill N., Schindelhauer D., Cooke H., Grimes B. Mammalian artificial chromosome formation from circular alphoid input DNA does not require telomere repeats. Hum. Mol. Genet. 2000;9:1623–1631. doi: 10.1093/hmg/9.11.1623. [DOI] [PubMed] [Google Scholar]
- 24.Mejia J.E., Alazami A., Willmott A., Marschall P., Levy E., Earnshaw W.C., Larin Z. Efficiency of de novo centromere formation in human artificial chromosomes. Genomics. 2002;79:297–304. doi: 10.1006/geno.2002.6704. [DOI] [PubMed] [Google Scholar]
- 25.Masumoto H., Ikeno M., Nakano M., Okazaki T., Grimes B., Cooke H., Suzuki N. Assay of centromere function using a human artificial chromosome. Chromosoma. 1998;107:406–416. doi: 10.1007/s004120050324. [DOI] [PubMed] [Google Scholar]
- 26.Muro Y., Masumoto H., Yoda K., Nozaki N., Ohashi M., Okazaki T. Centromere protein B assembles human centromeric alpha-satellite DNA at the 17-bp sequence, CENP-B box. J. Cell Biol. 1992;116:585–596. doi: 10.1083/jcb.116.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooke C.A., Bernat R.L., Earnshaw W.C. CENP-B: a major human centromere protein located beneath the kinetochore. J. Cell Biol. 1990;110:1475–1488. doi: 10.1083/jcb.110.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waye J.S., Willard H.F. Structure, organization, and sequence of alpha-satellite DNA from human chromosome 17: evidence for evolution by unequal crossing-over and an ancestral pentamer repeat shared with the human X chromosome. Mol. Cell. Biol. 1986;6:3156–3165. doi: 10.1128/mcb.6.9.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masumoto H., Masukata H., Muro Y., Nozaki N., Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 1989;109:1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Earnshaw W.C., Sullivan K.F., Machlin P.S., Cooke C.A., Kaiser D.A., Pollard T.D., Rothfield N.F., Cleveland D.W. Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J. Cell Biol. 1987;104:817–829. doi: 10.1083/jcb.104.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goryshin I.Y., Reznikoff W.S. Tn5 in vitro transposition. J. Biol. Chem. 1998;273:7367–7374. doi: 10.1074/jbc.273.13.7367. [DOI] [PubMed] [Google Scholar]
- 32.Shizuya H., Birren B., Kim U.J., Mancino V., Slepak T., Tachiiri Y., Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl Acad. Sci. USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullivan B.A., Schwartz S. Identification of centromeric antigens in dicentric Robertsonian translocations: CENP-C and CENP-E are necessary components of functional centromeres. Hum. Mol. Genet. 1995;4:2189–2197. doi: 10.1093/hmg/4.12.2189. [DOI] [PubMed] [Google Scholar]
- 34.Kouprina N., Ebersole T., Koriabine M., Pak E., Rogozin I.B., Katoh M., Oshimura M., Ogi K., Peredelchuk M., Solomon G., et al. Cloning of human centromeres by transformation-associated recombination in yeast and generation of functional human artificial chromosomes. Nucleic Acids Res. 2003;31:922–934. doi: 10.1093/nar/gkg182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okabe J., Eguchi A., Masago A., Hayakawa T., Nakanishi M. TRF1 is a critical trans-acting factor required for de novo telomere formation in human cells. Hum. Mol. Genet. 2000;9:2639–2650. doi: 10.1093/hmg/9.18.2639. [DOI] [PubMed] [Google Scholar]
- 36.Alexandrov I., Kazakov A., Tumeneva I., Shepelev V., Yurov Y. Alpha-satellite DNA of primates: old and new families. Chromosoma. 2001;110:253–266. doi: 10.1007/s004120100146. [DOI] [PubMed] [Google Scholar]
- 37.Kipling D., Warburton P.E. Centromeres, CENP-B and Tigger too. Trends Genet. 1997;13:141–145. doi: 10.1016/s0168-9525(97)01098-6. [DOI] [PubMed] [Google Scholar]
- 38.Goldberg I.G., Sawhney H., Pluta A.F., Warburton P.E., Earnshaw W.C. Surprising deficiency of CENP-B binding sites in African green monkey alpha-satellite DNA: implications for CENP-B function at centromeres. Mol. Cell. Biol. 1996;16:5156–5168. doi: 10.1128/mcb.16.9.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kapoor M., Montes de Oca Luna R., Liu G., Lozano G., Cummings C., Mancini M., Ouspenski I., Brinkley B.R., May G.S. The cenpB gene is not essential in mice. Chromosoma. 1998;107:570–576. doi: 10.1007/s004120050343. [DOI] [PubMed] [Google Scholar]
- 40.Perez-Castro A.V., Shamanski F.L., Meneses J.J., Lovato T.L., Vogel K.G., Moyzis R.K., Pedersen R. Centromeric protein B null mice are viable with no apparent abnormalities. Dev. Biol. 1998;201:135–143. doi: 10.1006/dbio.1998.9005. [DOI] [PubMed] [Google Scholar]
- 41.Hudson D.F., Fowler K.J., Earle E., Saffery R., Kalitsis P., Trowell H., Hill J., Wreford N.G., de Kretser D.M., Cancilla M.R., et al. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J. Cell Biol. 1998;141:309–319. doi: 10.1083/jcb.141.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Irelan J.T., Gutkin G.I., Clarke L. Functional redundancies, distinct localizations and interactions among three fission yeast homologs of centromere protein-B. Genetics. 2001;157:1191–1203. doi: 10.1093/genetics/157.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choo K.H. Centromere DNA dynamics: latent centromeres and neocentromere formation. Am. J. Hum. Genet. 1997;61:1225–1233. doi: 10.1086/301657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoda K., Ando S., Okuda A., Kikuchi A., Okazaki T. In vitro assembly of the CENP-B/alpha-satellite DNA/core histone complex: CENP-B causes nucleosome positioning. Genes Cells. 1998;3:533–548. doi: 10.1046/j.1365-2443.1998.00210.x. [DOI] [PubMed] [Google Scholar]
- 45.Warburton P.E., Waye J.S., Willard H.F. Nonrandom localization of recombination events in human alpha-satellite repeat unit variants: implications for higher-order structural characteristics within centromeric heterochromatin. Mol. Cell. Biol. 1993;13:6520–6529. doi: 10.1128/mcb.13.10.6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choo K.H. Engineering human chromosomes for gene therapy studies. Trends Mol. Med. 2001;7:235–237. doi: 10.1016/s1471-4914(01)01951-7. [DOI] [PubMed] [Google Scholar]
- 47.Prazeres D.M., Monteiro G.A., Ferreira G.N., Diogo M.M., Ribeiro S.C., Cabral J.M. Purification of plasmids for gene therapy and DNA vaccination. Biotechnol. Annu. Rev. 2001;7:1–30. doi: 10.1016/s1387-2656(01)07031-4. [DOI] [PubMed] [Google Scholar]
- 48.Somia N., Verma I.M. Gene therapy: trials and tribulations. Nature Rev. Genet. 2000;1:91–99. doi: 10.1038/35038533. [DOI] [PubMed] [Google Scholar]
- 49.Pannell D., Ellis J. Silencing of gene expression: implications for design of retrovirus vectors. Rev. Med. Virol. 2001;11:205–217. doi: 10.1002/rmv.316. [DOI] [PubMed] [Google Scholar]
- 50.Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E., et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 51.Kaname T., Huxley C. Isolation and subcloning of large fragments from BACs and PACs. Biotechniques. 2001;273:276–278. doi: 10.2144/01312bm07. [DOI] [PubMed] [Google Scholar]
- 52.Kaname T., Huxley C. Simple and efficient vectors for retrofitting BACs and PACs with mammalian neoR and EGFP marker genes. Gene. 2001;266:147–153. doi: 10.1016/s0378-1119(01)00375-4. [DOI] [PubMed] [Google Scholar]
- 53.Kim S.Y., Horrigan S.K., Altenhofen J.L., Arbieva Z.H., Hoffman R., Westbrook C.A. Modification of bacterial artificial chromosome clones using Cre recombinase: introduction of selectable markers for expression in eukaryotic cells. Genome Res. 1998;8:404–412. doi: 10.1101/gr.8.4.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mejia J.E., Larin Z. The assembly of large BACs by in vivo recombination. Genomics. 2000;70:165–170. doi: 10.1006/geno.2000.6372. [DOI] [PubMed] [Google Scholar]
- 55.Sosio M., Bossi E., Donadio S. Assembly of large genomic segments in artificial chromosomes by homologous recombination in Escherichia coli. Nucleic Acids Res. 2001;29:e37. doi: 10.1093/nar/29.7.e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jessen J.R., Meng A., McFarlane R.J., Paw B.H., Zon L.I., Smith G.R., Lin S. Modification of bacterial artificial chromosomes through chi-stimulated homologous recombination and its application in zebrafish transgenesis. Proc. Natl Acad. Sci. USA. 1998;95:5121–5126. doi: 10.1073/pnas.95.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]