Abstract
2-Hydroxy-2-deoxyadenosine triphosphate (2-OH-dATP), generated by the oxidation of dATP, can be misincorporated by DNA polymerases opposite guanine in template DNA during DNA replication, thus causing spontaneous mutagenesis. We demonstrated that mouse MUTYH (mMUTYH) has a DNA glycosylase activity excising not only adenine opposite 8-oxoguanine (8-oxoG) but also 2-hydroxyadenine (2-OH-A) opposite guanine, using purified recombinant thioredoxin-mMUTYH fusion protein. mMUTYH formed a stable complex with duplex oligonucleotides containing an adenine:8-oxoG pair, but the binding of mMUTYH to oligonucleotides containing a 2-OH-A:guanine pair was barely detectable, thus suggesting that mMUTYH recognizes and interacts with these two substrates in a different manner which may reflect the difference in the base excision repair process for each substrate. Mutant mMUTYH with G365D amino acid substitution, corresponding to a G382D germline mutation of human MUTYH found in familial adenomatous polyposis patients, almost completely retained its DNA glycosylase activity excising adenine opposite 8-oxoG; however, it possessed 1.5% of the wild-type activity excising 2-OH-A opposite guanine. Our results imply that the reduced repair capacity of the mutant hMUTYH(G382D), which inefficiently excises 2-OH-A opposite guanine, results in an increased occurrence of somatic G:C to T:A transversion mutations in the APC gene as well as tumorigenesis in the colon.
INTRODUCTION
Cellular DNA and its precursor nucleotides are at high risk of being oxidized by reactive oxygen species, which are inevitably generated by normal metabolic functions such as mitochondrial respiration or by environmental exposure to ionizing radiation and chemicals. The oxidation of DNA appears to result in either spontaneous mutagenesis or cell death and, as a result, it has been implicated in various age-related diseases such as cancer and neurodegeneration (1). Among various types of oxidized damage in DNA, 8-oxoguanine (8-oxoG) and 2-hydroxyadenine (2-OH-A), namely the oxidized forms of guanine and adenine, can pair with adenine or guanine in template DNA, respectively, during DNA replication. It is therefore considered to be one of the spontaneous causes of mutagenesis or cell death (2–4). The direct oxidation of DNA by a hydroxyradical has been reported to generate substantial amount of 8-oxoG, but little 2-OH-A. In contrast, 2-OH-A is exclusively generated by the oxidation of dATP in the nucleotide pool (5). The difference in the origins of each oxidized base suggests that 8-oxoG and 2-OH-A cause mutagenesis in a different manner, and therefore cells possess different error avoiding mechanisms for each.
In Escherichia coli, 8-oxo-dGTP generated in the nucleotide pool is hydrolyzed to the monophosphate by MutT protein, and 8-oxoG generated by the direct oxidation of guanine in DNA is excised by 8-oxoG DNA glycosylase, MutM (FPG) protein. Furthermore, adenine misincorporated opposite 8-oxoG in the template strand of DNA is removed by adenine DNA glycosylase, MutY protein. As a result of the cooperative action of MutT/MutM/MutY, E.coli efficiently protects the occurrence of spontaneous mutations such as A:T to C:G and G:C to T:A transversions, which are caused by 8-oxoG (6–8). Recently, it has been shown that the Orf135 gene in E.coli encodes 2-OH-dATPase and that the occurrence of G:C to T:A transversion in Orf135− mutants increased 2- to 3-fold when compared with wild type, thus suggesting that the oxidation of dATP in the nucleotide pool is one of the spontaneous causes of mutagenesis (9).
In eukaryotes, it was demonstrated that the human homolog of MutT (hMTH1) hydrolyses both 8-oxo-dGTP and 2-OH-dATP, thus preventing the spontaneous mutagenesis and cell death caused by these oxidized nucleotides (10–13). We previously reported that a partially purified human MutY homolog (hMUTYH) protein from Jurkat cells possesses DNA glycosylase activities excising 2-OH-A opposite guanine as well as adenine opposite 8-oxoG (14). E.coli MutY protein has also been reported to have a little or no 2-OH-A DNA glycosylase activity (15). It is therefore likely that the excision of 2-OH-A in DNA as well as the sanitization of 2-OH-dATP from the nucleotide pool is accomplished by different mechanisms in prokaryotes and mammals.
Recently, several germline mutations in the human MUTYH (hMUTYH) gene have been reported in familial adenomatous polyposis (FAP) patients (16,17). These patients have no germline mutation in the APC gene, frequently found in the major type of FAP patients, but somatic mutations in the APC gene were found in tumor tissue and most of them are G:C to T:A transversion mutations. These observations strongly suggest that a hMUTYH deficiency causes somatic mutations in the APC gene, thus resulting in multiple colorectal tumors. hMUTYH mutations found in the patients are mainly Y165C and/or G382D amino acid substitutions, and the patients carrying such homozygous or heterozygous mutations are highly susceptible to multiple colorectal tumors. The E.coli MutY(Y82C) mutant corresponding to the former markedly reduced the DNA glycosylase activity excising adenine opposite 8-oxoG, while the MutY(G253D) mutant corresponding to the latter exhibited no gross biochemical dysfunction but showed a subtle reduction in the DNA glycosylase activity which is apparent only at 2°C (16). We recently reported that the spontaneous mutation rate of mouse MUTYH-null embryonic stem cells is 2-fold higher than that of wild type and that the mMUTYH(G365D) mutant corresponding to the hMUTYH(G382D) mutant could not suppress the elevated mutation rate at all (18). Moreover, in OGG1-null mice which lack 8-oxoG DNA glycosylase activity, spontaneous lung tumorigenesis and the accumulation of 8-oxoG in DNA were elevated by several fold when compared with wild type, but no tumors in either the gastrointestinal tract or other organs were observed (19). These observations raise the question as to why the G:C to T:A transversion mutation or tumorigenesis occurs so frequently in the colon tissue of patients with germline mutations in the hMUTYH gene.
A biochemical analysis of the MUTYH function took an extremely long time to complete because of the difficulty in preparing a functional recombinant MUTYH protein. We recently obtained a purified recombinant thioredoxin-mMUTYH fusion protein, in which more than 90% of the residues were identical to those of hMUTYH (20). In the present study, we performed detailed biochemical analyses of the DNA glycosylases of mMUTYH and demonstrated that mMUTYH has the DNA glycosylase activity excising 2-OH-A opposite guanine in DNA as well as adenine opposite 8-oxoG, and that mMUTYH recognizes and interacts with each substrate in a different manner. In addition, the mMUTYH(G365D) mutant protein almost completely retained its DNA glycosylase activity excising adenine opposite 8-oxoG but possessed 1.5% of the wild-type activity excising 2-OH-A opposite guanine, thus suggesting that the reduced repair capacity of the mutant hMUTYH(G382D) excising 2-OH-A opposite guanine results in an increased occurrence of somatic G:C to T:A transversion mutations in the APC gene as well as tumorigenesis in the colon.
MATERIALS AND METHODS
Oligonucleotides
The oligonucleotides shown in Table 1 were obtained from Greiner Japan and the Hokkaido System Science.
Table 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea |
|---|---|
| *A | FAM-AGCGGCCATCGATACCGTCAACCTCGAGGAATTCC |
| *AO | FAM-AGCGGCCATCGATACCGTCAOACCTCGAGGAATTCC |
| G | GGAATTCCTCGAGGTGGACGGTATCGATGGCCGCT |
| GO | GGAATTCCTCGAGGTGO GACGGTATCGATGGCCGCT |
| T | GGAATTCCTCGAGGTT GACGGTATCGATGGCCGCT |
| 19-P | FAM-AGCGGCCATCGATACCGTC-phosphate |
| 19-OH | FAM-AGCGGCCATCGATACCGTC-OH |
FAM, 5′ end was labeled with FAM; GO, 8-oxoguanine; AO, 2-hydroxyadenine; Phosphate, the phosphate group was attached to the 3′-hydroxy group; OH, 3′ end was the hydroxy group.
*, base in the FAM-labeled strand.
aItalic letters indicate the target base of MUTYH (A, Ao) and its opposite base (G, Go, T).
Preparation of the recombinant mouse MUTYH proteins
To obtain soluble recombinant mMUTYH preparations, fusion proteins with thioredoxin (Trx), Trx-mMUTYH, Trx-mMUTYH(D207N), Trx-mMUTYH(R361A) and Trx-mMUTYH(G365D), were prepared as described previously (20,21).
Preparation of the MutY protein
The E.coli MutY protein was prepared as previously described by Miyako et al. (22). Briefly, the expression of E.coli MutY was induced by the addition of 1 mM isopropyl β-d-thiogalactoside (Wako Pure Chemicals) to the culture of E.coli SURE cells transformed with pU85Y following incubation at 37°C for 6 h. The cells were suspended with buffer T [50 mM Tris–HCl, pH 7.6, 0.1 mM EDTA, 0.5 mM DTT and 0.1 mM phenylmethylsulfonyl fluoride (PMSF)], disrupted by sonication and cell lysates were clarified by centrifugation. The lysates were treated with 25% streptomycin, and proteins precipitated by 40–75% ammonium sulfate were resuspended with buffer T and dialyzed against buffer A (20 mM potassium phosphate, pH 7.4, 0.5 mM DTT, 0.1 mM EDTA and 0.1 mM PMSF) containing 50 mM KCl. The lysates were then applied to a phosphocellulose column and eluted with a linear gradient of KCl (50–500 mM). The elutions were dialyzed against buffer S (50 mM potassium phosphate, pH 7.5, 0.5 mM DTT, 0.1 mM EDTA, 0.1 mM PMSF and 10% glycerol) containing 50 mM NaCl and applied to a Hitrap-SP (Amersham Biosciences) column, eluted with a linear gradient of NaCl (50–500 mM). The elutions, dialyzed against buffer S with 50 mM NaCl, were applied to a Hitrap-heparin (Amersham Biosciences) column and eluted with a linear gradient of NaCl (50–500 mM), 50% glycerol was added and stocked at −80°C.
Preparation of the recombinant mAPEX1 protein
The recombinant mAPEX1 protein was prepared as described previously (21).
Determination of the protein concentration
The protein concentration was determined using a Bio-Rad protein assay kit (Bio-Rad) and BSA as the standard.
Nicking assay
A fluorescence dye, 6-carboxy fluorescein-aminohexyl amidite (FAM) was attached to the 5′ end of the strand with a target base or damaged base and duplex oligonucleotide substrates for a nicking assay were prepared as previously described (23). The labeled strand was shown with an asterisk as *A and *AO as summarized in Table 1. A standard nicking assay was performed as previously described (21,23). Briefly, duplex oligonucleotide substrates were incubated in 12.5 μl of the reaction buffer (10 mM Tris–HCl, pH 7.6, 5 μM ZnCl2, 0.5 mM DTT, 0.5 mM EDTA, 1.5% glycerol and 100 μg/ml BSA) with either mMUTYH or MutY protein at 37°C for a given time. In the presence of mAPEX1, 0.2 mM MgCl2 was carried over from the buffer B in which mAPEX1 was stored. Next, the reaction mixture was heated in the presence of 0.17 N NaOH at 95°C for 4 min and added to 15 μl of 80% formamide containing 6 μg/ml BlueDextran (Sigma) and 20 mM EDTA. 3 μl of the mixture was fractionated on an 8% Long Ranger denatured gel (24 cm length) containing 7 M urea at 30 W for 2 h. Specifically, the nicked 19mer oligonucleotides labeled with FAM were detected using the model 373 automated DNA sequencer and quantified using the GeneScan 3.1 software package (Perkin Elmer), according to the manufacturer's instructions.
Gel shift assay
FAM-labeled duplex oligonucleotides (40 nM) were incubated in the reaction buffer with 80 nM Trx-MUTYH at 37°C for 60 min. Next, the reaction products were fractionated on a 4% native polyacrylamide gel (12 cm length) in 0.5× TBE at 4 W for 3 h. The oligonucleotides labeled with FAM were detected using the model 373 automated DNA sequencer and then analyzed using the GeneScan 3.1 software package. The results from one of several independent experiments are presented.
RESULTS
Mouse MUTYH protein but not E.coli MutY has 2-OH-A DNA glycosylase activity
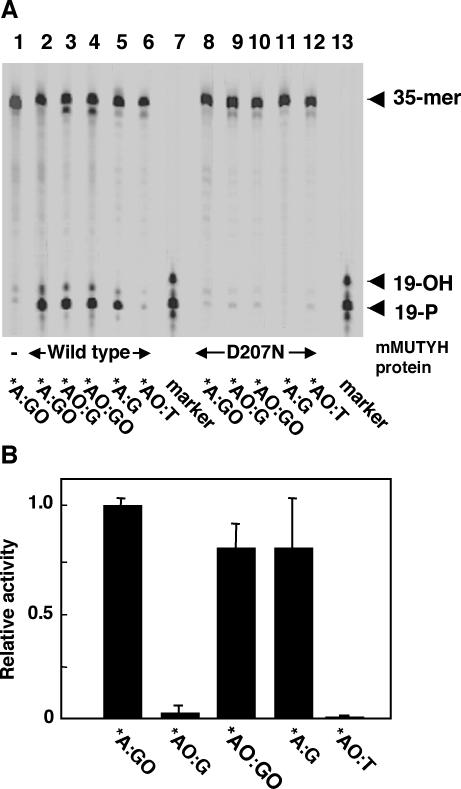
Duplex oligonucleotides containing adenine opposite 8-oxoG (*A:GO) or 2-OH-A opposite guanine (*AO:G), in which the asterisk indicates a base in the FAM-labeled strand, were incubated with the wild-type mMUTYH protein and the reaction products were fractionated by denatured-gel electrophoresis after alkaline treatment. Abasic sites produced by removal of adenine or 2-OH-A were cleaved by β/δ-elimination catalyzed by alkaline treatment and the cleaved products were detected as FAM-labeled 19mer oligonucleotides with phosphates at the 3′ termini (19-P). When duplex oligonucleotide substrates containing *A:GO or *AO:G pairs were incubated with mMUTYH, about 50% of the labeled strands were detected as cleaved 19mer fragments (19-P) (Figure 1A, lanes 2 and 3), indicating that mMUTYH has DNA glycosylase activity excising 2-OH-A opposite guanine as well as adenine opposite 8-oxoG. From duplex oligonucleotides containing *AO:GO and *A:G pairs, mMUTYH produced the 19-P product as much as from the substrates containing *A:GO or *AO:G pairs (Figure 1A, lanes 4 and 5). No cleaved band was produced from substrates containing the *AO:T pair by mMUTYH (Figure 1A, lane 6). Mutant mMUTYH(D207N) protein, in which Asp207, an active site residue for the adenine DNA glycosylase, was substituted with Asn, produced no 19-P product from any of the substrates examined (Figure 1A, lanes 8–12).
Figure 1.
MUTYH has a DNA glycosylase activity excising 2-OH-A opposite guanine as well as adenine opposite 8-oxoG, but E.coli MutY has a poor activity for excising 2-OH-A opposite guanine. (A) 20 nM duplex oligonucleotides containing *A:GO (lanes 1, 2 and 8), *AO:G (lanes 3 and 9), *AO:GO (lanes 4 and 10), *A:G (lanes 5 and 11) or *AO:T pairs (lanes 6 and 12) were incubated with 40 nM wild-type mMUTYH (lanes 2–6) or mutant mMUTYH(D207N) (lanes 8–12) at 37°C for 60 min. The reaction products were treated with NaOH and fractionated. Lane 1, no protein; lanes 7 and 13, marker oligonucleotides, 19-P and 19-OH. (B) 20 nM duplex oligonucleotides containing *A:GO, *AO:G, *AO:GO, *A:G or *AO:T pairs were incubated with 50 ng/μl E.coli MutY at 37°C for 60 min. The relative amount of the cleaved product for each duplex in comparison with that of the duplex oligonucleotides containing the *A:GO pair is shown as the relative activity, with SEM. *, base in the FAM-labeled strand.
In Figure 1A, two bands were detected in the reaction products by mMUTYH. The major one was the band corresponding to the 19-P product which was the product of δ-elimination at the abasic site. The other minor band which migrated between 19-P and 19-OH probably corresponds to the β-elimination product with a deoxyribose moiety at the 3′ terminus of the abasic site. In conclusion, both bands were certainly derived from the abasic site produced by the DNA glycosylase activity of mMUTYH.
Next, we examined the DNA glycosylase activity of E.coli MutY protein with the same substrates. Duplex substrates containing *A:GO or *A:G pairs were efficiently cleaved by E.coli MutY protein with alkaline treatment, while the relative cleavage of the duplex substrates containing *AO:G by MutY was less than 5% level of the substrate with the *A:GO pair (Figure 1B). These data were consistent with those of another report (15). Interestingly, duplex substrates containing the *AO:GO pair were cleaved by MutY as efficiently as were the substrates with the *A:G pair. Again, MutY did not cleave the substrates containing the *AO:T pair at all.
Neither mMUTYH nor E.coli MutY protein exhibited any DNA glycosylase activity excising 2-OH-A from the duplex substrates with the *AO:C pair (data not shown).
mMUTYH protein does not form a stable complex with duplex oligonucleotides containing the 2-OH-A:G pair
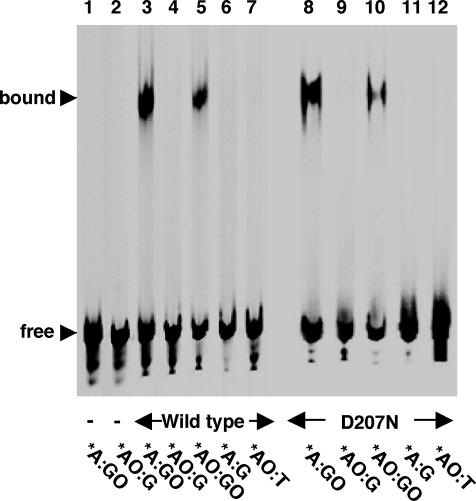
We previously reported that mMUTYH forms a stable complex with duplex oligonucleotides containing the adenine:8-oxoG pair and its reaction product (21). We thus examined the binding of mMUTYH to duplex oligonucleotides containing the 2-OH-A:G pair by a gel mobility shift assay. mMUTYH formed a stable complex with duplex oligonucleotides containing the *A:GO pair (Figure 2, lane 3), as previously reported (21), but no complex was formed with duplex oligonucleotides containing *AO:G, *A:G or *AO:T pairs (Figure 2, lanes 4, 6 and 7). In contrast, mMUTYH formed a substantial amount of complex with duplex oligonucleotides containing the *AO:GO pair (Figure 2, lane 5).
Figure 2.
MUTYH does not form a stable complex with duplex oligonucleotides containing 2-OH-A opposite guanine. 20 nM duplex oligonucleotides containing *A:GO (lanes 1, 3 and 8), *AO:G (lanes 2, 4 and 9), *AO:GO (lanes 5 and 10), *A:G (lanes 6 and 11) or *AO:T pairs (lanes 7 and 12) were incubated with 80 nM wild-type mMUTYH (lanes 3–7) or mutant mMUTYH(D207N) (lanes 8–12) at 37°C for 60 min. The reaction products were fractionated on a 4% native polyacrylamide gel. Lanes 1 and 2, no protein; free, unbound 35mer duplex; bound, MUTYH-bound duplex. *, base in the FAM-labeled strand.
Next, we examined the binding of mutant mMUTYH(D207N), which completely lacks any DNA glycosylase activity, to each duplex oligonucleotide. This mutant enables us to examine the formation of the mMUTYH–DNA complex without excising either adenine or the 2-OH-A base in the duplex. As shown in Figure 2 (lanes 8–12), essentially similar results were obtained with the mutant mMUTYH as with wild-type mMUTYH. A stable DNA–protein complex was formed between mMUTYH(D207N) and duplex oligonucleotides containing *A:GO, and to a lesser extent *AO:GO, but not *AO:G, *A:G and *AO:T. Neither the wild-type nor the mutant mMUTYH formed a stable complex with duplex oligonucleotides containing *AO:C (data not shown).
These results indicate that mMUTYH protein binds to DNA depending on the opposite base (8-oxoG or G), but not the base excised by itself (A or 2-OH-A).
mMUTYH protein does not protect the abasic site opposite guanine after excising 2-OH-A
We previously demonstrated that mMUTYH excises adenine opposite 8-oxoG while staying on its own reaction product containing the abasic site opposite 8-oxoG, thus protecting the abasic site from being attacked by AP endonuclease (APEX1) and the 8-oxoG base from being excised by 8-oxoG DNA glycosylase (OGG1) (21). As described above, mMUTYH does not form a stable complex with duplex oligonucleotides containing the 2-OH-A:G pair, thus suggesting that the abasic site produced by the excision of 2-OH-A opposite guanine can be immediately incised by APEX1.
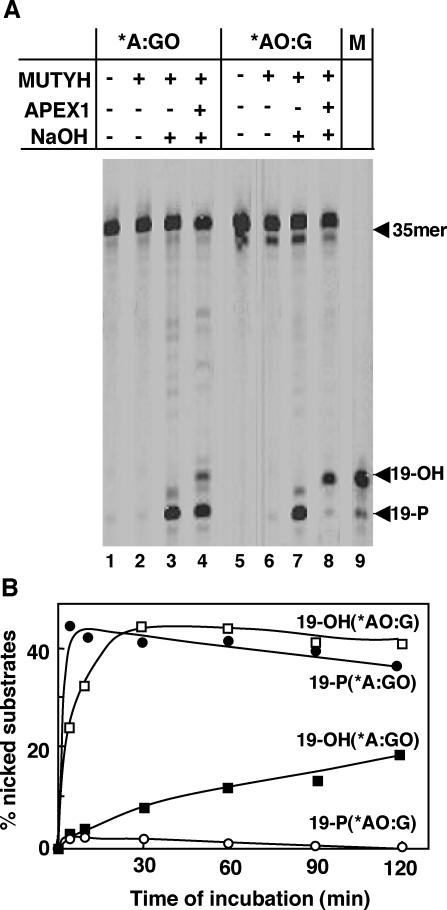
To examine this possibility, we incubated duplex oligonucleotides containing either the *A:GO or the *AO:G pair with mMUTYH and mAPEX1 simultaneously. If mAPEX1 incises the 5′ end of abasic sites generated by mMUTYH, then FAM-labeled 19mer oligonucleotides with hydroxy groups at the 3′ termini (19-OH) can be produced. On the other hand, the intact abasic sites can be detected as FAM-labeled 19mer oligonucleotides with phosphate groups at the 3′ termini (19-P) after δ-elimination by alkaline treatment. As shown in Figure 3A, a major product from duplex oligonucleotides containing the *A:GO pair was 19-P, thus confirming that the abasic sites opposite 8-oxoG generated by mMUTYH remained intact even in the presence of mAPEX1, as previously described (21). In contrast, most of the product from duplex oligonucleotides containing the *AO:G pair was 19-OH, thus indicating that the abasic sites generated through the removal of 2-OH-A opposite guanine by mMUTYH were efficiently incised by mAPEX1. Without alkaline treatment of the reaction product, neither 19-P nor 19-OH was produced by mMUTYH itself, thus confirming that mMUTYH lacks AP lyase activity (Figure 3A, lanes 2 and 6).
Figure 3.
MUTYH does not prevent APEX1 from incising the abasic site generated opposite guanine after excising 2-OH-A. (A) 20 nM duplex oligonucleotides containing *A:GO (lanes 1–4) or *AO:G pairs (lanes 5–8) were incubated with 40 nM mMUTYH (lanes 2–4 and lanes 6–8) and 200 nM mAPEX1 (lanes 4 and 8) at 37°C for 60 min. The reaction products were treated with NaOH (lanes 3, 4, 7 and 8) or without NaOH (lanes 1, 2, 5 and 6) and then fractioned. (B) As in (A), duplex oligonucleotides containing *A:GO or *AO:G pairs were incubated with mMUTYH and mAPEX1 at 37°C. The percentage of nicked substrates for the times noted are plotted. Closed circles, production of 19-P from duplex containing *A:GO; closed squares, production of 19-OH from the duplex containing *A:GO; open circles, production of 19-P from duplex containing *AO:G; open squares, production of 19-OH from duplex containing *AO:G. *, base in the FAM-labeled strand.
As shown in Figure 3B, more than 60% of the nicked products from the duplex oligonucleotides containing the *A:GO pair was detected as 19-P, even 2 h after incubation with mMUTYH and mAPEX1. On the other hand, from duplex oligonucleotides containing the *AO:G pair, only a little amount of 19-P was produced immediately after the duplex oligonucleotides were incubated with mMUTYH and mAPEX1, and thereafter almost all products were detected as 19-OH, products incised by mAPEX1. As a result, we concluded that mMUTYH does not protect the abasic site opposite guanine generated by the excision of 2-OH-A by itself.
DNA glycosylase activity of mMUTYH excising 2-OH-A opposite guanine does not turn over
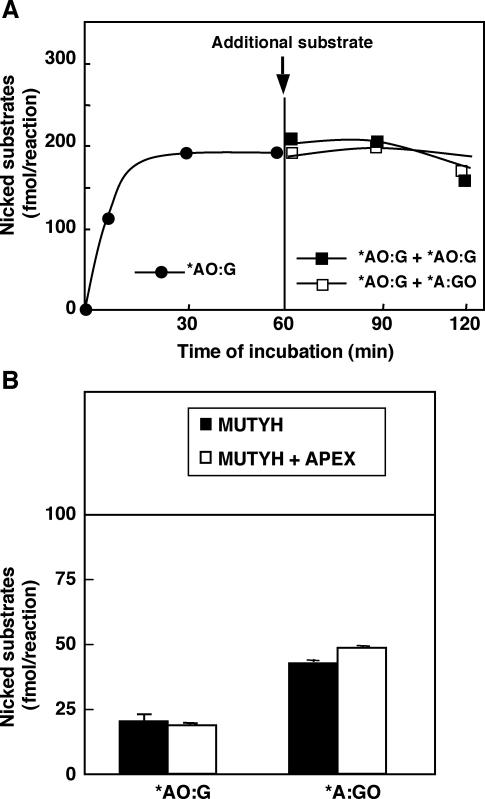
It is likely that the DNA glycosylase activity of mMUTYH excising 2-OH-A opposite guanine may turn over, because the mMUTYH protein does not form a stable complex with its reaction product containing the abasic site opposite guanine. To investigate this hypothesis, duplex oligonucleotides containing the *AO:G pair were incubated with mMUTYH for 60 min (when the reaction reached a plateau level) and then another duplex substrate containing the *AO:G or *A:GO pairs was added to the reaction. As shown in Figure 4A, an increased production of the 19-P product was not observed at all even after adding either of the substrates.
Figure 4.
The DNA glycosylase activity of MUTYH excising 2-OH-A opposite guanine does not turn over. (A) 20 nM duplex oligonucleotides containing the *A:GO pair were incubated with 70 nM mMUTYH at 37°C for 60 min (closed circles) and additional duplex oligonucleotides (20 nM) containing the *A:GO pair (closed squares) or the *A:GO pair (open squares) were added to the reaction mixture and were further incubated. The reaction products were treated with NaOH at the time noted and fractionated. The amounts of 19-P products (fmol/reaction) at the times noted are plotted. (B) 40 nM duplex oligonucleotides containing either the *A:GO or *AO:G pairs were incubated with 8 nM mMUTYH with or without 40 nM mAPEX1 at 37°C for 9 h. The reaction products were treated with NaOH and fractioned. Amounts of 19-P and 19-OH products (fmol/reaction) are shown with SEM. The amount of mMUTYH in the reaction (100 fmol) is indicated as a horizontal line. Closed bars, reaction with mMUTYH; open bars, reaction with mMUTYH and mAPEX1. *, base in the FAM-labeled strand.
It has been previously reported that APEX1 enhances the DNA glycosylase activity of MUTYH excising adenine opposite 8-oxoG (24). Therefore, we added mAPEX1 to the reaction in which a large amount of duplex oligonucleotides containing *AO:G or *A:GO pairs was incubated with a limited amount of mMUTYH. As shown in Figure 4B, essentially the same amount of reaction products was generated from the two substrates, in the absence or presence of mAPEX1. The amounts of the cleaved products from both substrates did not exceed those of mMUTYH in the reaction mixture even after 9 h incubation. As a result, we concluded that the DNA glycosylase activity of mMUTYH never turns over regardless of its substrates or the presence of mAPEX1.
Amino acid substitution in the NUDIX domain of mMUTYH reduces DNA glycosylase activity excising 2-OH-A opposite guanine
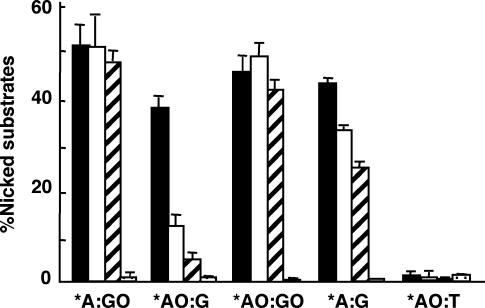
To identify the critical amino acid residue for DNA glycosylase activity of mMUTYH excising 2-OH-A opposite guanine, we examined the DNA glycosylase activity of three mutant mMUTYH with an amino acid substitution; G365D corresponding to hMUTYH(G382D) found in FAP patients, R361A in the NUDIX domain conserved among MUTYH, MutY, MutT and MTH1, and D207N as described above. Duplex oligonucleotides containing *A:GO, *AO:G, *AO:GO, *A:G or *AO:T pairs were incubated with wild-type or mutant (G365D, R361A or D207N) mMUTYH for 60 min, and the amounts of the cleaved fragment were measured after alkaline treatment (Figure 5). Wild-type mMUTYH produced an equivalent level of cleaved fragments from all substrates except for oligonucleotides containing the *AO:T pair, from which mMUTYH did not produce any product at all. As expected, mMUTYH(D207N) did not produce any product from all the substrates examined. On the other hand, two mutant proteins with G365D or R361A substitution produced essentially the same levels of cleaved fragments from duplex oligonucleotides containing *A:GO, *AO:GO and to a lesser extent the *A:G pair when compared with wild-type mMUTYH; however, only 15 or 9% of duplex oligonucleotides containing *AO:G were converted to the cleaved products by mMUTYH(G365D) or mMUTYH(R361A) after 60 min incubation, respectively.
Figure 5.
Amino acid substitution in the NUDIX domain of MUTYH reduces the DNA glycosylase activity excising 2-OH-A opposite guanine. 20 nM duplex oligonucleotides containing *A:GO, *AO:G, *AO:GO, *A:G or *AO:T pairs were incubated with 40 nM wild-type mMUTYH, mMUTYH(G365D), mMUTYH(R361A) or mMUTYH(D207N) at 37°C for 60 min. The reaction products were then treated with NaOH and fractionated. The percentages of the nicked substrates are shown with SEM. Closed bars, wild-type mMUTYH; open bars, mMUTYH(G365D); hatched bars, mMUTYH(R361A); dotted bars, mMUTYH(D207N). *, base in the FAM-labeled strand.
The G365D substitution selectively reduces DNA glycosylase activity of mMUTYH excising 2-OH-A opposite guanine
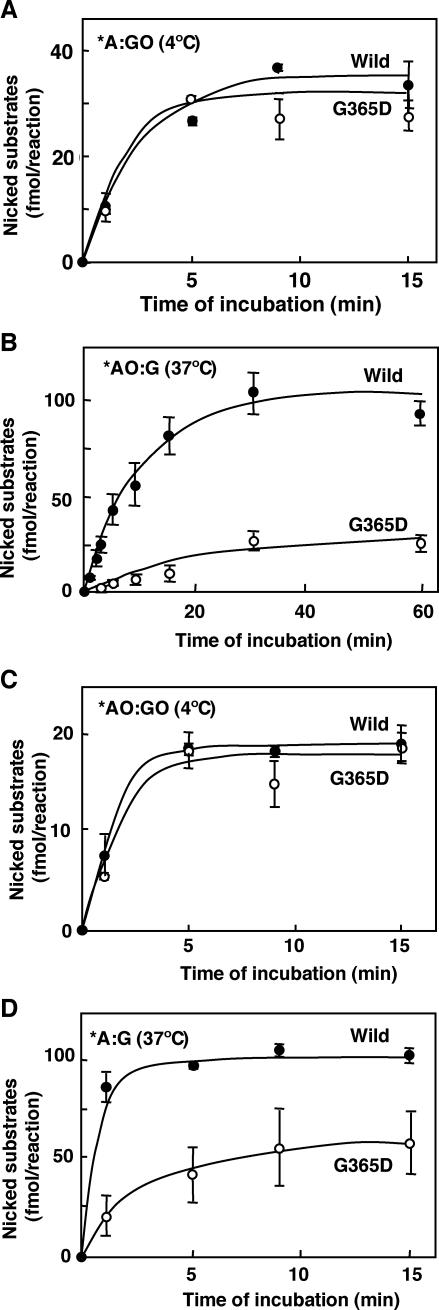
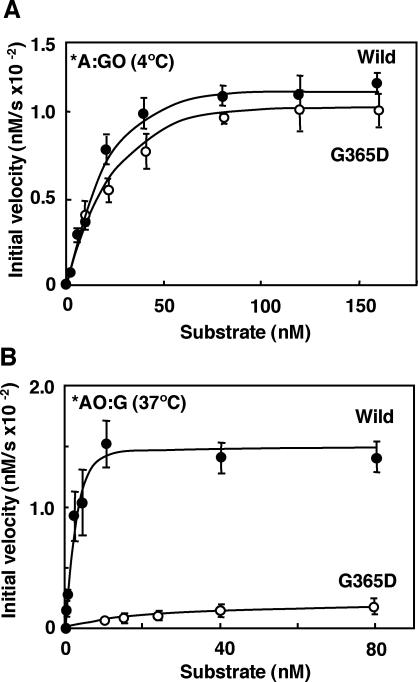
In order to delineate the biochemical dysfunction of the mMUTYH(G365D) mutant, we examined the time course of cleavage for each substrate oligonucleotide by mutant mMUTYH(G365D) and compared the findings with that by wild type (Figure 6). Reactions with substrates containing *A:GO or *AO:GO pairs were carried out at 4°C, because reactions with these substrates reached the plateau level within a minute at 37°C. Reactions with substrates containing *AO:G or *A:G pairs were carried out at 37°C as usual. There was no apparent difference in both the initial velocity and the plateau level of the reaction with the substrates containing *A:GO or *AO:GO pairs between the two mMUTYH (Figure 6A and C). However, mMUTYH(G365D) exhibited significant reductions in both the initial velocity and the plateau level of the reaction with substrates containing *AO:G or *A:G when compared with wild type mMUTYH (Figure 6B and D). We then determined the initial velocities of the glycosylase activity of wild-type mMUTYH and mMUTYH(G365D) with various concentrations of substrates containing *A:GO or *AO:G and plotted them against each concentration of the substrates (Figure 7). With increasing concentrations of the substrate containing *A:GO, mMUTYH(G365D) similarly increased its initial velocity but to slightly less a degree than for wild-type mMUTYH. Their apparent maximal velocities (Vmax) and apparent Michaelis–Menten constant (apparent Km) with the substrate were estimated from the curves shown in Figure 7 (Table 2). The catalytic efficiency (kcat/Km) of mMUTYH(G365D), calculated from these two parameters, was 85% of the level observed in wild-type mMUTYH, thus indicating that the catalytic power of mMUTYH (G365D) excising adenine opposite 8-oxoG was nearly equivalent to that of wild-type mMUTYH.
Figure 6.
Time course of the DNA glycosylase reaction of wild-type mMUTYH and mutant mMUTYH(G365D). Duplex oligonucleotides containing either the *A:GO (A) or *AO:GO pairs (C) were incubated with wild-type mMUTYH (closed circles) or mMUTYH(G365D) (open circles) at 4°C for the time noted, and those containing the *AO:G (B) or *A:G (D) pairs were incubated at 37°C. The concentrations of the oligonucleotides and mMUTYH in the reaction were the same as in Figure 5. The amounts of cleaved 19-P products (fmol/reaction) at the times noted are indicated with SEM. *, base in the FAM-labeled strand.
Figure 7.
Kinetics of the DNA glycosylase reaction of wild-type mMUTYH and mutant mMUTYH(G365D). Various concentrations of duplex oligonucleotides containing *A:GO (A) or *AO:G (B) were incubated with 40 nM wild-type mMUTYH (closed circles) or mutant mMUTYH(G365D) (open circles). The values of the initial velocities were plotted against the concentrations of the substrates. Reactions with duplex oligonucleotides containing the *A:GO pair were performed at 4°C, and those with duplex oligonucleotides containing the *AO:G pair were performed at 37°C as described in the legend for Figure 6. The mean values of the initial velocities were determined by more than three independent experiments and are shown with SEM. *, base in the FAM-labeled strand.
Table 2.
Kinetic parameters of wild-type mMUTYH and mutant mMUTYH(G365D)
| Apparent Vmax(nM s−1) × 10−2 | Apparent Km(nM) | kcat/Km (nM−1s−1) × 10−5 | |
|---|---|---|---|
| *A:GO (4°C) | |||
| Wild | 1.12 | 15.37 | 1.82 |
| G365D | 1.02 (0.91) | 16.50 (1.11) | 1.55 (0.85) |
| *AO:G (37°C) | |||
| Wild | 1.53 | 2.34 | 16.37 |
| G365D | 0.21 (0.14) | 21.95(9.38) | 0.24 (0.015) |
Values of apparent Vmax and apparent Km of wild-type mMUTYH and mutant mMUTYH(G365D) for each substrate containing *A:GO and *AO:G pairs were calculated from Figure 7. The values of kcat were calculated as the molecules of the product formed per molecule of MUTYH per second. The values in parentheses are the relative ratios to those of wild-type mMUTYH.
*, base in the FAM-labeled strand.
On the other hand, the initial velocity of mMUTYH(G365D) increased slightly with increasing concentrations of the substrate containing *AO:G, and reached only a 10% level of that observed for the wild-type mMUTYH in the presence of 80 nM of the substrate. As shown in Table 2, the apparent Vmax of mMUTYH(G365D) with the substrate was only 14% of the level observed in wild-type mMUTYH, and its apparent Km increased more than 9-fold when compared with that of wild-type mMUTYH. As a result, the catalytic efficiency (kcat/Km) of mMUTYH(G365D) was only about 1.5% of the level observed in wild type. However, the catalytic efficiency of mMUTYH(G365D) with the substrate containing *A:G was 10% of the level observed in the wild-type enzyme (data not shown). As a result, we concluded that the G365D substitution in mMUTYH significantly reduced the DNA glycosylase activity excising 2-OH-A opposite guanine but not that excising adenine opposite 8-oxoG.
DISCUSSION
In the present study, we demonstrated that the recombinant mMUTYH protein has a DNA glycosylase activity excising 2-OH-A opposite guanine as well as adenine opposite 8-oxoG and that residues R361 and G365 were critical for this activity. We previously reported that the fraction containing partially purified hMUTYH from Jurkat cells contains a DNA glycosylase activity which efficiently removes 2-OH-A opposite guanine (14). The present study finally proves that mammalian MUTYH proteins have DNA glycosylase activity excising 2-OH-A opposite guanine.
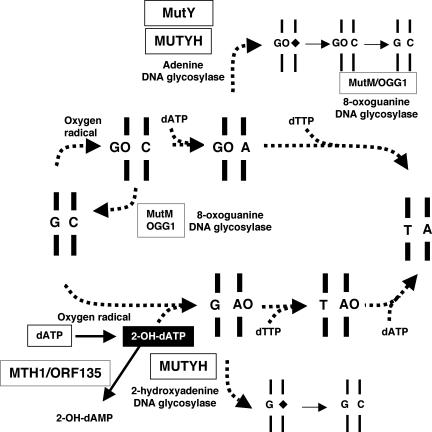
As shown in Figure 8, a G:C to T:A transversion mutation can be caused not only by the direct oxidation of guanine, generating 8-oxoG in DNA, but also by the misincorporation of 2-OH-dATP, formed by the oxidation of dATP in the nucleotide pool, during replication. The results in this study indicate that MUTYH recognizes both adenine misincorporated opposite 8-oxoG in the template strand and 2-OH-A misincorporated opposite guanine in the template strand and removes these premutagenic bases by its DNA glycosylase activity, thus preventing the occurrence of the G:C to T:A transversion mutation. Moreover, it has been previously reported that E.coli MutY hardly removes 2-OH-A opposite guanine (15), and we also confirmed this result with our own preparation of the MutY protein. These results suggest that there is a significant difference in the degree of accumulation of 2-OH-A in the genome of E.coli and mammals (25,26).
Figure 8.
The mutagenic pathways initiated 2-OH-dATP or 8-oxoG formation in DNA resulting in G:C to T:A transversion mutation and its protection by MutY and MUTYH. The direct oxidation of guanine in DNA generates 8-oxoG (GO), opposite to which dATP can be misinserted by DNA polymerase during replication, thus producing the 8-oxoG:A pair (GO:A). The 8-oxoG:A pair is converted to a thymine:adenine (T:A) pair during the next round of replication, thus resulting in a G:C to T:A transversion mutation. On the other hand, the oxidized form of dATP, 2-OH-dATP, was misinserted into the nascent strand opposite guanine in the template strand, thus producing a guanine:2-OH-A pair (G:AO). The guanine:2-OH-A pair is converted to a T:A pair after two more rounds of replication, thus resulting in a G:C to T:A transversion mutation. MutY and MUTYH excise adenine misinserted into nascent strand opposite 8-oxoG in the template, by their adenine DNA glycosylase activity, and thereafter MutM or OGG1 removes 8-oxoG opposite cytosine, thus avoiding the occurrence of G:C to T:A transversion mutation. In contrast, MUTYH has a 2-OH-A DNA glycosylase activity which is able to excise 2-OH-A opposite guanine, thus avoiding the occurrence of a G:C to T:A transversion mutation. 2-OH-dATP can be hydrolyzed to 2-OH-dAMP by MTH1 in mammals or by ORF135 in E.coli. A rhombus indicates an abasic site. *, base in the FAM-labeled strand.
We previously reported that E.coli MutT efficiently hydrolyzes 8-oxo-dGTP but not the oxidized forms of dATP, while the human MutT homolog (hMTH1) hydrolyzes 2-OH-dATP and 8-oxo-dATP as well as 8-oxo-dGTP (11,27). Kamiya et al. revealed that the protein coding Orf135 of E.coli hydrolyzes 2-OH-dATP and that disruption of the Orf135 gene resulted in a 2- to 3-fold increase in the spontaneous occurrence of G:C to T:A and A:T to C:G transversions when compared with the wild type (9). It was also demonstrated that E.coli DNA polymerase III inserts 2-OH-dATP opposite guanine or thymine in the template strand (4). It is thus likely that a disruption of the Orf135 gene increases such spontaneous mutations because MutY can not remove 2-OH-A misincorporated into the genome.
In mammals, it was reported that the occurrence of the A:T to C:G transversion mutation was slightly elevated in MTH1-deficient mice while that of the G:C to T:A transversion was significantly increased only in mice deficient for both of MSH2 and MTH1(28). A loss of the MTH1 function may cause an accumulation of 8-oxo-dGTP in the nucleotide pool, which increases the incorporation of 8-oxoG opposite adenine in the template strand of DNA during replication, thereby resulting in an increased occurrence of the A:T to C:G transversion. It has been demonstrated that MUTYH interacts with MSH2 and both of them are likely to be responsible for the post-replicative repair of adenine or 2-OH-A misinserted into the nascent strand during replication (29). Thus, we propose that both the accumulation of 2-OH-dATP in the nucleotide pool due to MTH1-deficiency and an inefficient post-replicative repair of 2-OH-A by MUTYH in the absence of MSH2 resulted in a significantly increased occurrence of the G:C to T:A transversion mutation in mice deficient for both of MSH2 and MTH1.
In the present study, we showed that E.coli MutY hardly excises 2-OH-A opposite guanine; however, it does efficiently excise 2-OH-A opposite 8-oxoG, thus indicating that MutY can excise 2-OH-A depending on the opposite base. In contrast, mMUTYH efficiently excises 2-OH-A regardless of the opposite base, guanine or 8-oxoG. Furthermore, the mMUTYH(G365D) mutant largely lost the capability to excise 2-OH-A opposite guanine but it did maintain its ability to excise adenine opposite guanine to some degree and also maintained its full activity to excise 2-OH-A opposite 8-oxoG. These observations strongly suggest that mMUTYH, but not E.coli MutY, recognizes 2-OH-A and adenine opposite guanine in a different manner. We also found that mMUTYH cannot excise the 2-OH-A paired with thymine. Considering the fact that the generation of 2-OH-A by the oxidation of dATP in the nucleotide pool is significantly higher than that by the direct oxidation of adenine in DNA (5), it seems highly likely that the 2-OH-A:T pair, a product of direct oxidation of the A:T pair in DNA, rarely exists in living cells. Consequently, it is expected that MUTYH has never acquired such a repair capacity.
mMUTYH protein formed a stable complex with duplex oligonucleotides containing the adenine:8-oxoG pair or its reaction product. But such a stable complex was not detected with duplex oligonucleotides containing the 2-OH-A:guanine pair, thus indicating that the mMUTYH protein interacts with duplex oligonucleotides containing adenine:8-oxoG and 2-OH-A:guanine pairs in a different manner. Reflecting these different interactions, duplex oligonucleotides containing 2-OH-A opposite guanine or adenine opposite 8-oxoG were differently processed by APEX1 following excision of 2-OH-A or adenine by MUTYH. The abasic sites generated in the former by MUTYH were immediately incised by APEX1 while most of those in the latter remained intact even in the presence of APEX1.
After the excision of adenine opposite 8-oxoG by MUTYH, the abasic site is likely to be incised by an AP endonuclease, and cytosine must be inserted opposite 8-oxoG during repair DNA synthesis. The remaining 8-oxoG opposite cytosine can be replaced by guanine through OGG1-initiated base excision repair (Figure 8). Since OGG1 efficiently excises the 8-oxoG opposite the abasic site as well as opposite cytosine, the excision of the 8-oxoG opposite the abasic site by OGG1, following the removal of adenine opposite 8-oxoG by MUTYH, generates an abasic site opposite the abasic site, thus causing a loss of genetic information. Moreover, such lesions can be easily converted to double-strand breaks by the AP lyase activity of OGG1 or by other AP lyases or AP endonucleases. As a result, MUTYH tightly binds and protects its own reaction product, the abasic site opposite 8-oxoG in DNA from inappropriate processing by OGG1 or APEX1 (21).
In contrast, after the excision of 2-OH-A opposite guanine by MUTYH, cytosine can be efficiently inserted opposite guanine during repair replication, thus completing the repair process. Since guanine, a normal base, is present opposite the abasic site resulting from the excision of 2-OH-A by MUTYH, MUTYH does not need to bind and protect the abasic site by itself, unlike the abasic site opposite 8-oxoG, otherwise binding of MUTYH may cause a delay in the repair of the abasic site opposite guanine.
The crystal structure of the MutY–DNA complex has revealed that tight hydrogen bonds are formed between MutY and 8-oxoG opposite adenine in DNA, and it was suggested that the presence of guanine opposite adenine in DNA disturbs this hydrogen bonding network, thus destabilizing the complex (30). In the case of 2-OH-A opposite guanine in DNA, it is likely that the guanine residue indeed destabilizes the interaction of MUTYH with its substrates or products, because MUTYH formed a stable complex with duplex oligonucleotides containing 2-OH-A opposite 8-oxoG.
It has been documented that the catalytic activity of MUTYH excising adenine opposite 8-oxoG does not turn over, and this is considered to be probably due to its forming a stable complex with its reaction product. In the present study, we found that the glycosylase activity of mMUTYH excising 2-OH-A opposite guanine also does not turn over at all. We suppose that MUTYH remains on the substrate even after excising 2-OH-A, but the stability of the complex is not strong enough to be detected by a gel mobility shift assay. It was reported that a high concentration of adenine inactivates the adenine DNA glycosylase activity of prokaryote MutY (31), thus raising another possibility for MUTYH turnover. MUTYH may remain bound with the excised adenine or 2-OH-A base and therefore it is inactivated after one round of reaction. Further elucidation is necessary to solve the issue.
An analysis of mutant mMUTYH with an amino acid substitution in the NUDIX domain revealed that R361A or G365D substitution reduces the DNA glycosylase activity of mMUTYH excising 2-OH-A opposite guanine, selectively. Since the G365D substitution corresponds to G382D germline mutation in hMUTYH found in FAP patients or multiple adenomatous polyposis patients (16,17), we carried out a detailed biochemical analysis of mMUTYH(G365D) and found the catalytic power of the mutant mMUTYH excising 2-OH-A opposite guanine to be 1.5% of the level observed in the wild-type enzyme, but the mutant retains equivalent catalytic activity excising adenine or 2-OH-adenine opposite 8-oxoG.
G365D substitution has the greatest influence on the elevation of the Km value on the substrates containing 2-OH-A opposite guanine, thus indicating that the Gly365 residue is responsible for its affinity to the substrate. According to the crystal structure of the MutY–DNA complex (30), the G260 residue corresponding to G365 in mMUTYH closely interacts with the phosphate backbone of the 8-oxoG-containing strand, but not directly with 8-oxoG itself or the adenine-containing strand. We assume that mMUTYH(G365D) forms tight hydrogen bonds with 8-oxoG directly through residues other than Asp365 and therefore an excision of adenine or 2-OH-A opposite 8-oxoG may not be affected by this substitution. However, in the case of 2-OH-A opposite guanine, it is expected that electrostatic repulsion is generated between the Asp365 residue and the phosphate backbone of the guanine-containing strand, and that this repulsion may prevent MUTYH and DNA from interacting closely enough for MUTYH to recognize and excise 2-OH-A. R361A substitution is also supposed to reduce the electrostatic interaction between the basic arginine residue and the phosphate backbone in DNA.
Because the DNA glycosylase activity of mMUTYH(G365D) excising 2-OH-A opposite guanine is reduced to 1.5% of the level observed in the wild-type enzyme, it is suggested that the accumulation of 2-OH-A in genomic DNA in the colon may be responsible for the increased somatic G:C to T:A transversion mutations in the APC gene, found in the adenomatous polyps of patients with a homozygous hMUTYH(G382D) mutation. A problem still remains as to why patients with mutant MUTYH exhibit a specific elevation of the spontaneous mutation rate in the colon. We reported that OGG1-deficient or MTH1-deficient mice did not exhibit increased tumorigenesis in the gastrointestinal tract (19); however, we recently observed that MUTYH-deficient mice exhibited an increased occurrence of spontaneous adenoma/adenocarcinoma in the small intestine and colon (K. Sakamoto, Y. Tominaga, K. Yamauchi, Y. Nakatsu, K. Sakumi, K. Yoshiyama, A. Asaeda, A. Egashira, S. Kura, T. Yao et al., in preparation), thus suggesting that a common cause(s) exists in both mice and humans which increases the spontaneous mutation rate selectively in the gastrointestinal tract. In patients with hereditary nonpolyposis colorectal cancer, the lack of a mismatch repair protein is known to selectively induce tumorigenesis in the colon (32). In these patients cancer develops without polyp formation, probably because of the significantly increased spontaneous mutation rate due to the loss of a mismatch repair. The mutation rate is higher by two orders of magnitudes than that in MUTYH-deficient patients (28). Since MUTYH and MSH2 interact during the repair process (29), we expect that 2-OH-A may be the common target for the repair by both MUTYH and MSH2, and it may therefore cause tumorigenesis in the gastrointestinal tract if it is not repaired appropriately.
Acknowledgments
We thank Drs Masato Furuichi and Yoshimichi Nakatsu for their helpful discussions; Akemi Matsuyama, Naomi Adachi, Setsuko Kitamura and Keiko Aiura for their technical assistance; Dr Hiroshi Honda for providing us with the opportunity to conduct this study; and Dr B. Quinn for comments on the manuscript. This work was supported by grants from CREST, Japan Science and Technology Agency, the Ministry of Education, Culture, Sports, Science, and Technology of Japan (grant number 16012248), and Japan Society for the Promotion of Science (grant numbers 15590347 and 16390119). Funding to pay the Open Access publication charges for this article was provided by Japan Society for Promotion of Science.
REFERENCES
- 1.Ames B.N., Shigenaga M.K., Hagen T.M. Oxidants, antioxidants, and the degenerative diseases of aging. Proc. Natl Acad. Sci. USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shibutani S., Takeshita M., Grollman A.P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 3.Nakabeppu Y., Tsuchimoto D., Furuichi M., Sakumi K. The defense mechanisms in mammalian cells against oxidative damage in nucleic acids and their involvement in the suppression of mutagenesis and cell death. Free Radic. Res. 2004;38:423–429. doi: 10.1080/10715760410001688348. [DOI] [PubMed] [Google Scholar]
- 4.Kamiya H., Kasai H. 2-Hydroxy-dATP is incorporated opposite G by Escherichia coli DNA polymerase III resulting in high mutagenicity. Nucleic Acids Res. 2000;28:1640–1646. doi: 10.1093/nar/28.7.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamiya H., Kasai H. Formation of 2-hydroxydeoxyadenosine triphosphate, an oxidatively damaged nucleotide, and its incorporation by DNA polymerases. Steady-state kinetics of the incorporation. J. Biol. Chem. 1995;270:19446–19450. doi: 10.1074/jbc.270.33.19446. [DOI] [PubMed] [Google Scholar]
- 6.Michaels M.L., Tchou J., Grollman A.P., Miller J.H. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry. 1992;31:10964–10968. doi: 10.1021/bi00160a004. [DOI] [PubMed] [Google Scholar]
- 7.Maki H., Sekiguchi M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature. 1992;355:273–275. doi: 10.1038/355273a0. [DOI] [PubMed] [Google Scholar]
- 8.Tajiri T., Maki H., Sekiguchi M. Functional cooperation of MutT, MutM and MutY proteins in preventing mutations caused by spontaneous oxidation of guanine nucleotide in Escherichia coli. Mutat. Res. 1995;336:257–267. doi: 10.1016/0921-8777(94)00062-b. [DOI] [PubMed] [Google Scholar]
- 9.Kamiya H., Murata-Kamiya N., Iida E., Harashima H. Hydrolysis of oxidized nucleotides by the Escherichia coli Orf135 protein. Biochem. Biophys. Res. Commun. 2001;288:499–502. doi: 10.1006/bbrc.2001.5781. [DOI] [PubMed] [Google Scholar]
- 10.Sakumi K., Furuichi M., Tsuzuki T., Kakuma T., Kawabata S., Maki H., Sekiguchi M. Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J. Biol. Chem. 1993;268:23524–23530. [PubMed] [Google Scholar]
- 11.Fujikawa K., Kamiya H., Yakushiji H., Fujii Y., Nakabeppu Y., Kasai H. The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein. J. Biol. Chem. 1999;274:18201–18205. doi: 10.1074/jbc.274.26.18201. [DOI] [PubMed] [Google Scholar]
- 12.Sakai Y., Furuichi M., Takahashi M., Mishima M., Iwai S., Shirakawa M., Nakabeppu Y. A molecular basis for the selective recognition of 2-hydroxy-dATP and 8-oxo-dGTP by human MTH1. J. Biol. Chem. 2002;277:8579–8587. doi: 10.1074/jbc.M110566200. [DOI] [PubMed] [Google Scholar]
- 13.Yoshimura D., Sakumi K., Ohno M., Sakai Y., Furuichi M., Iwai S., Nakabeppu Y. An oxidized purine nucleoside triphosphatase, MTH1, suppresses cell death caused by oxidative stress. J. Biol. Chem. 2003;278:37965–37973. doi: 10.1074/jbc.M306201200. [DOI] [PubMed] [Google Scholar]
- 14.Ohtsubo T., Nishioka K., Imaiso Y., Iwai S., Shimokawa H., Oda H., Fujiwara T., Nakabeppu Y. Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res. 2000;28:1355–1364. doi: 10.1093/nar/28.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamiya H., Kasai H. 2-hydroxyadenine in DNA is a very poor substrate of the Escherichia coli MutY protein. J. Radiat. Res. 2000;41:349–354. doi: 10.1269/jrr.41.349. [DOI] [PubMed] [Google Scholar]
- 16.Al-Tassan N., Chmiel N.H., Maynard J., Fleming N., Livingston A.L., Williams G.T., Hodges A.K., Davies D.R., David S.S., Sampson J.R., et al. Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nature Genet. 2002;30:227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- 17.Sieber O.M., Lipton L., Crabtree M., Heinimann K., Fidalgo P., Phillips R.K., Bisgaard M.L., Orntoft T.F., Aaltonen L.A., Hodgson S.V., et al. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N. Engl. J. Med. 2003;348:791–799. doi: 10.1056/NEJMoa025283. [DOI] [PubMed] [Google Scholar]
- 18.Hirano S., Tominaga Y., Ichinoe A., Ushijima Y., Tsuchimoto D., Honda-Ohnishi Y., Ohtsubo T., Sakumi K., Nakabeppu Y. Mutator phenotype of MUTYH-null mouse embryonic stem cells. J. Biol. Chem. 2003;278:38121–38124. doi: 10.1074/jbc.C300316200. [DOI] [PubMed] [Google Scholar]
- 19.Sakumi K., Tominaga Y., Furuichi M., Xu P., Tsuzuki T., Sekiguchi M., Nakabeppu Y. Ogg1 knockout-associated lung tumorigenesis and its suppression by Mth1 gene disruption. Cancer Res. 2003;63:902–905. [PubMed] [Google Scholar]
- 20.Ichinoe A., Behmanesh M., Tominaga Y., Ushijima Y., Hirano S., Sakai Y., Tsuchimoto D., Sakumi K., Wake N., Nakabeppu Y. Identification and characterization of two forms of mouse MUTYH proteins encoded by alternatively spliced transcripts. Nucleic Acids Res. 2004;32:477–487. doi: 10.1093/nar/gkh214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tominaga Y., Ushijima Y., Tsuchimoto D., Mishima M., Shirakawa M., Hirano S., Sakumi K., Nakabeppu Y. MUTYH prevents OGG1 or APEX1 from inappropriately processing its substrate or reaction product with its C-terminal domain. Nucleic Acids Res. 2004;32:3198–3211. doi: 10.1093/nar/gkh642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyako K., Takamatsu C., Umeda S., Tajiri T., Furuichi M., Nakabeppu Y., Sekiguchi M., Hamasaki N., Takeshige K., Kang D. Accumulation of adenine DNA glycosylase-sensitive sites in human mitochondrial DNA. J. Biol. Chem. 2000;275:12326–12330. doi: 10.1074/jbc.275.16.12326. [DOI] [PubMed] [Google Scholar]
- 23.Nishioka K., Ohtsubo T., Oda H., Fujiwara T., Kang D., Sugimachi K., Nakabeppu Y. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol. Biol. Cell. 1999;10:1637–1652. doi: 10.1091/mbc.10.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang H., Clendenin W.M., Wong D., Demple B., Slupska M.M., Chiang J.H., Miller J.H. Enhanced activity of adenine-DNA glycosylase (Myh) by apurinic/apyrimidinic endonuclease (Ape1) in mammalian base excision repair of an A/GO mismatch. Nucleic Acids Res. 2001;29:743–752. doi: 10.1093/nar/29.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nunoshiba T., Obata F., Boss A.C., Oikawa S., Mori T., Kawanishi S., Yamamoto K. Role of iron and superoxide for generation of hydroxyl radical, oxidative DNA lesions, and mutagenesis in Escherichia coli. J. Biol. Chem. 1999;274:34832–34837. doi: 10.1074/jbc.274.49.34832. [DOI] [PubMed] [Google Scholar]
- 26.Olinski R., Zastawny T., Budzbon J., Skokowski J., Zegarski W., Dizdaroglu M. DNA base modifications in chromatin of human cancerous tissues. FEBS Lett. 1992;309:193–198. doi: 10.1016/0014-5793(92)81093-2. [DOI] [PubMed] [Google Scholar]
- 27.Fujikawa K., Kamiya H., Yakushiji H., Nakabeppu Y., Kasai H. Human MTH1 protein hydrolyzes the oxidized ribonucleotide, 2-hydroxy-ATP. Nucleic Acids Res. 2001;29:449–454. doi: 10.1093/nar/29.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egashira A., Yamauchi K., Yoshiyama K., Kawate H., Katsuki M., Sekiguchi M., Sugimachi K., Maki H., Tsuzuki T. Mutational specificity of mice defective in the MTH1 and/or the MSH2 genes. DNA Repair. 2002;1:881–893. doi: 10.1016/s1568-7864(02)00113-1. [DOI] [PubMed] [Google Scholar]
- 29.Gu Y., Parker A., Wilson T.M., Bai H., Chang D.Y., Lu A.L. Human MutY homolog, a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins human MutS homolog 2/human MutS homolog 6. J. Biol. Chem. 2002;277:11135–11142. doi: 10.1074/jbc.M108618200. [DOI] [PubMed] [Google Scholar]
- 30.Fromme J.C., Banerjee A., Huang S.J., Verdine G.L. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature. 2004;427:652–656. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- 31.Guan Y., Manuel R.C., Arvai A.S., Parikh S.S., Mol C.D., Miller J.H., Lloyd S., Tainer J.A. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nature Struct. Biol. 1998;5:1058–1064. doi: 10.1038/4168. [DOI] [PubMed] [Google Scholar]
- 32.Fishel R., Lescoe M.K., Rao M.R., Copeland N.G., Jenkins N.A., Garber J., Kane M., Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]