Abstract
In-plane mobility of proteins in lipid membranes is one of the fundamental mechanisms supporting biological functionality. Here we use high-speed atomic force microscopy (HS-AFM) to show that a novel type of biomimetic channel—carbon nanotube porins (CNTPs)—is also laterally mobile in supported lipid membranes, mimicking biological protein behaviour. HS-AFM can capture real-time dynamics of CNTP motion in the supported lipid bilayer membrane, build long-term trajectories of the CNTP motion and determine the diffusion coefficients associated with this motion. Our analysis shows that diffusion coefficients of CNTPs fall into the same range as those of proteins in supported lipid membranes. CNTPs in HS-AFM experiments often exhibit ‘directed’ diffusion behaviour, which is common for proteins in live cell membranes.
This article is part of the themed issue ‘Membrane pores: from structure and assembly, to medicine and technology’.
Keywords: CNTPs, HS-AFM, supported lipid bilayer, 2D diffusion
1. Introduction
Lipid membranes represent one the fundamental components of the architecture of life because they provide a versatile matrix for a variety of membrane proteins that can perform a variety of tasks ranging from molecular recognition and signal transduction, to metabolite transport and membrane remodelling [1–3]. The 2D fluid nature of the lipid membrane not only allows it to adapt to a variety of shapes, but also permits membrane proteins to diffuse within this 2D plane, enabling a number of important biological processes [4]. For example, dimerization of receptor tyrosine kinases in the membrane allows auto-phosphorylation that acts as a switch in a number of cellular processes [5].
The heterogeneous and often crowded structure of biological membranes and membrane protein interactions with the cell motility apparatus causes membrane proteins to show complex dynamic behaviours that can span multiple length scales that can range from several nanometres to a few microns. Gross cell movement and evolution, protein confinement to lipid rafts, binding to extracellular matrix and other effects can complicate the membrane dynamics even further. As the result, diffusion coefficients of proteins in cellular membranes can differ from their mobility in model lipid membranes by orders of magnitude [6]. To understand the fundamental physics of protein motion in the lipid membrane, we need an approach that would combine simple and robust membrane protein models with imaging and tracking approaches that can follow membrane motion on the relevant length and time scales.
Most of the existing information about membrane protein motion came from experiments that used either gross ensemble-based assays, such as fluorescence recovery after photobleaching (FRAP) [7], or single-molecule techniques, such as single-particle optical tracking (SPT) [6]. Both of these techniques ultimately have to rely on tagging the proteins or membrane components either with fluorescent or scattering labels. These approaches have to contend with potential effects from additional drag caused by these often bulky attachments, as well as potential interactions of the labels with membrane and extracellular matrix components. In addition, photobleaching of fluorescent labels limits the duration of the tracking process. Recent development of high-speed AFM opened up the possibility of label-less tracking of membrane components at nm scale with 100 ms time resolution. Some of the pioneering examples included tracking membrane dynamics of OmpF proteins [8] and imaging the evolution of protein aggregates on living bacterial cell surfaces [9].
We have recently reported simple and versatile artificial membrane pore analogues—carbon nanotube porins (CNTPs)—short segments of single-wall carbon nanotubes that can self-insert into the lipid membrane and form a transmembrane pore [10]. Surprisingly, these very simple objects show a wealth of behaviours similar to membrane protein pores: they can transport water, ions and protons across the membrane [10] and exhibit ligand-gated blocking, and stochastic gating [10,11].
In this work, we demonstrate that CNTPs reproduce another key property of membrane proteins—their ability to diffuse in the lipid membrane. We show that high-speed AFM imaging can capture real-time dynamics of CNTP motion in the supported lipid bilayer membrane, build long-term trajectories of the CNTP motion and determine the diffusion coefficients associated with this motion. Our analysis also showed that CNTPs exhibit diffusion coefficients in the same range as other membrane proteins. Interestingly, CNTPs often exhibited ‘directed’ diffusion behaviour, common for proteins in live cell membranes, which we attributed to the interactions of the CNTPs with the HS-AFM probe.
2. Material and methods
(a). Carbon nanotube porin synthesis and incorporation into liposomes
The 0.8 nm diameter CNT were sourced from Sigma-Aldrich (Cat. No. 773735). 1,2-Dioleoyl-sn-glycero-3-phosphocholine lipid (DOPC) and 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) were purchased from Avanti Polar Lipids. The CNTP synthesis procedure followed the protocol that was described previously [12]. To incorporate CNTPs into the liposomes DOPC:DPPC (70 : 30 mass ratio), lipid was mixed in a glass vial and completely dried overnight in a vacuum desiccator chamber. An aliquot of CNTP stock solution was also dried overnight and then rehydrated with 20 mM HEPES buffer (pH 7.8). The resulting CNTP solution was used to hydrate the dried DOPC/DPPC mixture for 30 min at 40°C (the final lipid concentration was 1 mg ml−1) and then subjected to 10 cycles of freeze–thaw procedure to get rid of multi-lamellar structures. Subsequently, the liposomes with CNT were extruded 10 times through 200 nm polycarbonate membranes (Avanti Polar Lipids) to obtain liposomes with homogeneous size.
(b). Substrate preparation and liposome fusion
A mica disc with 1.5 mm in diameter was glued on the glass rod of the HS-AFM sample stage. All mica surfaces were freshly cleaved prior to sample deposition. To fuse the lipid bilayers onto the substrate surface, 4 μl of liposome solution was deposited on the mica surface and incubated for 2 h at 40°C. For this step, the sample was placed in a home-built humidity chamber and sealed with parafilm to reduce evaporation. After incubation, the unfused liposomes were rinsed away with 20 μl of 20 mM HEPES buffer (pH 7.8) five times.
(c). High-speed atomic force microscopy imaging
HS-AFM images of CNTPs were acquired in tapping mode at room temperature using an HS-AFM instrument (RIBM, Japan) equipped with ultra-short AFM cantilevers with high-density carbon/diamond-like carbon (HDC/DLC) tips (USC-F1.2-k0.15, NanoWorld, tip radius <10 nm). This instrument uses dynamic proportional-integral-differential (PID) controller [13,14] to eliminate probe ‘parachuting’ artefacts from images and reduce the tip-sample forces. The HS-AFM fluid cell was filled with 120 μl of 20 mM HEPES buffer. In a typical experiment, we collected 128 × 128 pixel images from a 200 × 200 nm area at a scan rate of 2 frames s−1. Images of CNTPs adsorbed on the lipid bilayer surface were recorded from smaller areas (typically 50 × 50 nm).
(d). Data processing and analysis
Raw HS-AFM image data were converted to ImageJ (developed by Wayne Rasband, National Institutes of Health, Bethesda, MD) stacks using custom software built with Matlab2015 (MathWorks, Natick, MA). The motion trajectories of CNTPs were extracted using TrackMate Plugin package in ImageJ (http://imagej.net/TrackMate). Image drift was corrected using the macro developed by Nicholas M. Schneider (https://github.com/NMSchneider/fixTranslation-Macro-for-ImageJ). The MSD values were calculated by a custom IgorPro 6 script (WaveMetrics, Lake Oswego, OR, USA), and the diffusion coefficients were obtained by the fitting the MSD trajectories to equation (3.1). The lipid and CNT models were created by the VMD software package [15].
3. Results and discussion
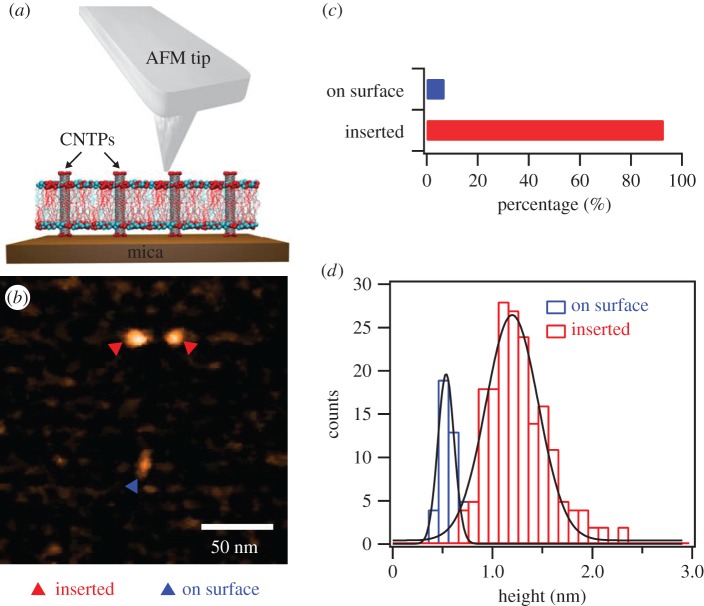
AFM imaging of CNTPs inserted in the bilayer is only possible in supported lipid bilayer geometry where the bilayer rests on a flat surface of mica (figure 1a). To create these supported bilayers, we have fused lipid vesicles, which contained pre-inserted 0.8 nm diameter CNTPs, onto the mica disc surface of the HS-AFM sample stage. After this procedure, AFM images showed that most of the mica surface was covered with the lipid bilayer (figure 1b). Occasionally, the coverage of the surface was incomplete, showing small regions of underlying mica surface (electronic supplementary material, figure S1a). Cross-sectional analysis of some of these areas showed that the thickness of the lipid bilayer was ca. 4 nm (electronic supplementary material, figure S1b), which agrees well with previous AFM investigations of the supported lipid bilayer morphology [16,17].
Figure 1.
(a) Schematics of the HS-AFM measurement showing the AFM tip scanning over CNTPs inserted into a lipid bilayer matrix supported on mica surface. (b) Representative AFM image showing CNTPs inserted into the lipid bilayer (red triangles) and adsorbed on the bilayer surface (blue triangle). (c) Histogram showing the fractions of CNTPs inserted into the bilayer (n = 184) and adsorbed on the bilayer surface (n = 14). (d) Histograms of the height of the CNTP features relative to the bilayer surface level (solid lines represent Gaussian fits to the data).
AFM images also reveal that CNTPs can adopt two main conformations in this system (figure 1b,c). A significant portion (93%) of the CNTPs appeared as sharp point-like protrusions on the surface of the bilayer. We assigned these features to the CNTPs that were inserted in the membrane and had their ends protrude above the bilayer by on average 1.3 ± 0.3 nm (figure 1d). Given the average length of the CNTPs of ca. 10 nm, which we reported in earlier publication [10,12], we believe that the images show strong evidence that CNTPs adopt a strongly tilted configuration in the bilayer due to hydrophobic mismatch, which also agrees with the observations from MD simulations [18–20]. A much smaller fraction (7%) of the CNTPs appeared as oblong objects on the bilayer surface that protruded only 0.6 ± 0.1 nm above the bilayer surface (figure 1d), which is consistent with the CNT diameter of 0.8 nm (we assume that the difference is due to the deformation of the lipid bilayer). On the basis of the statistical analysis (electronic supplementary material, figure S2a), the average height of the oblong features was significantly different from the height of the point-like features (Wilcoxon–Mann–Whitney test, p < 0.001), strongly suggesting that the oblong features represent the CNTPs non-specifically adsorbed on the surface of the lipid bilayer. Interestingly, the average length of these adsorbed CNTPs measured from the AFM images 24.3 ± 6.8 nm (electronic supplementary material, figure S2b) was significantly longer than our reported length of the membrane-inserted CNTPs (10.6 ± 0.9 nm) [12], supporting the conclusion that only a limited range of the CNTP lengths is capable of inserting into the lipid bilayer.
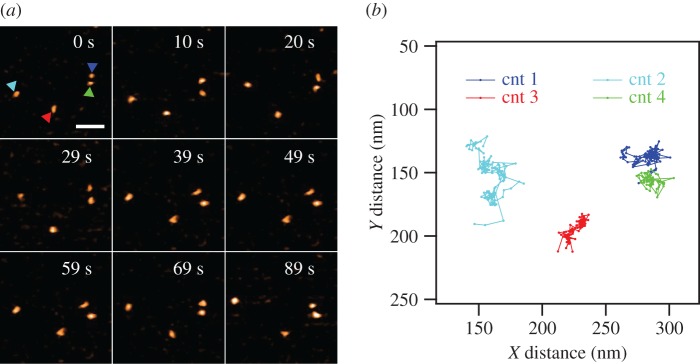
Significantly, HS-AFM movies collected at a rate of 2 fps showed clear evidence of CNTP lateral movement in the membrane (figure 2a). Most CNTPs showed continuous lateral motion with only a small percentage of CNTPs remaining stationary in the membrane. We note that we did not observe any significant signs of CNTP clustering. This behaviour probably originates from the presence of negative charges at the CNTPs ends that form when carboxylic acid groups, created during the sonication cutting of longer CNTs, ionize at neutral pH [10]. These charges create electrostatic repulsion that prevents the CNTP association and subsequent bundle formation.
Figure 2.
(a) Selected frames from a HS-AFM movie of CNTP diffusion in the supported lipid bilayer (see electronic supplementary information for the full movie). CNTPs are marked by coloured triangles. (b) Motion trajectories extracted from the HS-AFM movies of the four CNTPs shown in (a). The time interval between each step was 500 ms. The colour of each trajectory corresponds to the colour of the triangle marking in (a). The scale bar is 50 nm.
We used HS-AFM to follow the motion of more than 100 CNTPs in the lipid bilayer and then applied particle tracking algorithms (see Material and methods) to extract the time trajectories of CNTP motion (figure 2b). The CNTPs appear to follow a series of stochastic steps highly reminiscent of Brownian motion, which is expected for objects inserted into a 2D liquid environment of the lipid bilayer. However, the analysis of mean square displacement (MSD) reveals the somewhat more complicated character of the motion kinetics. MSD represents the measure of the distance that can be explored by a random walk process, and for an isotropic random walk the MSD expected to increase proportionally to the duration of the walk [21].
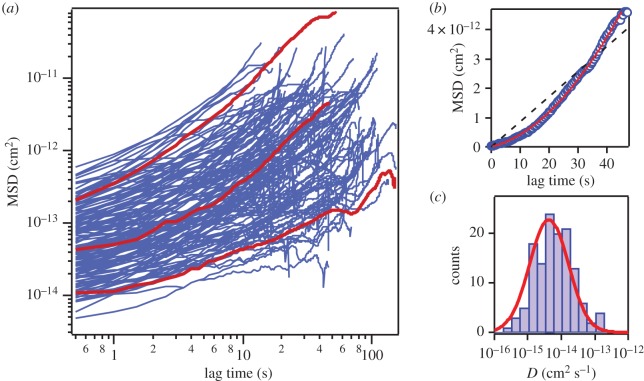
Curiously, a logarithmic scale plot of the large number of the MSD traces for CNTPs (figure 3a) shows clear systematic deviations from the expected linear dependence. Instead, the CNTPs follow the kinetics characteristic for the ‘directed’ diffusion type of motion, which combines isotropic random walk with a directed motion, where the MSD values follow a different dependence:
| 3.1 |
Equation (1) provides an excellent fit to the experimentally derived MSD traces (figure 3b, red solid line). In contrast, the classic Brownian motion dependence provides a universally poor fit (figure 3b, black dashed line). Interestingly, this directed diffusion pattern is reminiscent of the behaviour that is often exhibited by proteins residing in the cytoplasmic membranes of live cells [6,21]. However, in those systems the directed motion component is almost always associated with cell motility, or with protein interactions with other transport systems localized to the cytoskeleton. Clearly, none of those mechanisms are present in our system, thus the similarity is coincidental. We can attribute the directed motion component to the possible presence of small gross flow in the supported lipid bilayer effects of the AFM instrument drift (HS-AFM instrument does not allow closed-loop feedback scanning and thus cannot compensate for drift effectively). Even though we used a dynamic PID controller [13,14] in our work, we note that our sample can be challenging for AFM imaging. CNTPs have high aspect ratios and protrude above the bilayer plane (and often are tilted) [10,22]. Furthermore, we have reported that incorporation of CNTPs into the liposome can alter the thickness of the lipid bilayer [23], introducing additional structural perturbations.
Figure 3.
(a) A logarithmic scale plot of the MSD versus time traces (blue solid lines) for a large number (n = 127) of CNTPs. Red solid lines highlight three representative individual trajectories. (b) A representative MSD versus time trace from (a) plotted on a linear scale. The red solid and black dashed lines represent the fits to the ‘directed’ diffusion model (equation (3.1)) and normal diffusion (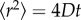 ), respectively. (c) A histogram of the CNTPs diffusion coefficient values extracted from fitting the trajectories to equation (3.1). Red solid line indicates Gaussian fit to the data.
), respectively. (c) A histogram of the CNTPs diffusion coefficient values extracted from fitting the trajectories to equation (3.1). Red solid line indicates Gaussian fit to the data.
Fitting of the MSD curves to equation (3.1) allowed us to extract values for the CNTP diffusion coefficient, D, and the CNTP drift speed, v. The statistical test of the correlation between these two values (see electronic supplementary material, figure S3, for the scatterplot of both values) shows only weak correlation (with correlation coefficient, C = 0.59), giving us confidence that the diffusion coefficients that we extracted from the MSD trace analysis represent the intrinsic mobility of the CNTPs in supported lipid bilayers. Diffusion coefficient values follow a log-normal distribution (figure 3c) that centres at 6.1 ± 3.6 × 10−15 cm2 s−1. This behaviour is consistent with the motion characteristics of biological proteins in lipid membranes as observed by HS-AFM [8,9,24] and other techniques [25,26]. In general, diffusion coefficients of protein in lipid membranes are in the order of 10−10 cm2 s−1 [27], with values showing considerable variations depending on the protein size and structure. For instance, the diffusion coefficient of ion-driven rotors of bacterial ATP synthase in supported POPC lipid membrane is in a range of 10−14 cm2 s−1 [24]. In a supported membrane, diffusion of proteins slows down considerably [4,24,28]. We note that despite the relatively small size of the CNTPs, their diffusion coefficients are roughly 10 times lower than the values obtained for membrane proteins in previous studies [8,9,24]. We believe that the bilayer composition (DOPC:DPPC, 70 : 30) used in our study could be partially responsible for these differences. In contrast to DOPC, DPPC lacks double bonds on the fatty acid tails, and its phase transition temperature (41°C) is much higher than DOPC (−17°C), pointing to an added degree of structural order, which could slow down the CNTP diffusion. In addition, computer simulations showed that different interactions between embedded nanopores and annular lipids can also affect the diffusion behaviour [29]. In our case, besides the hydrophobic interaction of CNT wall and lipid tail, the negatively charged CNT ends can interact with the positively charged lipid head groups.
We have also explored the possibility of controlling the motion characteristics of the CNTPs by changing the nature of their interactions with the mica substrate underneath the lipid bilayer. Normally, the negatively charged CNTP ends, which, as we mentioned earlier, are due to the presence of ionized carboxyl groups at the CNTP rims, cannot interact strongly with the negatively charged mica and thus should not restrict the CNTP mobility. In contrast, when we pre-treated the mica surface with a divalent ion salt solution, which gave the surface a slight positive charge, most of the CNTPs in our supported lipid bilayers became stationary, indicating that the negatively charged ends interacted with the positively charged surface and pinned the CNTPs.
4. Conclusion
In this work, we used the HS-AFM to observe and characterize real-time diffusion of CNTPs in supported lipid bilayers on mica. In our experiments, CNTPs exhibited the directed diffusion pattern with the experimentally determined diffusion coefficients following the log-normal distribution centred at the value consistent with the diffusion coefficients of membrane proteins observed in supported lipid bilayer membranes. This study demonstrates that the similarities between CNTPs and biological membrane pores include not only similar transport properties, but also the ability to move laterally in the membrane. Our study opens up the possibility for researchers to use CNTPs as convenient models to study membrane protein physics, as well as versatile and mobile components for artificial cells and hybrid systems that combine biological cells and man-made components.
Supplementary Material
Acknowledgements
P.-O.C. acknowledges MARA summer internship programme support.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
Y.Z. and A.N. designed the experiments, Y.Z. and R.H.T. prepared samples, Y.Z. performed HS-AFM imaging, Y.Z., P.-O.C. and A.N. analysed the data, A.N. and Y.Z. wrote the manuscript and all authors commented on it.
Competing interests
The authors declare no competing financial interest.
Funding
This work was supported by the US Department of Energy, Office of Basic Energy Sciences, Division of Materials Sciences and Engineering under Award SCW0972. Work at the Lawrence Livermore National Laboratory was performed under the auspices of the US Department of Energy under Contract DE-AC52-07NA27344.
References
- 1.van Meer G, Voelker DR, Feigenson GW. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124. ( 10.1038/nrm2330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lemmon MA. 2008. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111. ( 10.1038/nrm2328) [DOI] [PubMed] [Google Scholar]
- 3.McMahon HT, Gallop JL. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438, 590–596. ( 10.1038/nature04396) [DOI] [PubMed] [Google Scholar]
- 4.Sackmann E. 1996. Supported membranes: scientific and practical applications. Science 271, 43–48. ( 10.1126/science.271.5245.43) [DOI] [PubMed] [Google Scholar]
- 5.Nair PM, Salaita K, Petit RS, Groves JT. 2011. Using patterned supported lipid membranes to investigate the role of receptor organization in intercellular signaling. Nat. Protocols 6, 523–539. ( 10.1038/nprot.2011.302) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saxton MJ, Jacobson K. 1997. Single-particle tracking: applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 26, 373–399. ( 10.1146/annurev.biophys.26.1.373) [DOI] [PubMed] [Google Scholar]
- 7.Reits EAJ, Neefjes JJ. 2001. From fixed to FRAP: measuring protein mobility and activity in living cells. Nat. Cell Biol. 3, E145–E147. ( 10.1038/35078615) [DOI] [PubMed] [Google Scholar]
- 8.Casuso I, Khao J, Chami M, Paul-Gilloteaux P, Husain M, Duneau J-P, Stahlberg H, Sturgis JN, Scheuring S. 2012. Characterization of the motion of membrane proteins using high-speed atomic force microscopy. Nat. Nano 7, 525–529. ( 10.1038/nnano.2012.109) [DOI] [PubMed] [Google Scholar]
- 9.Yamashita H, Taoka A, Uchihashi T, Asano T, Ando T, Fukumori Y. 2012. Single-molecule imaging on living bacterial cell surface by high-speed AFM. J. Mol. Biol. 422, 300–309. ( 10.1016/j.jmb.2012.05.018) [DOI] [PubMed] [Google Scholar]
- 10.Geng J, et al. 2014. Stochastic transport through carbon nanotubes in lipid bilayers and live cell membranes. Nature 514, 612–615. ( 10.1038/nature13817) [DOI] [PubMed] [Google Scholar]
- 11.Kim K, Geng J, Tunuguntla R, Comolli LR, Grigoropoulos CP, Ajo-Franklin CM, Noy A. 2014. Osmotically-driven transport in carbon nanotube porins. Nano Lett. 14, 7051–7056. ( 10.1021/nl5034446) [DOI] [PubMed] [Google Scholar]
- 12.Tunuguntla RH, Allen FI, Kim K, Belliveau A, Noy A. 2016. Ultrafast proton transport in sub-1-nm diameter carbon nanotube porins. Nat. Nano 11, 639–644. ( 10.1038/nnano.2016.43) [DOI] [PubMed] [Google Scholar]
- 13.Uchihashi T, Kodera N, Ando T. 2012. Guide to video recording of structure dynamics and dynamic processes of proteins by high-speed atomic force microscopy. Nat. Protocols 7, 1193–1206. ( 10.1038/nprot.2012.047) [DOI] [PubMed] [Google Scholar]
- 14.Kodera N, Sakashita M, Ando T. 2006. Dynamic proportional-integral-differential controller for high-speed atomic force microscopy. Rev. Sci. Instrum. 77, 083704 ( 10.1063/1.2336113) [DOI] [Google Scholar]
- 15.Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J. Mol. Graph. Model. 14, 33–38. ( 10.1016/0263-7855(96)00018-5) [DOI] [PubMed] [Google Scholar]
- 16.Giocondi M-C, Yamamoto D, Lesniewska E, Milhiet P-E, Ando T, Le Grimellec C. 2010. Surface topography of membrane domains. Biochim. Biophys. Acta 1798, 703–718. ( 10.1016/j.bbamem.2009.09.015) [DOI] [PubMed] [Google Scholar]
- 17.Attwood S, Choi Y, Leonenko Z.. 2013. Preparation of DOPC and DPPC supported planar lipid bilayers for atomic force microscopy and atomic force spectroscopy. Int. J. Mol. Sci. 14, 3514 ( 10.3390/ijms14023514) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen SO, Ensing B, Ortiz V, Moore PB, Klein ML. 2005. Lipid bilayer perturbations around a transmembrane nanotube: a coarse grain molecular dynamics study. Biophys. J. 88, 3822–3828. ( 10.1529/biophysj.104.057703) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B, Li X, Li B, Xu B, Zhao Y. 2009. Carbon nanotube based artificial water channel protein: membrane perturbation and water transportation. Nano Lett. 9, 1386–1394. ( 10.1021/nl8030339) [DOI] [PubMed] [Google Scholar]
- 20.Lelimousin M, Sansom MSP. 2013. Membrane perturbation by carbon nanotube insertion: pathways to internalization. Small 9, 3639–3646. ( 10.1002/smll.201202640) [DOI] [PubMed] [Google Scholar]
- 21.Saxton MJ. 2007. Modeling 2D and 3D diffusion. In Methods in membrane lipids (ed. Dopico AM.), pp. 295–321. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- 22.Jensen MØ, Mouritsen OG. 2004. Lipids do influence protein function—the hydrophobic matching hypothesis revisited. Biochim. Biophys. Acta 1666, 205–226. ( 10.1016/j.bbamem.2004.06.009) [DOI] [PubMed] [Google Scholar]
- 23.Tran IC, Tunuguntla RH, Kim K, Lee JRI, Willey TM, Weiss TM, Noy A, van Buuren T. 2016. Structure of carbon nanotube porins in lipid bilayers: an In Situ small-angle X-ray scattering (SAXS) study. Nano Lett. 16, 4019–4024. ( 10.1021/acs.nanolett.6b00466) [DOI] [PubMed] [Google Scholar]
- 24.Müller DJ, Engel A, Matthey U, Meier T, Dimroth P, Suda K. 2003. Observing membrane protein diffusion at subnanometer resolution. J. Mol. Biol. 327, 925–930. ( 10.1016/S0022-2836(03)00206-7) [DOI] [PubMed] [Google Scholar]
- 25.Dahan M, Lévi S, Luccardini C, Rostaing P, Riveau B, Triller A. 2003. Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science 302, 442–445. ( 10.1126/science.1088525) [DOI] [PubMed] [Google Scholar]
- 26.Manley S, Gillette JM, Patterson GH, Shroff H, Hess HF, Betzig E, Lippincott-Schwartz J. 2008. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Meth. 5, 155–157. ( 10.1038/nmeth.1176) [DOI] [PubMed] [Google Scholar]
- 27.TE C. 1993. Proteins—structures and molecular properties, 2nd edn New York, NY: WH. Freeman. [Google Scholar]
- 28.Sonnleitner A, Schütz GJ, Schmidt T. 1999. Free Brownian motion of individual lipid molecules in biomembranes. Biophys. J. 77, 2638–2642. ( 10.1016/S0006-3495(99)77097-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Fandiño R, Piñeiro Á, Trick JL, Sansom MSP. 2016. Lipid bilayer membrane perturbation by embedded nanopores: a simulation study. ACS Nano 10, 3693–3701. ( 10.1021/acsnano.6b00202) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material.