Abstract
Listeria monocytogenes (Lm) is a Gram-positive facultative intracellular pathogen. Infections in humans can lead to listeriosis, a systemic disease with a high mortality rate. One important mechanism of Lm dissemination involves cell-to-cell spread after bacteria have entered the cytosol of host cells. Listeriolysin O (LLO; encoded by the hly gene) is a virulence factor present in Lm that plays a central role in the cell-to-cell spread process. LLO is a member of the cholesterol-dependent cytolysin (CDC) family of toxins that were initially thought to promote disease largely by inducing cell death and tissue destruction—essentially acting like a ‘bazooka’. This view was supported by structural studies showing CDCs can form large pores in membranes. However, it is now appreciated that LLO has many subtle activities during Lm infection of host cells, and many of these likely do not involve large pores, but rather small membrane perforations. It is also appreciated that membrane repair pathways of host cells play a major role in limiting membrane damage by LLO and other toxins. LLO is now thought to represent a ‘Swiss army knife’, a versatile tool that allows Lm to induce many membrane alterations and cellular responses that promote bacterial dissemination during infection.
This article is part of the themed issue ‘Membrane pores: from structure and assembly, to medicine and technology’.
Keywords: bacterial infection, cholesterol-dependent cytolysin, Listeria monocytogenes, listeriolysin O
1. The many faces of listeriolysin O function
As our understanding of listeriolysin O (LLO) expands, so too does the number of functions ascribed to this toxin. LLO was previously thought to be ‘phagosome-specific lysin’, since it plays a key role in phagosome escape by bacteria [1]. However, it is now appreciated that LLO can impact host cells from several locations: the extracellular medium, the phagosome and the cytosol. In all environments, LLO has important functions that are thought to promote infection of the host by Listeria monocytogenes (Lm).
(a). Extracellular listeriolysin O activities
(i). Bacterial internalization
Automated fluorescence-based assays revealed that LLO was sufficient to direct bacterial internalization into HepG2 cells. Coating non-invasive bacteria or polystyrene beads with LLO promoted their internalization into phagosomes, which rapidly acquired the early endosome marker EEA1 [2]. Calcium and potassium fluxes across the plasma membrane were shown to be required for this LLO-dependent internalization [3,4].
(ii). Activation of host signalling pathways
The transient cytosolic calcium elevation following LLO pore formation is responsible for the induction of multiple signalling pathways in the host cell [5–7]. To date these include activation of ERK-1, ERK-2, p38, c-Jun and Raf-MEK-MAP kinases pathways [8–10], phosphatidylinositol metabolism [11,12], nuclear translocation of NF-κB and secretion of the proinflammatory cytokines IL-6, IL-8, GM-CSF and IL-1α [5–7,13]. Activation of tyrosine kinases was shown to promote internalization of bacteria via LLO [2].
(iii). Apoptosis
Lm has been shown to induce apoptosis of several cell types during infection [14–16]. Relevant to the immune response, LLO was found to mediate rapid apoptosis of lymphocytes both in vitro and in vivo [17]. Treatment with subnanomolar doses of LLO was sufficient (in the absence of bacteria) to induce apoptosis by caspase-dependent and -independent pathways. LLO-mediated apoptosis increases susceptibility to Lm infection due to upregulation of IL-10, an anti-inflammatory cytokine [18].
(b). Listeriolysin O functions in the phagosome
Lm pathogenesis requires escape from the phagosome/vacuole (these terms are used interchangeably) and entry into the host cell cytosol. Early electron microscopy (EM) studies demonstrated that Lm mutants lacking LLO (Δhly) were restricted to the vacuole and avirulent in vivo [19–23]. Since then, phagosome escape has become the most well-established function of LLO. It is now clear that phagosome escape is a dynamic process accompanied by multiple host cell responses and is accomplished by only a minority of internalized bacteria (estimated 14%) [24]. During cell-to-cell spread, LLO also plays a role in escape from double-membrane compartments, referred to as spreading vacuoles, in neighbouring cells.
(i). Graded perforations of the phagosomal membrane
The previous model for LLO function was based on its homology and predicted structural similarity to other cholesterol-dependent cytolysin (CDC) toxins that generated 23–26 nm pores [25]. However, live cell imaging studies by Joel Swanson and colleagues showed that LLO can induce a graded series of much smaller membrane perforations in the phagosomal membrane during infection [26–28]. In these studies, small fluorescent molecules were internalized passively during Lm phagocytosis by macrophages. Leakage of the fluorescent molecules from phagosomes into the cytosol occurred in a size-dependent manner (e.g. leakage of Lucifer Yellow (522 Da) preceded Dextran Texas Red (10 000 Da) during phagosome maturation). Remarkably, leakage of protons and calcium was observed under conditions where small molecules were retained in phagosomes. Since proton and calcium accumulation in phagosomes is required for their fusion with lysosomes, Dr Swanson's group proposed a model whereby small perforations of the phagosome (with channel-like activity) create a ‘window of opportunity’ for other bacterial and host factors to promote phagosome escape.
(ii). Managing reactive oxygen species
Production of reactive oxygen species (ROS) by the phagocyte NOX2 NADPH oxidase is a well-established anti-microbial defence system against Lm. The virulence attenuation seen in Δhly infections can be partially recovered in a NADPH oxidase knockout (Cybb/NOX2−/−) background [29], demonstrating a role for LLO in managing ROS. In agreement, Δhly mutants had elevated levels of intracellular ROS localized to the phagosome [29]. It is unclear how LLO pore formation limits ROS production in Lm-containing phagosomes.
(iii). Growth in phagosomes and establishment of chronic infection
Lm was shown to induce a chronic infection in severe combined immunodeficiency (SCID) mice [30]. At 28 days post-infection, the host was able to contain the bacteria within spacious vacuolar structures, limiting their cytosolic growth and cell-to-cell spread in liver phagocytes. This landmark study by Unanue and colleagues established that Lm, previously considered a ‘cytosol-adapted pathogen’, could also colonize vacuoles during infection of host cells in vivo.
Using SCID mice and macrophage cell lines in vitro, we characterized the population of Lm that can grow in spacious Listeria-containing phagosomes (SLAPs) [31]. We observed that SLAPs are large, non-degradative compartments with a neutral pH and a single delimiting membrane that stains positively for several markers, including LAMP1, LC3-B and the proton ATPase. SLAPs contain multiple Lm, and bacteria were found to grow in these compartments with a doubling time of approximately 8 h (versus 40 min for Lm in the cytosol).
LLO production was both necessary and sufficient for the formation of SLAPs during infection. We found that a bacterial mutant expressing lower amounts of LLO (approximately one-third of the normal haemolytic activity) did not escape phagosomes but were able to grow in SLAPs over a delayed time course [31]. The mechanism by which LLO facilitates SLAP formation requires further investigation. Other pathogens may also use pore-forming toxins to colonize phagosomes during infection of host cells.
(iv). Inducing autophagy
Damaged cellular compartments can act as intracellular danger signals and trigger autophagy [32,33]. It is known that components of the autophagy pathway can limit Lm infection in vivo in both mice and flies [34,35] and that autophagy is activated during Lm infection of host cells in vitro under some conditions [36]. LLO is sufficient for lipidation of the autophagy protein LC3 and its recruitment to Lm [37]. Thus, LLO-mediated damage of phagosomes may be sufficient to induce an autophagic response. Despite this, autophagy does not seem to impact bacterial growth in host cells, and several strategies for bacterial evasion of autophagic killing have been described [38–42]. The relationship between Lm and autophagy continues to be explored. Moving forward, it must be borne in mind that Lm interactions with host autophagy are strain-specific [43].
(c). Listeriolysin O functions in the cytosol of host cells
(i). Mitochondrial fragmentation
Infection with LLO-competent Lm leads to a transient calcium-dependent burst of mitochondrial network fragmentation [44]. This corresponded to a drop in respiration and cellular ATP levels. Disrupting mitochondrial fission or fusion was found to inhibit intracellular growth of Lm [44]. The mechanism for how mitochondrial fission and fusion events impact Lm pathogenesis remains to be explored.
(ii). Endoplasmic reticulum stress and the unfolded protein response
LLO-mediated damage to the endoplasmic reticulum (ER), the main site for intracellular calcium storage, was shown to be a source of calcium elevation during infection [45]. The presence of LLO also led to induction of the unfolded protein response (UPR) [46]. Activation of the UPR does not benefit Lm, and artificial induction of ER stress reduced bacterial intracellular growth [46].
(iii). Protein degradation
Through unknown mechanisms, LLO pore formation promotes degradation of several host proteins [47–49]. During Lm infection there is a reduction in the number of proteins undergoing SUMOlyation; the reversible addition of SUMO, a ubiquitin-like polypeptide [49]. This was due to LLO-dependent degradation of Ubc9, a key enzyme in the SUMO pathway. Although Ubc9 degradation was not calcium dependent, degradation of the DNA breaks sensor Mre11 and human telomerase reverse transcriptase (hTERT) were [47,48]. It is likely that other yet unidentified host proteins are targeted for degradation by LLO-dependent mechanisms.
(iv). Inflammasome activation
Lm infection activates the NLRP3, AIM2, NALP3, IPAF and NLRC4 inflammasomes leading to activation of caspase-1, maturation of IL-1β and IL-18, and pyroptosis [50–54]. It is unclear whether the reduced caspase-1 activity observed with Δhly infection is the result of pore formation or the absence of Lm in the cytosol [50,54,55]. In support of the latter possibility, the presence of Lm DNA and flagellin in the cytosol triggers caspase-1 activation [56] as does cytosolic bacterial lysis [57]. Both models could exist simultaneously as purified LLO and cytosolic Lm could separately activate the NLRP3 inflammasome in human peripheral blood mononuclear cells [53].
(v). Cell-to-cell spread
Lm that enter the cytosol express ActA, a cell-surface protein that interacts with actin-regulatory factors from the host cell to promote actin-based motility [58]. Motile bacteria can induce cell surface filopodia-like structures (called protrusions) that can lead to subsequent spread to neighbouring cells. We recently showed that LLO activity in protrusions can cause localized plasma membrane damage, visualized by the exofacial exposure of phosphatidylserine (PS); normally localized exclusively to the inner leaflet of the plasma membrane [59]. In macrophages, this loss of membrane asymmetry promoted association of protrusions with neighbouring cells through the PS-binding receptor TIM-4 and enhanced cell-to-cell spread by bacteria. TIM-4 plays an important role in efferocytosis, the clearance of dead/dying cells in tissues, and was linked to innate immunity to Mycobacterium tuberculosis [60]. Therefore, Lm can exploit efferocytosis through LLO-mediated plasma membrane damage to promote its own cell-to-cell spread.
(d). Pore-independent functions of listeriolysin O
Most of the functions attributed to LLO appear to be by-products of either pore formation or entry of Lm into the cytosol. However, there are some functions of LLO that seem to be independent of its role in pore formation.
(i). Listeriolysin O as an immune antigen
LLO acts as an immune antigen: LLO-specific CD8+ T cells are protective against Lm infection [61,62]. In support of this activity being distinct from its role in pore formation, mutations that render LLO non-haemolytic do not affect its antigenicity [63]. In fact, non-haemolytic forms of LLO have proved useful as adjuvants in tumour immunotherapy [64].
(ii). Histone modification
Lm infection causes dephosphorylation of histone H3 and deacetylation of histone H4, impacting the expression of 146 genes [65]. The effect was dependent on LLO membrane binding but not pore formation. Other CDCs, but not membrane permeabilizing detergents, could similarly induce histone modifications, indicating the effect is specific to CDC–membrane interactions.
(e). Summary of listeriolysin O functions
The diversity of LLO-dependent effects on the host cell is remarkable and highlights the importance of this virulence factor to Lm pathogenesis. Many of LLO's biological impacts stem from its ability to drive an influx of calcium across the plasma membrane and/or release of calcium from intracellular stores during Lm infection [45]. LLO-dependent vacuolar escape elicits activation of immune and host signalling pathways as bacteria move from one intracellular niche to another. As described above (§1a–d), LLO also has functions that are not linked to its pore-forming activity. Importantly, LLO activity is not unrestricted—the ‘bazooka’ does not just kill everything in its vicinity. Otherwise, host cells would not be able to survive infection by hundreds of bacteria, which is routinely observed. Instead, LLO activity is highly regulated by both Lm and host cellular processes.
2. Regulation of listeriolysin O activity during infection
(a). Regulation of listeriolysin O by Listeria monocytogenes
(i). Transcription of hly
Expression of hly was quantified in bacteria trapped at each stage of infection: the primary vacuole (using a Δhly mutant), the cytosol (using a non-spreading ΔactA mutant), and in spreading vacuoles (using a ΔplcB mutant that cannot escape spreading vacuoles) [66]. This and other studies revealed robust hly expression regardless of the infection stage [66–69]. However, these studies are limited in that they use population-based measurements. Infection of J774 and Caco2 cells revealed heterogeneous hly expression that was not seen with actA, iap or inlC reporters [66], indicating that Lm may exploit heterogeneous expression of LLO during its interaction with host cells.
PrfA is the most well-characterized transcription factor required for hly expression [70,71]. prfA is transcribed from three promoters (P1–3). Basal transcription from P1 and P2 appears sufficient to drive primary vacuole escape [72]. Cell adherence is sufficient to induce prfA expression which is further amplified once intracellular [73,74]. P3, a bi-cistronic plcA-prfA promoter, is part of a positive auto-regulatory loop that increases PrfA levels in the host cytosol [72]. The PrfA regulon is most strongly activated following entry into the host cell cytosol [66].
Environmental cues regulating prfA (and by extension hly) expression include ROS [75], pH [76], sugar availability [72,77] and branched chain amino acids [78]. PrfA translation is thermally regulated. At non-permissive temperatures (30°C), the prfA 5′ untranslated region (UTR) forms a non-permissive secondary structure that is relieved following a shift to 37°C [79–81]. The UTR is also constrained by two S-adenosylmethionine (SAM) riboswitches SreA and SreB [82].
PrfA exists in two functional states: a state of low activity and of high activity following interaction with an unidentified cofactor [83]. Bacterial and host glutathione were recently shown to activate PrfA activity during infection of host cells [84]. A PrfA mutant (PrfA*) locked in the high activity state was sufficient to bypass the requirement of glutathione during infection. PrfA* mutants do not cause a virulence defect and, when used in vitro, more closely resemble expression seen in vivo [72]. Using a cell-wall binding domain fluorescent probe and Rab7 localization, it was shown that PrfA* did not affect phagosome escape for up to 8 h post-infection in J774 cells [85]. Overall, these findings demonstrate that expression of hly is not limited to the vacuole and, given its membership in the PrfA regulon, is actively transcribed in the cytosol.
(ii). Secretion of listeriolysin O
LLO is transported across the bacterial membrane by the Sec secretion system. In line with the need for LLO activity during phagosome escape, phagosome trapped (Δhly) Lm showed high expression of Sec secretion system components [86]. The Sec accessory proteins SecD and SecF and the post-translational secretion chaperone PrsA2 are required for both proper secretion and activity of LLO [86–89]. Proper LLO secretion and function also rely on cleavage of the Sec secretion signal after translocation by the signal peptidase SipZ. Expression of sipZ increases in the phagosome and, albeit at lower levels, continues to be expressed in the cytosol [90]. Deletion of sipZ decreased LLO secretion and reduced haemolytic activity by fivefold [91]. Secretion of LLO during cell-to-cell spread has yet to be investigated.
(b). Regulation of listeriolysin O by the host cell
(i). Reduction/oxidation of listeriolysin O
LLO requires reduction at Cys485 for activation in vitro [92]. The thiol oxidoreductase GILT has been shown to reduce LLO in the phagosome, promoting its activity and the subsequent entry of Lm into the cytosol [93]. Lm has a reduced ability to escape the phagosome in GILT−/− bone marrow-derived macrophages [93]. To our knowledge, no thiol oxidoreductases in the cytosol have been identified as regulating LLO activity, though the naturally reducing environment provided by the glutaredoxin and thioredoxin systems of the mammalian cytosol may negate the need for additional regulators [94,95].
Oxidation of LLO may also limit its activity in the phagosome. ROS (produced by the NOX2 NADPH oxidase) and reactive nitrogen species (produced by iNOS) have been proposed to inactivate LLO, thereby limiting phagosome escape by Lm [96]. Whether LLO is oxidized by host cellular factors in the cytosol has not been investigated (figure 1).
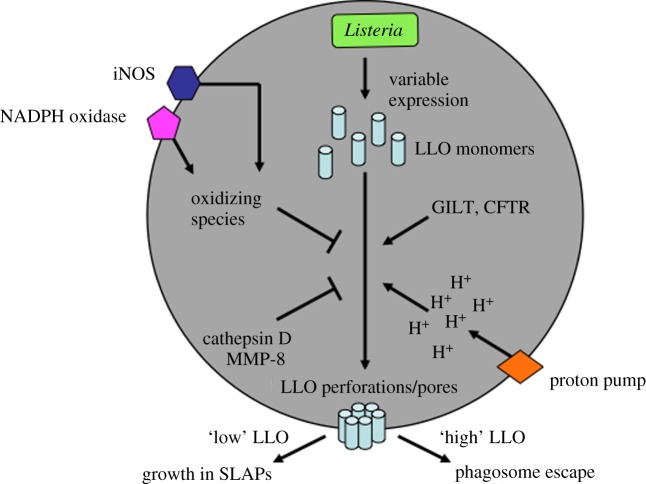
Figure 1.
Factors impacting LLO activity in the phagosome. Individual Listeria monocytogenes produce and secrete variable amounts of LLO monomers, depending on many intrinsic and extrinsic factors they encounter prior to uptake by host cells. Thus, in the lumen of the phagosome, variable amounts of LLO have the capability of binding to cholesterol (and possibly other factors) on the luminal face of the phagosomal membrane. Before that binding can occur, many factors in the phagosome impact LLO. Reduction of LLO by GILT, chloride delivery to the phagosome by CFTR and acidification of the phagosome by the vacuolar ATPase (proton pump) all promote LLO activity. In contrast, oxidizing species generated by iNOS and the NOX2 NADPH oxidase and proteolytic cleavage of LLO by cathepsin D and MMP-8 all inhibit LLO activity in the phagosome. Together, all of these factors impact the amount (and likely also the type) of LLO activity at the phagosomal membrane. Relatively ‘high’ activity of LLO is thought to promote escape of bacteria from the phagosome, in combination with other bacterial and host factors (e.g. phospholipases). In contrast, ‘low’ activity of LLO has been shown to allow bacterial growth in phagosomes (SLAPs).
(ii). Chloride promotes listeriolysin O oligomerization
LLO oligomerization depends on chloride availability [97–100]. Although cellular chloride levels vary from cell to cell, chloride levels are higher in phagosomes relative to the cytosol, suggesting a tendency to form higher-order LLO oligomers in the vacuole. In neutrophils, it was shown that there is a chloride influx into the phagosome lumen such that the concentration approaches 70 mM in contrast to the 40–50 mM seen in the cytosol [101,102]. Inhibition of the chloride transporter cystic fibrosis transmembrane conductance regulator (CFTR) decreased Lm vacuole escape [100]. The role of other chloride transporters in Lm pathogenesis has not been explored.
(iii). Proteolytic degradation of listeriolysin O in the phagosome
Cathepsin D was found to cleave LLO during Lm infection of fibroblasts and macrophages [103,104]. In neutrophils, LLO can also be degraded prior to phagosome closure at the plasma membrane by the matrix metalloproteinase-8 [105]. Also, some α-defenins appear to limit CDC activity in the phagosome [106] though their expression is cell-type dependent.
(iv). The proline, glutamic acid, serine and threonine sequence and listeriolysin O degradation by the ubiquitin–proteasome system
LLO stability in the host cell cytosol is impacted by proteolytic degradation mechanisms which impact the ability of Lm to cause infection. While sufficient levels of LLO are required to infect host cells and promote cell-to-cell spread, abnormally high levels of LLO (caused by loss of intrinsic regulatory mechanisms described below in §2b(v–vii)) are linked to cellular toxicity and clearance of extracellular bacteria by innate immune mechanisms, including killing by neutrophils and inflammatory monocytes.
The N-terminus of LLO, unlike other CDCs, is rich in proline, glutamate, serine and threonine (PEST) residues [1,107]. Deletion of this PEST sequence does not affect LLO haemolytic activity but does attenuate Lm virulence [108,109]. Although there are conflicting reports on whether ΔPEST mutants can escape the phagosome [108,109], studies agree that ΔPEST mutants have greater cytotoxicity. Changes to the PEST nucleotide sequence in LLO (but not the amino acid sequence) were found to increase LLO expression, potentially contributing to this greater toxicity [110].
The LLO PEST domain contains three putative mitogen-activated protein kinase phosphorylation sites of which mutation of Ser44 phenocopies ΔPEST (increased cytosolic toxicity) [108,111]. Inhibition of the proteasome leads to an accumulation of LLO, indicating that LLO is degraded in the host cell. This accumulation was reversed with phosphatase treatment, suggesting phosphorylation was linked to degradation [111]. However, neither the PEST sequence nor Ser44 were necessary for proteasomal degradation of LLO. Immunoprecipitation experiments have further demonstrated that LLO is ubiquitinated, which may contribute to protein turnover [111]. Although it is clear that the PEST sequence plays a role in limiting LLO activity in the phagosome, our understanding of the mechanism remains incomplete. Structural studies indicated that the PEST sequence interacts with the adjacent symmetry-related molecule in the crystal lattice which could point to a role in oligomerization [112]. Transmission EM showed that wild-type and ΔPEST mutant pores looked different with an increase in the number of incomplete arcs in crowded rows in the latter [112].
LLO has been shown to be degraded by the N-end rule pathway. Mutation of the N-terminal Lys in LLO led to increased cellular toxicity in J774 macrophages, but overall had only a minor impact on virulence [113].
(v). pH dependence
The role of pH in limiting LLO pore formation is the best recognized form of LLO activity modulation [1,97,114]. Blocking acidification of the phagosome has been shown to completely block phagosome escape by Lm [26]. Structural studies have revealed an inhibition of LLO activity at neutral pH. In addition to the acidic residues E247, D208 and D320 previously identified as the pH sensor, the structure of LLO revealed multiple pH-sensitive clusters that interact both directly and indirectly through bound Na+ and H2O [97,112]. Deprotonation of these critical residues leads to charge repulsion, unfolding and aggregation of regions within D3, a region involved in oligomerization and ring formation [112,115]. However, pH regulation of LLO appears more complex than originally thought and many groups do report LLO membrane binding at neutral pH [2,26,116]. Haemolytic activity of LLO is also normal after a rapid shift from acidic to neutral pH, albeit this activity is short-lived as the protein begins to aggregate [116–119]. These findings are consistent with the ability of LLO to impact cellular membranes in the cytosol of host cell (discussed in §1), but in a limited manner that typically does not cause host cell rupture.
(vi). Monomer diffusion
LLO activity requires assembly of monomers into oligomeric structures on membranes. In the confines of a phagosome, this process would be relatively efficient since monomers are released into a single compartment with high access to their target membrane. In the cytosol, monomer diffusion is expected to be higher in the larger volume of this gel-like, expansive compartment and potentially limited further by movement of the bacteria through actin-based motility. While this is not a specific host regulation of LLO, it is nonetheless a factor impacting LLO activity in the cytosol that must be considered.
(vii). Plasma membrane repair pathways
A number of cellular pathways promote integrity of the plasma membrane and mediate resistance to bacterial toxins [120,121]. LLO mediates damage to the plasma membrane during Lm infection, and this damage was shown to be limited by caspase-7 activity [122]. Macrophages deficient in caspase-7 had increased plasma membrane permeability and deficient intracellular growth. Additionally, members of the annexin family of membrane repair proteins were found to limit plasma membrane damage by LLO during Lm infection of HeLa cells [59].
3. Structural insights into listeriolysin O activity
Lm pathogenicity relies heavily on LLO pore formation and the aforementioned host responses to this toxin. For many years our mechanistic understanding of LLO relied on extrapolation from related CDCs but recent structural and atomic force microscopy (AFM) analysis has provided new insight into LLO function as it relates to membrane perforation.
(a). Structure of listeriolysin O
X-Ray crystallography studies revealed that the LLO monomer is an elongated, four domain structure (D1–D4) with strong structural resemblance to other CDCs [123–126]. D1, D2 and D3 form the LLO core, whereas D4 extends away from the core. D4 contains the highly conserved undecapeptide sequence (ECTGLAWEWWR) and loops (L1–L3) and is required for cholesterol recognition and membrane binding [112]. Despite strong conservation of D4, LLO carries more polar residues in L2 and a more neutral charge, compared to the negatively charged PFO. The transmembrane pore constitutes a β-barrel where each monomer contributes two transmembrane β-hairpins derived from α-helices in D3 [112,127,128].
(b). Membrane binding and pore formation
(i). Cholesterol recognition
Mutagenesis analysis on perfringolysin O (PFO), streptolysin O and pneumolysin (PLY) revealed that two conserved residues in loop 1 of D4, corresponding to Thr515 and Leu516 on LLO, were essential for cholesterol binding and haemolytic activity [129]. Although the insolubility of cholesterol has limited our structural understanding of its recognition by CDCs, the 3-β-hydroxyl group appears important since epicholesterol, an isomer that differs only in the orientation of the 3-β-hydroxyl group, was not bound by PFO [130]. Subsequent to cholesterol recognition, PFO becomes anchored to the membrane by insertion of the undecapeptide, and loops L2 and L3 [115].
(ii). Listeriolysin O oligomerization
Following membrane attachment, CDC monomers oligomerize into a pre-pore complex that extends 113 Å above the membrane [131,132]. The oligomeric interface is located in D3, with β1 of one monomer binding to β4 of the neighbouring monomer. PFO has poor spontaneous aggregation because β4 is normally shielded by β5 [115]. Membrane binding caused a conformational shift that reoriented β5, exposing the oligomeric interface [115]. LLO shows strong charge complementarity between D1 and D3 of neighbouring monomers. Inverting residue charges along these regions abolished LLO activity and prevented oligomeric ring formation; instead granular protein complexes or discontinuous protein fibres were formed [112].
AFM has recently enabled visualization of LLO oligomers. LLO formed arcs, slits and rings in a cholesterol- and time-dependent manner that progressively fused to form larger rings [133]. Other groups failed to observe ring structures using high-speed AFM but instead visualized rapid arc formation that stalled after incorporation of 20 monomers. Multiple arcs then annealed to form larger, ring-like, oligomers [134].
(iii). The pre-pore to pore transition
Formation of ring-like LLO pre-pores precedes membrane insertion. There are conflicting reports on the degree of LLO oligomerization required for transition from the pre-pore to pore. One group observed membrane insertion of arcs and slits followed by further oligomerization [133]. Others observed membrane insertion only with higher-order oligomers with no further oligomerization following pore formation [134].
The transition from the pre-pore to pore state involves a vertical collapse of 40 Å with large conformational changes as the central α-helices convert to transmembrane β-hairpins [25,135]. Cryo-EM maps of the PLY pore show a variable 320–430 Å pore diameter [25]. CDC oligomers appear to have variable pore sizes, with 35–47 subunits forming the pore. These size differentials support the observation that multiple arcs appear to anneal to form a ring-like structure rather than a single oligomeric ring [134].
(c). Listeriolysin O lineactivity
Recent AFM observed that in addition to its membrane pore formation activity, LLO had subsequent lineactivity: LLO could cause further, large-scale membrane damage from the membrane edge [134]. Such an activity could be unique to CDCs as lineactant activity was not observed with the aerolysin-like pore-forming toxin lysenin. Lineactant activity requires a membrane edge and may act to enlarge existing LLO pores. This is consistent with the previously discussed finding of graded pore formation where small fluorescent dextrans moved across membranes rapidly following LLO treatment and, over time, larger dextrans subsequently transversed the membrane.
4. Conclusion
It is clear from the wealth of studies on LLO that its activity cannot be simplified as an on–off switch. Based on the research discussed above, it is becoming clear that LLO has at least two modes of action: membrane perforation/lineactivity and the formation of large pores. This versatility allows LLO to promote its many functions in different environments during infection of its host. The ability of LLO (and other CDC) monomers to undergo conformational changes upon membrane binding appears to be the key feature that links these activities. In other words, the ‘moving parts’ of LLO allow it to do many things in different places. The bacterial and host factors that control LLO activities are now recognized as critical determinants in both the initiation of LLO-associated phenotypes and the survival of host cells during the infection. Membrane repair pathways in particular are likely to be important determinants of the outcome of infection by Lm and other bacteria expressing CDC toxins. LLO is clearly not behaving as an unrestricted ‘bazooka’, even under the most severe (and artificial) infection conditions seen in vitro. Rather, LLO is now recognized as a precise tool used by Lm to modulate host cellular pathways in a manner that promotes infection. Indeed, Cossart and colleagues have referred to LLO as a ‘Swiss army knife’, a fitting analogy based on our new appreciation of LLO as a multi-functional tool used by Lm to promote infection. We anticipate that LLO has a great deal more to teach us about bacterial pathogenesis and host innate immune responses to infection.
Competing interests
We declare we have no competing interests.
Funding
Our studies of Listeria pathogenesis are supported by an operating grant (MOP no. 136973) and postdoctoral fellowship (to S.E.O.) from the Canadian Institutes of Health Research.
References
- 1.Schnupf P, Portnoy DA. 2007. Listeriolysin O: a phagosome-specific lysin. Microbes Infect. 9, 1176–1187. ( 10.1016/j.micinf.2007.05.005) [DOI] [PubMed] [Google Scholar]
- 2.Vadia S, Arnett E, Haghighat AC, Wilson-Kubalek EM, Tweten RK, Seveau S. 2011. The pore-forming toxin listeriolysin O mediates a novel entry pathway of L. monocytogenes into human hepatocytes. PLoS Pathog. 7, e1002356 ( 10.1371/journal.ppat.1002356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vadia S, Seveau S. 2014. Fluxes of Ca2+ and K+ are required for the listeriolysin O-dependent internalization pathway of Listeria monocytogenes. Infect. Immun. 82, 1084–1091. ( 10.1128/IAI.01067-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dramsi S, Cossart P. 2003. Listeriolysin O-mediated calcium influx potentiates entry of Listeria monocytogenes into the human Hep-2 epithelial cell line. Infect. Immun. 71, 3614–3618. ( 10.1128/IAI.71.6.3614-3618.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dewamitta SR, et al. 2010. Listeriolysin O-dependent bacterial entry into the cytoplasm is required for calpain activation and interleukin-1α secretion in macrophages infected with Listeria monocytogenes. Infect. Immun. 78, 1884–1894. ( 10.1128/IAI.01143-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rose F, Zeller SA, Chakraborty T, Domann E, Machleidt T, Kronke M, Seeger W, Grimminger F, Sibelius U. 2001. Human endothelial cell activation and mediator release in response to Listeria monocytogenes virulence factors. Infect. Immun. 69, 897–905. ( 10.1128/IAI.69.2.897-905.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsuchiya K, Kawamura I, Takahashi A, Nomura T, Kohda C, Mitsuyama M. 2005. Listeriolysin O-induced membrane permeation mediates persistent interleukin-6 production in Caco-2 cells during Listeria monocytogenes infection in vitro. Infect. Immun. 73, 3869–3877. ( 10.1128/IAI.73.7.3869-3877.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiglein I, Goebel W, Troppmair J, Rapp UR, Demuth A, Kuhn M. 1997. Listeria monocytogenes infection of HeLa cells results in listeriolysin O-mediated transient activation of the Raf-MEK-MAP kinase pathway. FEMS Microbiol. Lett. 148, 189–195. ( 10.1111/j.1574-6968.1997.tb10287.x) [DOI] [PubMed] [Google Scholar]
- 9.Tang P, Sutherland CL, Gold MR, Finlay BB. 1998. Listeria monocytogenes invasion of epithelial cells requires the MEK-1/ERK-2 mitogen-activated protein kinase pathway. Infect. Immun. 66, 1106–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang P, Rosenshine I, Cossart P, Finlay BB. 1996. Listeriolysin O activates mitogen-activated protein kinase in eucaryotic cells. Infect. Immun. 64, 2359–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sibelius U, et al. 1996. Listeriolysin is a potent inducer of the phosphatidylinositol response and lipid mediator generation in human endothelial cells. Infect. Immun. 64, 674–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sibelius U, et al. 1996. The listerial exotoxins listeriolysin and phosphatidylinositol-specific phospholipase C synergize to elicit endothelial cell phosphoinositide metabolism. J. Immunol. 157, 4055–4060. [PubMed] [Google Scholar]
- 13.Kayal S, Lilienbaum A, Poyart C, Memet S, Israel A, Berche P. 1999. Listeriolysin O-dependent activation of endothelial cells during infection with Listeria monocytogenes: activation of NF-κB and upregulation of adhesion molecules and chemokines. Mol. Microbiol. 31, 1709–1722. ( 10.1046/j.1365-2958.1999.01305.x) [DOI] [PubMed] [Google Scholar]
- 14.Rogers HW, Callery MP, Deck B, Unanue ER. 1996. Listeria monocytogenes induces apoptosis of infected hepatocytes. J. Immunol. 156, 679–684. [PubMed] [Google Scholar]
- 15.Guzmán CA, Domann E, Rohde M, Bruder D, Darji A, Weiss S, Wehland J, Chakraborty T, Timmis KN. 1996. Apoptosis of mouse dendritic cells is triggered by listeriolysin, the major virulence determinant of Listeria monocytogenes. Mol. Microbiol. 20, 119–126. ( 10.1111/j.1365-2958.1996.tb02494.x) [DOI] [PubMed] [Google Scholar]
- 16.Valenti P, Greco R, Pitari G, Rossi P, Ajello M, Melino G, Antonini G. 1999. Apoptosis of Caco-2 intestinal cells invaded by Listeria monocytogenes: protective effect of lactoferrin. Exp. Cell Res. 250, 197–202. ( 10.1006/excr.1999.4500) [DOI] [PubMed] [Google Scholar]
- 17.Carrero JA, Calderon B, Unanue ER. 2004. Listeriolysin O from Listeria monocytogenes is a lymphocyte apoptogenic molecule. J. Immunol. 172, 4866–4874. ( 10.4049/jimmunol.172.8.4866) [DOI] [PubMed] [Google Scholar]
- 18.Carrero JA, Unanue ER. 2012. Mechanisms and immunological effects of apoptosis caused by Listeria monocytogenes. Adv. Immunol. 113, 157–174. ( 10.1016/B978-0-12-394590-7.00001-4) [DOI] [PubMed] [Google Scholar]
- 19.Gaillard JL, Berche P, Sansonetti P. 1986. Transposon mutagenesis as a tool to study the role of hemolysin in the virulence of Listeria monocytogenes. Infect. Immun. 52, 50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaillard JL, Berche P, Mounier J, Richard S, Sansonetti P. 1987. In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect. Immun. 55, 2822–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Portnoy DA, Jacks PS, Hinrichs DJ. 1988. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J. Exp. Med. 167, 1459–1471. ( 10.1084/jem.167.4.1459) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cossart P, Vicente MF, Mengaud J, Baquero F, Perez-Diaz JC, Berche P. 1989. Listeriolysin O is essential for virulence of Listeria monocytogenes: direct evidence obtained by gene complementation. Infect. Immun. 57, 3629–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kathariou S, Metz P, Hof H, Goebel W. 1987. Tn916-induced mutations in the hemolysin determinant affecting virulence of Listeria monocytogenes. J. Bacteriol. 169, 1291–1297. ( 10.1128/jb.169.3.1291-1297.1987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Chastellier C, Berche P. 1994. Fate of Listeria monocytogenes in murine macrophages: evidence for simultaneous killing and survival of intracellular bacteria. Infect. Immun. 62, 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tilley SJ, Orlova EV, Gilbert RJ, Andrew PW, Saibil HR. 2005. Structural basis of pore formation by the bacterial toxin pneumolysin. Cell 121, 247–256. ( 10.1016/j.cell.2005.02.033) [DOI] [PubMed] [Google Scholar]
- 26.Beauregard KE, Lee KD, Collier RJ, Swanson JA. 1997. pH-dependent perforation of macrophage phagosomes by listeriolysin O from Listeria monocytogenes. J. Exp. Med. 186, 1159–1163. ( 10.1084/jem.186.7.1159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henry R, Shaughnessy L, Loessner MJ, Alberti-Segui C, Higgins DE, Swanson JA. 2006. Cytolysin-dependent delay of vacuole maturation in macrophages infected with Listeria monocytogenes. Cell. Microbiol. 8, 107–119. ( 10.1111/j.1462-5822.2005.00604.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaughnessy LM, Hoppe AD, Christensen KA, Swanson JA. 2006. Membrane perforations inhibit lysosome fusion by altering pH and calcium in Listeria monocytogenes vacuoles. Cell. Microbiol. 8, 781–792. ( 10.1111/j.1462-5822.2005.00665.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam GY, Fattouh R, Muise AM, Grinstein S, Higgins DE, Brumell JH. 2011. Listeriolysin O suppresses phospholipase C-mediated activation of the microbicidal NADPH oxidase to promote Listeria monocytogenes infection. Cell Host Microbe 10, 627–634. ( 10.1016/j.chom.2011.11.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhardwaj V, Kanagawa O, Swanson PE, Unanue ER. 1998. Chronic Listeria infection in SCID mice: requirements for the carrier state and the dual role of T cells in transferring protection or suppression. J. Immunol. 160, 376–384. [PubMed] [Google Scholar]
- 31.Birmingham CL, Canadien V, Kaniuk NA, Steinberg BE, Higgins DE, Brumell JH. 2008. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles. Nature 451, 350–354. ( 10.1038/nature06479) [DOI] [PubMed] [Google Scholar]
- 32.Kloft N, Neukirch C, Bobkiewicz W, Veerachato G, Busch T, von Hoven G, Boller K, Husmann M. 2010. Pro-autophagic signal induction by bacterial pore-forming toxins. Med. Microbiol. Immunol. 199, 299–309. ( 10.1007/s00430-010-0163-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. 2012. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482, 414–418. ( 10.1038/nature10744) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yano T, et al. 2008. Autophagic control of Listeria through intracellular innate immune recognition in Drosophila. Nat. Immunol. 9, 908–916. ( 10.1038/ni.1634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Z, et al. 2008. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 4, 458–469. ( 10.1016/j.chom.2008.10.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rich KA, Burkett C, Webster P. 2003. Cytoplasmic bacteria can be targets for autophagy. Cell. Microbiol. 5, 455–468. ( 10.1046/j.1462-5822.2003.00292.x) [DOI] [PubMed] [Google Scholar]
- 37.Meyer-Morse N, et al. 2010. Listeriolysin O is necessary and sufficient to induce autophagy during Listeria monocytogenes infection. PLoS ONE 5, e8610 ( 10.1371/journal.pone.0008610) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Birmingham CL, Canadien V, Gouin E, Troy EB, Yoshimori T, Cossart P, Higgins DE, Brumell JH. 2007. Listeria monocytogenes evades killing by autophagy during colonization of host cells. Autophagy 3, 442–451. ( 10.4161/auto.4450) [DOI] [PubMed] [Google Scholar]
- 39.Mitchell G, Ge L, Huang Q, Chen C, Kianian S, Roberts MF, Schekman R, Portnoy DA. 2015. Avoidance of autophagy mediated by PlcA or ActA is required for Listeria monocytogenes growth in macrophages. Infect. Immun. 83, 2175–2184. ( 10.1128/IAI.00110-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tattoli I, Sorbara MT, Yang C, Tooze SA, Philpott DJ, Girardin SE. 2013. Listeria phospholipases subvert host autophagic defenses by stalling pre-autophagosomal structures. EMBO J. 32, 3066–3078. ( 10.1038/emboj.2013.234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshikawa Y, et al. 2009. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat. Cell Biol. 11, 1233–1240. ( 10.1038/ncb1967) [DOI] [PubMed] [Google Scholar]
- 42.Dortet L, et al. 2011. Recruitment of the major vault protein by InlK: a Listeria monocytogenes strategy to avoid autophagy. PLoS Pathog. 7, e1002168 ( 10.1371/journal.ppat.1002168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cemma M, Lam GY, Stöckli M, Higgins DE, Brumell JH. 2015. Strain-specific interactions of Listeria monocytogenes with the autophagy system in host cells. PLoS ONE. 10, e0125856 ( 10.1371/journal.pone.0125856) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stavru F, Bouillaud F, Sartori A, Ricquier D, Cossart P. 2011. Listeria monocytogenes transiently alters mitochondrial dynamics during infection. Proc. Natl Acad. Sci. USA 108, 3612–3617. ( 10.1073/pnas.1100126108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gekara NO, Westphal K, Ma B, Rohde M, Groebe L, Weiss S. 2007. The multiple mechanisms of Ca2+ signalling by listeriolysin O, the cholesterol-dependent cytolysin of Listeria monocytogenes. Cell. Microbiol. 9, 2008–2021. ( 10.1111/j.1462-5822.2007.00932.x) [DOI] [PubMed] [Google Scholar]
- 46.Pillich H, Loose M, Zimmer KP, Chakraborty T. 2012. Activation of the unfolded protein response by Listeria monocytogenes. Cell. Microbiol. 14, 949–964. ( 10.1111/j.1462-5822.2012.01769.x) [DOI] [PubMed] [Google Scholar]
- 47.Samba-Louaka A, Stavru F, Cossart P. 2012. Role for telomerase in Listeria monocytogenes infection. Infect. Immun. 80, 4257–4263. ( 10.1128/IAI.00614-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Samba-Louaka A, Pereira JM, Nahori MA, Villiers V, Deriano L, Hamon MA, Cossart P, Valdivia RH. 2014. Listeria monocytogenes dampens the DNA damage response. PLoS Pathog. 10, e1004470 ( 10.1371/journal.ppat.1004470) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ribet D, et al. 2010. Listeria monocytogenes impairs SUMOylation for efficient infection. Nature 464, 1192–1195. ( 10.1038/nature08963) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim S, Bauernfeind F, Ablasser A, Hartmann G, Fitzgerald KA, Latz E, Hornung V. 2010. Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. Eur. J. Immunol. 40, 1545–1551. ( 10.1002/eji.201040425) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rathinam VA, et al. 2010. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 11, 395–402. ( 10.1038/ni.1864) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eitel J, Suttorp N, Opitz B. 2010. Innate immune recognition and inflammasome activation in Listeria monocytogenes infection. Front Microbiol. 1, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meixenberger K, et al. 2010. Listeria monocytogenes-infected human peripheral blood mononuclear cells produce IL-1β, depending on listeriolysin O and NLRP3. J. Immunol. 184, 922–930. ( 10.4049/jimmunol.0901346) [DOI] [PubMed] [Google Scholar]
- 54.Warren SE, Mao DP, Rodriguez AE, Miao EA, Aderem A. 2008. Multiple Nod-like receptors activate caspase 1 during Listeria monocytogenes infection. J. Immunol. 180, 7558–7564. ( 10.4049/jimmunol.180.11.7558) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hara H, Tsuchiya K, Nomura T, Kawamura I, Shoma S, Mitsuyama M. 2008. Dependency of caspase-1 activation induced in macrophages by Listeria monocytogenes on cytolysin, listeriolysin O, after evasion from phagosome into the cytoplasm. J. Immunol. 180, 7859–7868. ( 10.4049/jimmunol.180.12.7859) [DOI] [PubMed] [Google Scholar]
- 56.Wu J, Fernandes-Alnemri T, Alnemri ES. 2010. Involvement of the AIM2, NLRC4, and NLRP3 inflammasomes in caspase-1 activation by Listeria monocytogenes. J. Clin. Immunol. 30, 693–702. ( 10.1007/s10875-010-9425-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sauer JD, Witte CE, Zemansky J, Hanson B, Lauer P, Portnoy DA. 2010. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe 7, 412–419. ( 10.1016/j.chom.2010.04.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Portnoy DA, Auerbuch V, Glomski IJ. 2002. The cell biology of Listeria monocytogenes infection: the intersection of bacterial pathogenesis and cell-mediated immunity. J. Cell Biol. 158, 409–414. ( 10.1083/jcb.200205009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Czuczman MA, et al. 2014. Listeria monocytogenes exploits efferocytosis to promote cell-to-cell spread. Nature 509, 230–234. ( 10.1038/nature13168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin CJ, et al. 2012. Efferocytosis is an innate antibacterial mechanism. Cell Host Microbe 12, 289–300. ( 10.1016/j.chom.2012.06.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pamer EG, Harty JT, Bevan MJ. 1991. Precise prediction of a dominant class I MHC-restricted epitope of Listeria monocytogenes. Nature 353, 852–855. ( 10.1038/353852a0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harty JT, Bevan MJ. 1992. CD8+ T cells specific for a single nonamer epitope of Listeria monocytogenes are protective in vivo. J. Exp. Med. 175, 1531–1538. ( 10.1084/jem.175.6.1531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carrero JA, Vivanco-Cid H, Unanue ER. 2012. Listeriolysin O is strongly immunogenic independently of its cytotoxic activity. PLoS ONE 7, e32310 ( 10.1371/journal.pone.0032310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wallecha A, Wood L, Pan ZK, Maciag PC, Shahabi V, Paterson Y. 2013. Listeria monocytogenes-derived listeriolysin O has pathogen-associated molecular pattern-like properties independent of its hemolytic ability. Clin. Vaccine Immunol. 20, 77–84. ( 10.1128/CVI.00488-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hamon MA, et al. 2007. Histone modifications induced by a family of bacterial toxins. Proc. Natl Acad. Sci. USA 104, 13 467–13 472. ( 10.1073/pnas.0702729104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bubert A, Sokolovic Z, Chun SK, Papatheodorou L, Simm A, Goebel W. 1999. Differential expression of Listeria monocytogenes virulence genes in mammalian host cells. Mol. Gen. Genet. 261, 323–336. [DOI] [PubMed] [Google Scholar]
- 67.Chatterjee SS, et al. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74, 1323–1338. ( 10.1128/IAI.74.2.1323-1338.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Joseph B, Przybilla K, Stühler C, Schauer K, Slaghuis J, Fuchs TM. 2006. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J. Bacteriol. 188, 556–568. ( 10.1128/JB.188.2.556-568.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Camejo A, et al. 2009. In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog. 5, e1000449 ( 10.1371/journal.ppat.1000449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chakraborty T, Leimeister-Wächter M, Domann E, Hartl M, Goebel W, Nichterlein T, Notermans S. 1992. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 174, 568–574. ( 10.1128/jb.174.2.568-574.1992) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.de las Heras A, Cain RJ, Bielecka MK, Vázquez-Boland JA. 2011. Regulation of Listeria virulence: PrfA master and commander. Curr. Opin. Microbiol. 14, 118–127. ( 10.1016/j.mib.2011.01.005) [DOI] [PubMed] [Google Scholar]
- 72.Scortti M, Monzó HJ, Lacharme-Lora L, Lewis DA, Vázquez-Boland JA. 2007. The PrfA virulence regulon. Microbes Infect. 9, 1196–1207. ( 10.1016/j.micinf.2007.05.007) [DOI] [PubMed] [Google Scholar]
- 73.Renzoni A, Cossart P, Dramsi S. 1999. PrfA, the transcriptional activator of virulence genes, is upregulated during interaction of Listeria monocytogenes with mammalian cells and in eukaryotic cell extracts. Mol. Microbiol. 34, 552–561. ( 10.1046/j.1365-2958.1999.01621.x) [DOI] [PubMed] [Google Scholar]
- 74.Moors MA, Levitt B, Youngman P, Portnoy DA. 1999. Expression of listeriolysin O and ActA by intracellular and extracellular Listeria monocytogenes. Infect. Immun. 67, 131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Makino M, Kawai M, Kawamura I, Fujita M, Gejo F, Mitsuyama M. 2005. Involvement of reactive oxygen intermediate in the enhanced expression of virulence-associated genes of Listeria monocytogenes inside activated macrophages. Microbiol. Immunol. 49, 805–811. ( 10.1111/j.1348-0421.2005.tb03661.x) [DOI] [PubMed] [Google Scholar]
- 76.Behari J, Youngman P. 1998. A homolog of CcpA mediates catabolite control in Listeria monocytogenes but not carbon source regulation of virulence genes. J. Bacteriol. 180, 6316–6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ripio MT, Brehm K, Lara M, Suárez M, Vázquez-Boland JA. 1997. Glucose-1-phosphate utilization by Listeria monocytogenes is PrfA dependent and coordinately expressed with virulence factors. J. Bacteriol. 179, 7174–7180. ( 10.1128/jb.179.22.7174-7180.1997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sonenshein AL. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 8, 203–207. ( 10.1016/j.mib.2005.01.001) [DOI] [PubMed] [Google Scholar]
- 79.Loh E, Memarpour F, Vaitkevicius K, Kallipolitis BH, Johansson J, Sondén B. 2012. An unstructured 5'-coding region of the prfA mRNA is required for efficient translation. Nucleic Acids Res. 40, 1818–1827. ( 10.1093/nar/gkr850) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110, 551–561. ( 10.1016/S0092-8674(02)00905-4) [DOI] [PubMed] [Google Scholar]
- 81.Leimeister-Wächter M, Domann E, Chakraborty T. 1992. The expression of virulence genes in Listeria monocytogenes is thermoregulated. J. Bacteriol. 174, 947–952. ( 10.1128/jb.174.3.947-952.1992) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Loh E, et al. 2009. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell 139, 770–779. ( 10.1016/j.cell.2009.08.046) [DOI] [PubMed] [Google Scholar]
- 83.Renzoni A, Klarsfeld A, Dramsi S, Cossart P. 1997. Evidence that PrfA, the pleiotropic activator of virulence genes in Listeria monocytogenes, can be present but inactive. Infect. Immun. 65, 1515–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reniere ML, Whiteley AT, Hamilton KL, John SM, Lauer P, Brennan RG. 2015. Glutathione activates virulence gene expression of an intracellular pathogen. Nature 517, 170–173. ( 10.1038/nature14029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Deshayes C, et al. 2012. Allosteric mutants show that PrfA activation is dispensable for vacuole escape but required for efficient spread and Listeria survival in vivo. Mol. Microbiol. 85, 461–477. ( 10.1111/j.1365-2958.2012.08121.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Burg-Golani T, Pozniak Y, Rabinovich L, Sigal N, Nir Paz R, Herskovits AA. 2013. Membrane chaperone SecDF plays a role in the secretion of Listeria monocytogenes major virulence factors. J. Bacteriol. 195, 5262–5272. ( 10.1128/JB.00697-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alonzo F, Freitag NE. 2010. Listeria monocytogenes PrsA2 is required for virulence factor secretion and bacterial viability within the host cell cytosol. Infect. Immun. 78, 4944–4957. ( 10.1128/IAI.00532-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alonzo F, Xayarath B, Whisstock JC, Freitag NE. 2011. Functional analysis of the Listeria monocytogenes secretion chaperone PrsA2 and its multiple contributions to bacterial virulence. Mol. Microbiol. 80, 1530–1548. ( 10.1111/j.1365-2958.2011.07665.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Forster BM, Zemansky J, Portnoy DA, Marquis H. 2011. Posttranslocation chaperone PrsA2 regulates the maturation and secretion of Listeria monocytogenes proprotein virulence factors. J. Bacteriol. 193, 5961–5970. ( 10.1128/JB.05307-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Raynaud C, Charbit A. 2005. Regulation of expression of type I signal peptidases in Listeria monocytogenes. Microbiology 151, 3769–3776. ( 10.1099/mic.0.28066-0) [DOI] [PubMed] [Google Scholar]
- 91.Bonnemain C, Raynaud C, Réglier-Poupet H, Dubail I, Frehel C, Lety MA, Berche P, Charbit A. 2004. Differential roles of multiple signal peptidases in the virulence of Listeria monocytogenes. Mol. Microbiol. 51, 1251–1266. ( 10.1111/j.1365-2958.2004.03916.x) [DOI] [PubMed] [Google Scholar]
- 92.Michel E, Reich KA, Favier R, Berche P, Cossart P. 1990. Attenuated mutants of the intracellular bacterium Listeria monocytogenes obtained by single amino acid substitutions in listeriolysin O. Mol. Microbiol. 4, 2167–2178. ( 10.1111/j.1365-2958.1990.tb00578.x) [DOI] [PubMed] [Google Scholar]
- 93.Singh R, Jamieson A, Cresswell P. 2008. GILT is a critical host factor for Listeria monocytogenes infection. Nature 455, 1244–1247. ( 10.1038/nature07344) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Berndt C, Lillig CH, Holmgren A. 2007. Thiol-based mechanisms of the thioredoxin and glutaredoxin systems: implications for diseases in the cardiovascular system. Am. J. Physiol. Heart Circ. Physiol. 292, H1227–H1236. ( 10.1152/ajpheart.01162.2006) [DOI] [PubMed] [Google Scholar]
- 95.Holmgren A. 2000. Antioxidant function of thioredoxin and glutaredoxin systems. Antioxid. Redox Signal. 2, 811–820. ( 10.1089/ars.2000.2.4-811) [DOI] [PubMed] [Google Scholar]
- 96.Myers JT, Tsang AW, Swanson JA. 2003. Localized reactive oxygen and nitrogen intermediates inhibit escape of Listeria monocytogenes from vacuoles in activated macrophages. J. Immunol. 171, 5447–5453. ( 10.4049/jimmunol.171.10.5447) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schuerch DW, Wilson-Kubalek EM, Tweten RK. 2005. Molecular basis of listeriolysin O pH dependence. Proc. Natl Acad. Sci. USA 102, 12 537–12 542. ( 10.1073/pnas.0500558102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Walton CM, Wu CH, Wu GY. 1999. A method for purification of listeriolysin O from a hypersecretor strain of Listeria monocytogenes. Protein Expr. Purif. 15, 243–245. ( 10.1006/prep.1998.1022) [DOI] [PubMed] [Google Scholar]
- 99.Myers ER, Dallmier AW, Martin SE. 1993. Sodium chloride, potassium chloride, and virulence in Listeria monocytogenes. Appl. Environ. Microbiol. 59, 2082–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Radtke AL, Anderson KL, Davis MJ, DiMagno MJ, Swanson JA, O'Riordan MX. 2011. Listeria monocytogenes exploits cystic fibrosis transmembrane conductance regulator (CFTR) to escape the phagosome. Proc. Natl Acad. Sci. USA 108, 1633–1638. ( 10.1073/pnas.1013262108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Aiken ML, Painter RG, Zhou Y, Wang G. 2012. Chloride transport in functionally active phagosomes isolated from human neutrophils. Free Radic. Biol. Med. 53, 2308–2317. ( 10.1016/j.freeradbiomed.2012.10.542) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Painter RG, Marrero L, Lombard GA, Valentine VG, Nauseef WM, Wang G. 2010. CFTR-mediated halide transport in phagosomes of human neutrophils. J. Leukoc. Biol. 87, 933–942. ( 10.1189/jlb.1009655) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.del Cerro-Vadillo E, et al. 2006. Cutting edge: a novel nonoxidative phagosomal mechanism exerted by cathepsin-D controls Listeria monocytogenes intracellular growth. J. Immunol. 176, 1321–1325. ( 10.4049/jimmunol.176.3.1321) [DOI] [PubMed] [Google Scholar]
- 104.Carrasco-Marín E, et al. 2009. The innate immunity role of cathepsin-D is linked to Trp-491 and Trp-492 residues of listeriolysin O. Mol. Microbiol. 72, 668–682. ( 10.1111/j.1365-2958.2009.06673.x) [DOI] [PubMed] [Google Scholar]
- 105.Arnett E, Vadia S, Nackerman CC, Oghumu S, Satoskar AR, McLeish KR, Uriarte SM, Seveau S. 2014. The pore-forming toxin listeriolysin O is degraded by neutrophil metalloproteinase-8 and fails to mediate Listeria monocytogenes intracellular survival in neutrophils. J. Immunol. 192, 234–244. ( 10.4049/jimmunol.1301302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lehrer RI, et al. 2009. Human α-defensins inhibit hemolysis mediated by cholesterol-dependent cytolysins. Infect. Immun. 77, 4028–4040. ( 10.1128/IAI.00232-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rogers S, Wells R, Rechsteiner M. 1986. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science 234, 364–368. ( 10.1126/science.2876518) [DOI] [PubMed] [Google Scholar]
- 108.Decatur AL, Portnoy DA. 2000. A PEST-like sequence in listeriolysin O essential for Listeria monocytogenes pathogenicity. Science 290, 992–995. ( 10.1126/science.290.5493.992) [DOI] [PubMed] [Google Scholar]
- 109.Lety MA, Frehel C, Dubail I, Beretti JL, Kayal S, Berche P, Charbit A. 2001. Identification of a PEST-like motif in listeriolysin O required for phagosomal escape and for virulence in Listeria monocytogenes. Mol. Microbiol. 39, 1124–1139. ( 10.1111/j.1365-2958.2001.02281.x) [DOI] [PubMed] [Google Scholar]
- 110.Schnupf P, Hofmann J, Norseen J, Glomski IJ, Schwartzstein H, Decatur AL. 2006. Regulated translation of listeriolysin O controls virulence of Listeria monocytogenes. Mol. Microbiol. 61, 999–1012. ( 10.1111/j.1365-2958.2006.05286.x) [DOI] [PubMed] [Google Scholar]
- 111.Schnupf P, Portnoy DA, Decatur AL. 2006. Phosphorylation, ubiquitination and degradation of listeriolysin O in mammalian cells: role of the PEST-like sequence. Cell. Microbiol. 8, 353–364. ( 10.1111/j.1462-5822.2005.00631.x) [DOI] [PubMed] [Google Scholar]
- 112.Köster S, van Pee K, Hudel M, Leustik M, Rhinow D, Kühlbrandt W, Chakraborty T, Yildiz Ö. 2014. Crystal structure of listeriolysin O reveals molecular details of oligomerization and pore formation. Nat. Commun. 5, 3690 ( 10.1038/ncomms4690) [DOI] [PubMed] [Google Scholar]
- 113.Schnupf P, Zhou J, Varshavsky A, Portnoy DA. 2007. Listeriolysin O secreted by Listeria monocytogenes into the host cell cytosol is degraded by the N-end rule pathway. Infect. Immun. 75, 5135–5147. ( 10.1128/IAI.00164-07) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Podobnik M, Marchioretto M, Zanetti M, Bavdek A, Kisovec M, Lunelli L, Dalla Serra M, Anderluh G. 2015. Corrigendum: plasticity of listeriolysin O pores and its regulation by pH and unique histidine. Acad. Emerg. Med. 5, 15690 ( 10.1038/srep15690) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ramachandran R, Tweten RK, Johnson AE. 2004. Membrane-dependent conformational changes initiate cholesterol-dependent cytolysin oligomerization and intersubunit β-strand alignment. Nat. Struct. Mol. Biol. 11, 697–705. ( 10.1038/nsmb793) [DOI] [PubMed] [Google Scholar]
- 116.Bavdek A, et al. 2007. Sterol and pH interdependence in the binding, oligomerization, and pore formation of listeriolysin O. Biochemistry 46, 4425–4437. ( 10.1021/bi602497g) [DOI] [PubMed] [Google Scholar]
- 117.Nomura T, Kawamura I, Kohda C, Baba H, Ito Y, Kimoto T, Watanabe I, Mitsuyama M. 2007. Irreversible loss of membrane-binding activity of Listeria-derived cytolysins in non-acidic conditions: a distinct difference from allied cytolysins produced by other Gram-positive bacteria. Microbiology. 153, 2250–2258. ( 10.1099/mic.0.2007/005843-0) [DOI] [PubMed] [Google Scholar]
- 118.Bavdek A, Kostanjšek R, Antonini V, Lakey JH, Dalla Serra M, Gilbert RJ, Anderluh G. 2012. pH dependence of listeriolysin O aggregation and pore-forming ability. FEBS J. 279, 126–141. ( 10.1111/j.1742-4658.2011.08405.x) [DOI] [PubMed] [Google Scholar]
- 119.Podobnik M, Marchioretto M, Zanetti M, Bavdek A, Kisovec M, Cajnko MM. 2015. Plasticity of listeriolysin O pores and its regulation by pH and unique histidine [corrected]. Sci Rep. 5, 9623 ( 10.1038/srep09623) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Andrews NW, Almeida PE, Corrotte M. 2014. Damage control: cellular mechanisms of plasma membrane repair. Trends Cell Biol. 24, 734–742. ( 10.1016/j.tcb.2014.07.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Draeger A, Schoenauer R, Atanassoff AP, Wolfmeier H, Babiychuk EB. 2014. Dealing with damage: plasma membrane repair mechanisms. Biochimie. 107(Pt A), 66–72. ( 10.1016/j.biochi.2014.08.008) [DOI] [PubMed] [Google Scholar]
- 122.Cassidy SK, Hagar JA, Kanneganti TD, Franchi L, Nuñez G, O'Riordan MX. 2012. Membrane damage during Listeria monocytogenes infection triggers a caspase-7 dependent cytoprotective response. PLoS Pathog. 8, e1002628 ( 10.1371/journal.ppat.1002628) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rossjohn J, Feil SC, McKinstry WJ, Tweten RK, Parker MW. 1997. Structure of a cholesterol-binding, thiol-activated cytolysin and a model of its membrane form. Cell 89, 685–692. ( 10.1016/S0092-8674(00)80251-2) [DOI] [PubMed] [Google Scholar]
- 124.Bourdeau RW, et al. 2009. Cellular functions and X-ray structure of anthrolysin O, a cholesterol-dependent cytolysin secreted by Bacillus anthracis. J. Biol. Chem. 284, 14645–14656. ( 10.1074/jbc.M807631200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Polekhina G, Giddings KS, Tweten RK, Parker MW. 2005. Insights into the action of the superfamily of cholesterol-dependent cytolysins from studies of intermedilysin. Proc. Natl Acad. Sci. USA 102, 600–605. ( 10.1073/pnas.0403229101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Feil SC, Ascher DB, Kuiper MJ, Tweten RK, Parker MW. 2014. Structural studies of Streptococcus pyogenes streptolysin O provide insights into the early steps of membrane penetration. J. Mol. Biol. 426, 785–792. ( 10.1016/j.jmb.2013.11.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shatursky O, Heuck AP, Shepard LA, Rossjohn J, Parker MW, Johnson AE, Tweten RK. 1999. The mechanism of membrane insertion for a cholesterol-dependent cytolysin: a novel paradigm for pore-forming toxins. Cell 99, 293–299. ( 10.1016/S0092-8674(00)81660-8) [DOI] [PubMed] [Google Scholar]
- 128.Shepard LA, et al. 1998. Identification of a membrane-spanning domain of the thiol-activated pore-forming toxin Clostridium perfringens perfringolysin O: an α-helical to β-sheet transition identified by fluorescence spectroscopy. Biochemistry 37, 14563–14574. ( 10.1021/bi981452f) [DOI] [PubMed] [Google Scholar]
- 129.Farrand AJ, LaChapelle S, Hotze EM, Johnson AE, Tweten RK. 2010. Only two amino acids are essential for cytolytic toxin recognition of cholesterol at the membrane surface. Proc. Natl Acad. Sci. USA 107, 4341–4346. ( 10.1073/pnas.0911581107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Heuck AP, Savva CG, Holzenburg A, Johnson AE. 2007. Conformational changes that effect oligomerization and initiate pore formation are triggered throughout perfringolysin O upon binding to cholesterol. J. Biol. Chem. 282, 22 629–22 637. ( 10.1074/jbc.M703207200) [DOI] [PubMed] [Google Scholar]
- 131.Hotze EM, Wilson-Kubalek EM, Rossjohn J, Parker MW, Johnson AE, Tweten RK. 2001. Arresting pore formation of a cholesterol-dependent cytolysin by disulfide trapping synchronizes the insertion of the transmembrane β-sheet from a prepore intermediate. J. Biol. Chem. 276, 8261–8268. ( 10.1074/jbc.M009865200) [DOI] [PubMed] [Google Scholar]
- 132.Olofsson A, Hebert H, Thelestam M. 1993. The projection structure of perfringolysin O (Clostridium perfringens theta-toxin). FEBS Lett. 319, 125–127. ( 10.1016/0014-5793(93)80050-5) [DOI] [PubMed] [Google Scholar]
- 133.Mulvihill E, van Pee K, Mari SA, Müller DJ, Yildiz Ö. 2015. Directly observing the lipid-dependent self-assembly and pore-forming mechanism of the cytolytic toxin listeriolysin O. Nano Lett. 15, 6965–6973. ( 10.1021/acs.nanolett.5b02963) [DOI] [PubMed] [Google Scholar]
- 134.Ruan Y, Rezelj S, Bedina Zavec A, Anderluh G, Scheuring S. 2016. Listeriolysin O membrane damaging activity involves arc formation and lineaction—implication for Listeria monocytogenes escape from phagocytic vacuole. PLoS Pathog. 12, e1005597 ( 10.1371/journal.ppat.1005597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Czajkowsky DM, Hotze EM, Shao Z, Tweten RK. 2004. Vertical collapse of a cytolysin prepore moves its transmembrane β-hairpins to the membrane. EMBO J. 23, 3206–3215. ( 10.1038/sj.emboj.7600350) [DOI] [PMC free article] [PubMed] [Google Scholar]