Abstract
The hepatitis C virus (HCV) has a positive single-stranded RNA genome, and translation starts within the internal ribosome entry site (IRES) in a cap-independent manner. The IRES is well conserved among HCV subtypes and has a unique structure consisting of four domains. We used an in vitro selection procedure to isolate RNA aptamers capable of binding to the IRES domains III–IV. The aptamers that were obtained shared the consensus sequence ACCCA, which is complementary to the apical loop of domain IIId that is known to be a critical region of IRES-dependent translation. This convergence suggests that domain IIId is preferentially selected in an RNA–RNA interaction. Mutation analysis showed that the aptamer binding was sequence and structure dependent. One of the aptamers inhibited translation both in vitro and in vivo. Our results indicate that domain IIId is a suitable target site for HCV blockage and that rationally designed RNA aptamers have great potential as anti-HCV drugs.
INTRODUCTION
The hepatitis C virus (HCV) is a member of the Flaviviridae family and is the major etiological agent of post-transfusion non-A, non-B hepatitis. HCV infection leads to chronic hepatitis, liver cirrhosis and hepatocellular carcinoma. The number of HCV carriers has increased to approximately 300 million worldwide. Although treatments involving a combination of interferon and the antiviral drug rivabirin offer a more favorable outcome than interferon alone, patients are unable to bear the risk of further liver disease after each therapy [reviewed in (1)]. More effective therapeutic drugs are therefore required.
HCV has a positive single-stranded RNA genome of ∼9.6 kb that encodes a large polyprotein consisting of 3010 amino acids (2). Translation initiation occurs via the cap-independent mechanism and requires a highly conserved structure that is located at the 5′-untranslated region [5′-UTR; (3,4)], which is known as the internal ribosome entry site (IRES; Figure 1A). The ternary interaction of the IRES, the 40S ribosomal subunit and eIF3 is essential for translation initiation (5–8). The IRES sequence is well-conserved among HCV isolates and is therefore a good target for anti-HCV drugs (9–15). The IRES is composed of four domains (I–IV) and the tertiary structure of the IRES has been studied by cryo-electron microscopy (16). The structure–function relationship of domain III–IV is particularly well-studied from the viewpoint of its importance during translation initiation [reviewed in (17)]. The domain structures of IIIb, IIId and IIIe were solved by NMR (18–20). Furthermore, the four-way junction structure formed by domains IIIa, IIIb and IIIc was solved by X-ray crystallography (21). The four-way structure and domain IIIb are critical for binding to eIF3. Several substitutions in domain III entirely abolish IRES activity (18,22–24).
Figure 1.
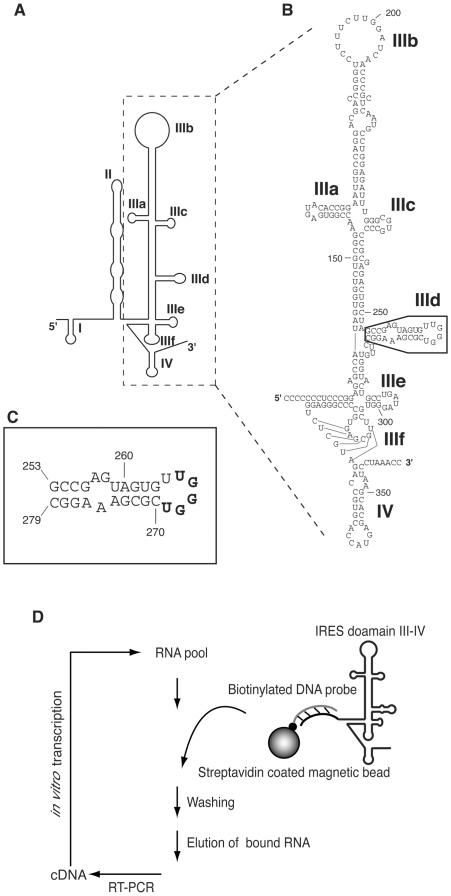
(A) Schematic structure of the HCV IRES RNA [adapted from (35)]. Structural domains are shown as I–IV. (B) Detailed nucleotide sequence of domain III and IV RNA used in this study. Boxed region indicates domain IIId. (C) Secondary structure of domain IIId. In this report, the sequence bound to the consensus sequence of aptamer is shown in bold letters. (D) Schematic of the in vitro selection procedure to isolate RNAs that bind to the HCV IRES.
An in vitro selection procedure can generate functional nucleic acids with affinities to various target molecules (25–27). The RNA or DNA obtained during selection is known as an aptamer. RNA aptamers have been isolated that bind to tRNA (28), HIV TAR (29), DIS (30), 16S ribosomal RNA (31) and HCV IRES domain IV (32) via RNA–RNA interactions. In our previous work, we isolated RNA aptamers against HCV IRES domain II to develop a method of isolating aptamers against long RNA molecules, in order to find a protruding site of domain II and to investigate the effect of blocking such a site (33). In this paper, we isolated aptamers against domain IIId of the IRES, and found that one had a highly inhibitory effect on HCV translation.
MATERIALS AND METHODS
Oligonucleotides, RNA pools and target RNA
Template DNA for RNA pool, PCR primers and 3′-biotinylated deoxyoligonucleotide were obtained from Espec Oligo Service. Phosphoramidites for RNA chemical synthesis were purchased from Glen Research. The templates and PCR primers were as follows (the T7 promoter region is underlined and N denotes A, G, C or T): template DNA for N30V pool, 5′-AGTAATACGACTCACTATAGGGAGAATTCCGACCAGAAG-(N30)-CCTTTCCTCTCTCCTTCCTCTTCT-3′; 5′ end primer of N30V pool, 5′-AGTAATACGACTCACTATAGGGAGAATTCCGACCAGAAG-3′; 3′ end primer of N30V pool, 5′-CCTTTCCTCTCTCCTTCCTCTTCT-3′; biotinylated DNA probe, 5′-TCCTGGAGGCTGCACGACACTCATACC. The 3′-biotinylated apical region of domain IIId, 5′-GCCGAGUAGUGUUGGGUCGCGAAAGGC-Bio-3′, was chemically synthesized using a DNA/RNA synthesizer (model 394; Applied Biosystems). Products were purified as described in the user bulletin ABI (no. 53; 1989).
The preparation of random RNA pool has been described previously (33). HCV IRES domain III–IV RNA (nt 119–361), additional sequences for the hybridization tag and a G base to improve transcription efficiency were generated by in vitro transcription with T7 RNA polymerase and a PCR fragment from an IRES-encoding vector, p5′IL-U3′X (from Prof. Shimotohno of Kyoto University, Japan).
In vitro selection of aptamers
The selection procedure was carried out similarly as described previously (33). Biotinylated DNA probe (0.5 μM) was hybridized to domain III–IV of IRES RNA (2 μM) and then mixed with streptavidin magnetic beads (0.1 mg of Streptavidin MagneSphere Paramagnetic Particles, Promega, or 0.4 mg of MAGNOTEX-SA, Takara) in 100 μl of buffer E (20 mM HEPES–KOH, pH 7.9, 200 mM KCl and 5 mM MgCl2), and was washed with buffer E. At the time of preparing the beads, we used excess amount of target molecules to fill up all probes. Ten nanomoles of RNA pool was pretreated with biotinylated DNA probe immobilized to streptavidin beads for 5 min to remove non-specific binders. Free RNAs were recovered and then mixed with probe-IRES immobilized to streptavidin beads. The reaction mixture (900 μl) was incubated for 5 min in buffer E at room temperature. To minimize contamination of non-specific binders, we alternatively used two kinds of streptavidin beads from different suppliers. The IRES RNA–aptamer complexes were magnetically separated and washed with buffer E. To increase the stringency of selection conditions, the amount of RNA was decreased from 10 nmol in the first generation to 2 nmol in the fourth generation, and the number of washing times was increased from one to three (Table 1). The IRES-bound RNAs were eluted at 94°C with 7 M Urea and recovered by ethanol precipitation. The RNA pool was reversed-transcribed using avian myeloblastosis virus reverse transcriptase (MBI) at 42°C for 1 h. IRES RNA was also eluted by this procedure, but not reverse-transcribed with the specific primers. The dsDNA product was amplified by PCR (Nippon Gene), transcribed in vitro by T7 RNA polymerase (T7 Ampliscribe kit, Epicentre Technology) and purified on an 8% PAGE containing 7 M urea. Obtained RNAs were used for the next cycle of the selection. The PCR product after the fourth selection cycle was introduced into pGEM-T Easy (Promega) and cloned in Escherichia coli JM109 strain. Plasmid DNA was isolated from individual clones and sequenced (Big Dye Terminator Sequencing kit, Applied Biosystems) on a 377 DNA sequencer (Applied Biosystems). The secondary structure models of selected aptamers were drawn with the MulFold program based on the Zuker algorithm (34).
Table 1.
Selection condition
| Cycle | RNA pool (nmol) | Target (pmol) | Volume (μl) | Wash (μl × times) |
|---|---|---|---|---|
| 1 | 10 | 50 | 900 | 1000 × 1 |
| 2 | 4 | 40 | 800 | 600 × 1 |
| 3 | 3 | 30 | 800 | 800 × 2 |
| 4 | 2 | 30 | 1100 | 600 × 3 |
Binding and kinetic analyses of aptamers using surface plasmon-resonance (SPR) technology
SPR experiments were performed with a BIACORE 2000 apparatus (Biacore). All procedures were carried out at 20°C using the surface of a streptavidin-coated sensor chip SA (Biacore). About 250 response units (RU) of domain IIId were captured on the chip. Various concentrations of aptamer (3.125–1600 nM) were applied. The collected response data were analyzed with the BIA evaluation program version 3.2. Kinetic parameters were determined using a simple model. The equations used to describe the reaction model are as follows: dR/dt = konC(Rmax − R) − koffR, dR/dt = −koffR, where R is the signal response, Rmax is the maximum response level, C is the molar concentration of the injected sample RNA, and kon and koff are the association and dissociation rate constants, respectively.
Inhibition assay of IRES-dependent translation with the aptamer
Inhibition of firefly luciferase mRNA in vitro translation (encoding IRES-Core-luciferase-3′-UTR) was performed using rabbit reticulocyte (Flexi Rabbit Reticulocyte Lysates kit, Promega) according to the manufacturer's protocol. First, 4 μg of aptamer and 0.5 μg of luciferase mRNA (transcribed from p5′IL-U3′X) were mixed and incubated for 5 min on ice. Next, 16 μl of reticulocyte lysates was added to the mixture, and the translation reaction was carried out in the presence of potassium chloride (120 mM) and magnesium acetate (2.5 mM) for 90 min at 30°C. Luciferase activity was measured using the PicaGene kit (Toyo-ink).
RNase mapping analysis of 3-07 aptamer and complexes of aptamer/IRES domain III–IV
RNAs (3-07 or domain III–IV) were labeled at 5′ end with [γ-32P]ATP by T4 polynucleotide kinase (Takara), and purified on an 8% PAGE containing 7 M urea. They were denatured at 90°C for 2 min and refolded at room temperature for 15 min. The labeled RNA (50 000 c.p.m.) was used for all RNase mapping reactions and partially digested by either RNase A, T1 or U2. For the alkaline ladder marker, probe RNAs were hydrolyzed for 4 min at 90°C in 6 μl reaction mixture containing 3.5 μl of alkaline solution (500 mM NaHCO3, pH 9.25) and 5 μg of E.coli total tRNA (Sigma).
To analyze the structure of aptamer 3-07, it was digested with 0.5 ng of RNase A (Sigma) in 10 μl selection buffer containing 5 μg of E.coli total tRNA (Sigma) at room temperature for 1 min. For the A-specific marker of 3-07, it was digested with 0.001 U of RNase U2 in the 10 μl RNase U2 optimal buffer (16.5 mM sodium citrate, pH 3.5 and 0.85 mM EDTA) containing 5 μg E.coli total tRNA at room temperature for 1 min.
To investigate the interaction of IRES domain III–IV and aptamers, IRES was partially digested with 0.1 U of RNase T1 (NEB) in 10 μl selection buffer containing 5 μg of E.coli total tRNA in the presence or the absence of 20 pmol of aptamers at room temperature for 2 min. Two kinds of markers, which were A-specific cleavage (A marker) and G-specific cleavage (G marker) of IRES domain III–IV, were prepared in the absence of any aptamer. For the A marker, the labeled IRES was digested with 0.001 U of RNase U2 in 10 μl RNase U2 optimal buffer containing 5 μg E.coli total tRNA. For the G marker, the labeled IRES was digested with 1.0 U RNase T1 in the 10 μl RNase T1 optimal buffer (16.5 mM sodium citrate, pH 5.0 and 0.85 mM EDTA) containing 5 μg E.coli total tRNA at room temperature for 1 min.
Digestion reactions were stopped by addition of equal volume of urea dye loading buffer (7 M urea, 0.083% xylen cyanol FF and 0.083% bromophenol blue), then cooled on ice. After ethanol precipitation, all samples were resuspended in half-concentrated urea dye loading buffer and denatured at 90°C for 2 min. These samples were subsequently analyzed on 8% denaturing polyacrylamide gel containing 7 M urea.
Transfection of aptamers or plasmids into HeLa cells
HeLa cells were grown in DMEM (Sigma) containing 10% heat-inactivated fetal bovine serum (Life Technologies). The day before transfection, 105 cells were plated per well of a 24-well plate in growth medium. Cells were 90–95% confluent at the time of transfection. Reporter plasmid DNA or RNA 3-07, 3-07(GGG), 3-07(GGAAAG) or N30H (60mer unrelated RNA pool containing random 30 nt) were transfected by Lipofectamine 2000 (Invitrogen) in two steps. In the first, 0.8 μg of the reporter plasmid p5′IL-U3′X (encoding firefly luciferase translated in an IRES-dependent manner) and 0.8 ng of the internal control plasmid pRL-CMV (Promega; encoding Renilla luciferase translated in a cap-dependent manner) were co-transfected. After 5 h, the media was replaced and 0.05 pmol or 0.5 pmol RNAs were transfected in the second step. Five hours after the second transfection, the media was changed again. Twenty-four hours after the first transfection, the cells were harvested and the luciferase activities of the lysates were measured using the Dual-Luciferase Reporter Assay System (Promega). Results were normalized against Renilla luciferase activity.
RESULTS
In vitro selection of RNAs that bind to HCV IRES domain III–IV
In vitro selection is a useful strategy to survey RNA–RNA interactions (28–33). As long target sequences of ∼100 nt usually present difficulties in chemical synthesis, we previously developed a simple selection method that used a biotinylated DNA probe to fix long target RNA onto beads (33). Using this method, we could obtain aptamers with strong binding affinities, although their inhibition of IRES-dependent translation was only moderate. Here, we aimed to generate aptamers that bind to HCV IRES domain III–IV (Figure 1B), which is much longer, at 250 nt, than domain II. These aptamers were expected to strongly inhibit translation, because domain III–IV is extremely important in HCV translation. The target domain III–IV was hybridized to a biotinylated DNA probe (27 nt), which was complementary to the 5′ extension of domain III (corresponding to domain II), and were fixed to streptavidin magnetic beads (Figure 1D). The 10 nmol randomized pool of 30 nt lengths of RNA was used in the initial selection step. The selection cycle was repeated four times (G4).
Sequence analysis of selected aptamers
To survey convergence, RNAs from the fourth generation were cloned and 18 RNA clones were sequenced (Figure 2). Nine clones of group A in Figure 2 had a 5 nt consensus sequence, 5′-ACCCA-3′, which is complementary to U265–U269 in the apical loop of domain IIId (Figure 1C). This convergence suggests that aptamers could interact with the apical loop of IRES domain IIId and that this domain seems to be preferentially selected as the target region of RNA–RNA interactions. Eight clones contained complementary sequences to the single-stranded 3′ end of domain IV (group B in Figure 2). These clones did not inhibit IRES-dependent translation (as described later). The remaining clone had a sequence that was complementary to the loop region of domain IIIb (group C in Figure 2). We stopped to do more selection cycles after fourth generation, because we could obtain one major consensus sequence and further continuing selections may result accumulating of group B clones. Hereafter, we focused on clones containing the 5′-ACCCA-3′ consensus sequence.
Figure 2.
Nucleotide sequences of a parental N30V RNA pool and G4 RNA aptamers. N in the RNA pool sequence denotes A, G, C or U. The RNA sequences obtained are classified into three categories: (A) is a group that contains the consensus ‘ACCCA’ sequence complementary to domain IIId and (2) indicates that two identical sequences were obtained; (B) is a group that contains the ‘GUUU’ sequence complementary to the single-stranded region between domain II and III of the IRES; and (C) is a group that contains complementary sequences to domain IIIb.
Binding analysis of aptamers to IRES domain IIId by SPR
The binding affinities between aptamers and domain IIId were determined using SPR technology. To immobilize on a streptavidin chip, we used chemically synthesized 3′-biotinylated domain IIId (nt 253–279). To collect binding data, various concentrations (3.125–1600 nM) of aptamers were loaded onto the chip. The responses obtained were analyzed using the BIA evaluation program 3.2. All aptamers containing the consensus sequence ACCCA showed similar KD values, except for 3-17 (Table 2). The difference between the KD values mainly depended on the values of kon. Aptamer 3-15, which did not have the consensus sequence, showed no response. This indicates that the consensus sequence is essential for binding between aptamers and domain IIId.
Table 2.
Rate constants and dissociation constants of RNA aptamers against domain IIId of HCV IRES
| RNA aptamer | kon (×104 M−1s−1) | koff (×10−4 s−1) | KD (nM) |
|---|---|---|---|
| 3-01 | 19 ± 10 | 5.4 ± 1 | 4.4 ± 3 |
| 3-03 | 1.1 ± 0.6 | 3.5 ± 1 | 43 ± 30 |
| 3-04 | 6.0 ± 1 | 7.2 ± 2 | 12 ± 2 |
| 3-05 | 10 ± 0.9 | 7.2 ± 0.1 | 7.2 ± 0.7 |
| 3-06 | 6.1 ± 3 | 6.5 ± 3 | 13 ± 9 |
| 3-07 | 5.9 ± 2 | 5.1 ± 2 | 9.6 ± 4 |
| 3-15 | ND | ND | ND |
| 3-17 | 1.8 ± 0.4 | 23 ± 2 | 130 ± 30 |
| 3-18 | 1.3 ± 1 | 2.7 ± 2 | 22 ± 4 |
ND: binding response was not detected.
Inhibition of IRES-dependent translation in vitro by aptamers
In order to survey the effect of aptamers on IRES-dependent translation activity, we performed an in vitro translation assay of IRES–luciferase mRNA containing the 3′-UTR of the HCV genome after the luciferase gene. Translation was examined using rabbit reticulocyte lysate and 0.5 μg of mRNA in the presence of aptamers (Figure 3). Aptamer 3-07 strongly decreased the luciferase activity to 10% of the control levels. Aptamers 3-03 and 3-04 reduced activity to 30–40%, and other aptamers had almost no effect. Aptamers 3-15 and 3-08 without the consensus sequence did not have an inhibitory effect. This result indicates that translation inhibition is dependent on the presence of the consensus sequence and inhibition efficiency is not related to the binding efficiency. Accordingly, we focused on aptamer 3-07 and we detected dose-dependent inhibition by aptamer 3-07 (IC50 ∼0.6 μM) using 0.5 μg of mRNA. No effect was observed using luciferase mRNA without the IRES (data not shown). This result shows that translation inhibition is dependent on aptamer 3-07 and is a specific effect on the IRES. As the inhibitory effect varied among aptamers, the aptamer structure seems to be important for translation inhibition. Recently, antisense RNA-targeted domain IIId showed more strong inhibition (IC50 < 10 nM) compared to our aptamer (13), but experimental conditions are different.
Figure 3.
Inhibition analysis of IRES-mediated translation in vitro by aptamers. Luciferase activity in the absence of an aptamer was used as a control (100%). Values represent results from at least three independent experiments. A, B and C are equivalent to the groups in Figure 2.
Secondary structure analysis of aptamer 3-07
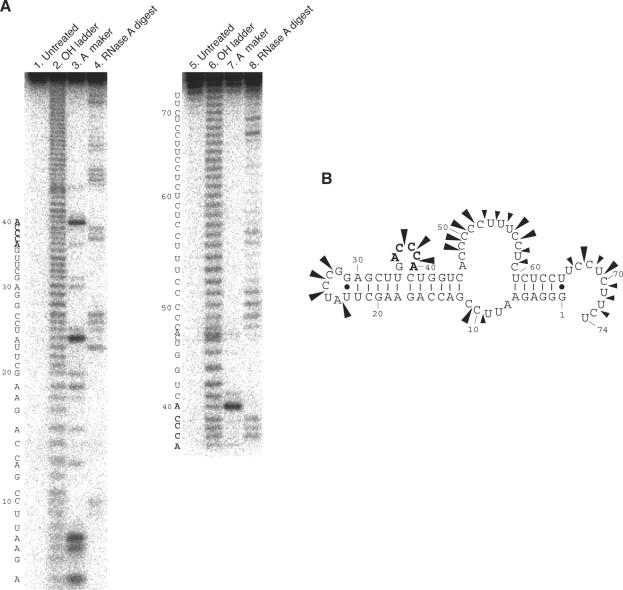
To analyze the secondary structure of aptamer 3-07 predicted by Mulfold program, we carried out RNase A mapping in the selection buffer. The cleavage pattern was analyzed on a denaturing PAGE (Figure 4A) and cleavages mainly occurred at single-stranded regions (Figure 4B). This result shows that the predicted structure by Mulfold program is correct and the consensus sequence forms an internal loop (Figure 4B).
Figure 4.
(A) RNase A mapping of aptamer 3-07. 5′ end-labeled aptamer 3-07 was digested with RNase A. Left and right figures show 5′ and 3′side cleavage pattern of 3-07 aptamer, respectively. End-labeled aptamer was applied to a PAGE without further reactions (lanes 1 and 5). Alkaline ladder (lanes 2 and 6) and RNase U2 digestion (lanes 3 and 7) were used as markers. RNase U2 digestion carried out under the U2 buffer. Aptamer 3-07 was digested with RNase A (C and U specific cleavage) under the selection buffer (lanes 4 and 8). (B) Predicted secondary structure of aptamer 3-07. Arrowheads and their size indicate cleavage site by RNase A and cleavage intensity, respectively.
Mutation analysis of aptamer 3-07
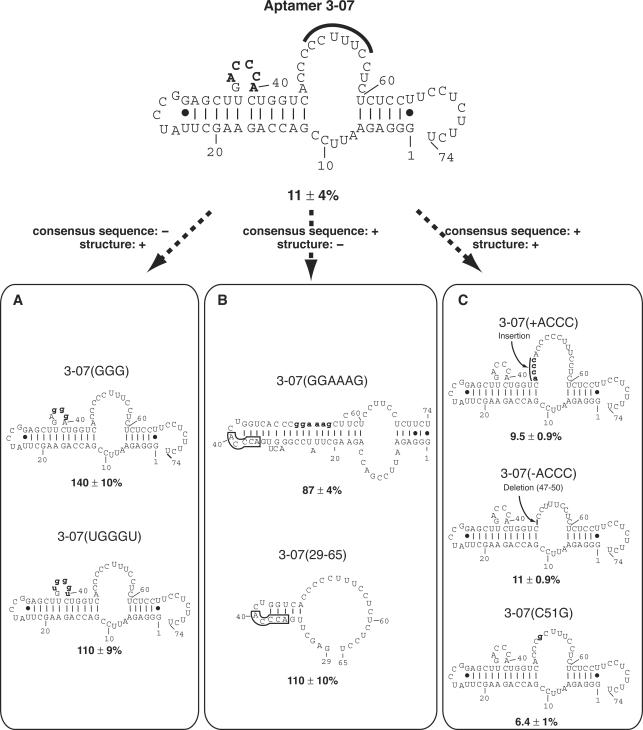
To investigate whether the consensus sequence or the whole structure of 3-07 aptamer is important for the inhibition of IRES-dependent translation, we constructed several mutants and analyzed the relationship between their inhibition ability and secondary structure (Figure 5). The secondary structures of these aptamers are based on Mulfold program. The consensus sequence mutants, 3-07(GGG) and 3-07(UGGGU) were drastically decreased in the inhibition efficiency even though they had similar structure to the original 3-07 (Figure 5A). Whereas 3-07(GGAAAG) and 3-07 (29–65) that have different structures from the original were also decreased in their inhibition ability even though they had the consensus sequence (Figure 5B). These results suggest that first the consensus sequence, 5′-ACCCA-3′ is important and second the whole structure of aptamer 3-07 is important for the inhibition of IRES-dependent translation.
Figure 5.
Structure–inhibition relationships of aptamer 3-07 and its mutants in vitro. Relative luciferase activity in the absence of aptamer was set at 100% and is shown below each structure. Numbering is based on the original aptamer 3-07. (Top) Secondary structure of aptamer 3-07. The consensus sequence is shown in bold letters. The sequence CCUUUUC that is complementary to the stem region of IRES domain IIId is marked by bold line. (Bottom) Mutants are classified by sequence and structure; the consensus sequence and secondary structure described as retaining (+) or not (−). All secondary structures were drawn by the Mulfold program and numbering is based on the original aptamer 3-07. In (B), the boxed regions indicate the consensus sequence. Mutated sequences are indicated by bold and lowercase letters.
The sequence of nt 51–56 of aptamer 3-07, which is a primer-binding region for RT–PCR, is complementary to the stem region of IRES domain IIId (nt 273–278, 5′-GAAAGG-3′). To analyze the effect of this sequence against the translation, three 3-07 mutants were constructed in addition to 3-07(GGAAAG) that was substituted at nt 51–56 region for 5′-GAAAGG-3′ (Figure 5B and C). The 3-07(+ACCC) mutant with the sequence 5′-ACCC-3′ inserted at the 5′ side of nt 51, the 3-07(−ACCC) mutant with the 5′-ACCC-3′ nt 51–56 sequence deleted and the 3-07(C51G) mutant carrying a C51 to G mutation. Of these mutants, 3-07(−ACCC), 3-07(+ACCC) and 3-07(C51G) retained the same strong inhibitory effect as the 3-07 aptamer (Figure 5C).
The dissociation constants of dysfunctional mutants in Figure 5 were more than ten times higher than that of the 3-07 aptamer, and we could not detect the binding of 3-07(GGG) mutant (Table 3). These findings demonstrate again that consensus sequence 5′-ACCCA-3′ and structure of 3-07 are significant for binding to IRES and inhibition of IRES-dependent translation.
Table 3.
Rate constants and dissociation constants of aptamer 3-07 mutants against domain IIId of HCV IRES
| 3-07 mutant | kon (×104 M−1 s−1) | koff (×10−4 s−1) | KD (nM) |
|---|---|---|---|
| 3-07(GGG) | ND | ND | ND |
| 3-07(GGAAAG) | 0.53 ± 0.2 | 5.5 ± 2 | 110 ± 40 |
| 3-07(C51G) | 4.0 ± 0.8 | 3.7 ± 0.6 | 9.5 ± 0.6 |
| 3-07(29-65) | 0.42 ± 0.1 | 13 ± 1 | 330 ± 100 |
ND: binding response was not detected.
Identification of binding regions between IRES domain III–IV and aptamer 3-07
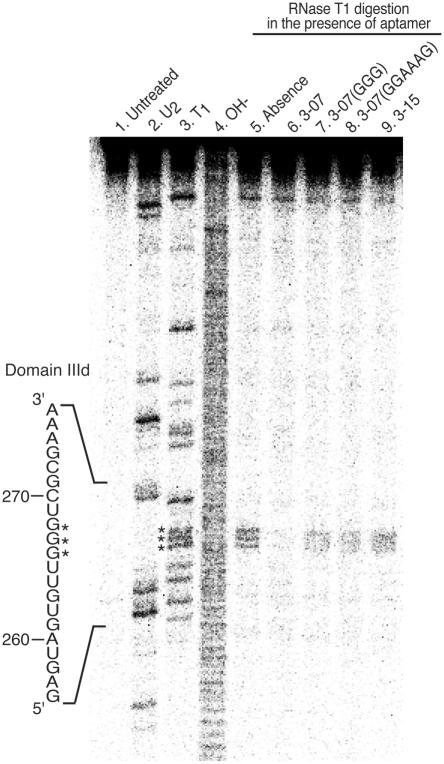
To identify the binding regions of IRES domain III–IV and aptamer 3-07, we carried out RNase T1 mapping analysis of IRES domain III–IV in the presence or absence of aptamers, 3-07, 3-07(GGG), 3-07(GGAAAG) and aptamer 3-15, which does not have the consensus sequence (Figure 6).
Figure 6.
RNase T1 mapping of IRES domain III–IV in the presence of aptamer 3-07. 5′ end-labeled IRES RNA was digested with RNase U2, RNase T1 and an alkaline condition, respectively, were used as markers (lanes 2, 3 and 4). Cleavage reactions for markers were carried out under optimum buffer conditions for respective RNases. RNase T1 digestion with selection buffer was performed on 5′ end-labeled IRES (lane 5), the aptamer indicated (lanes 6 and 9) or mutants of aptamer 3-07 (lanes 7 and 8). Reactions were stopped and run on an 8% denaturing PAGE. Asterisks represent G triplets of domain IIId. Note that cleavage of G triplets was only protected in the presence of aptamer 3-07.
We detected cleavages only at the G triplet (266–268) of the apical loop near domain IIId in the absence of aptamers (lane 5). These cleavages were protected in the presence of aptamer 3-07 (lane 6), but not protected in the presence of mutants, 3-07(GGG) and 3-07(GGAAAG), and also aptamer 3-15 (lanes 7, 8 and 9, respectively). No other difference of cleavage pattern was detected. This result indicates that the apical loop of IRES domain IIId predominantly interacts with aptamer 3-07 at its internal loop. Therefore, it is considered that internal loop, ACCCA, and whole structure of 3-07 are important for binding to IRES domain IIId.
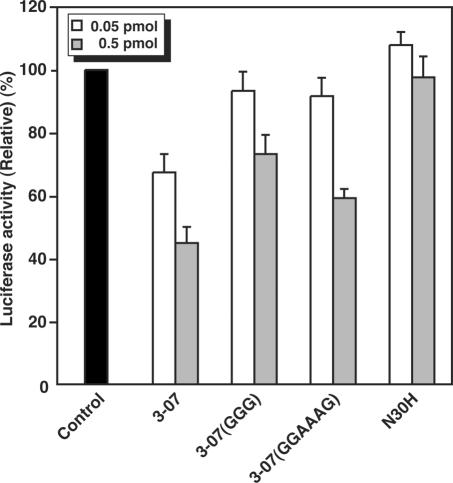
Inhibition of IRES-dependent translation in HeLa cells
To evaluate the effect of the 3-07 aptamer in mammalian cells, RNA aptamers 3-07, 3-07(GGG), 3-07(GGAAAG) and N30H RNA pool (as a negative control) were transfected into HeLa cells transiently expressing firefly and Renilla luciferases, which were translated via IRES- and cap-dependent manners, respectively. Results were normalized against Renilla luciferase activity. Transfection of either 0.05 or 0.5 pmol of aptamer 3-07 decreased firefly luciferase activity to 67 and 45%, respectively (Figure 7). Transfecting 0.5 pmol of mutants 3-07(GGG) and 3-07(GGAAAG) also reduced firefly luciferase activity, although to a lesser extent (73 and 59%, respectively). This result suggests that the consensus sequence 5′-ACCCA-3′ is the most important for inhibiting IRES function in HeLa cells and that the whole structure also has some inhibitory effect.
Figure 7.
Inhibition of IRES-dependent translation initiation in mammalian cells. RNAs 0.05 pmol (white bar) or 0.5 pmol (gray bar) were co-transfected into HeLa cells transiently expressing firefly and Renilla luciferase. The measured values of firefly luciferase activity driven by HCV IRES were normalized to Renilla luciferase activities. The graph shows relative values of luciferase activities against the non-transfected control. Each value is the average of three independent experiments.
DISCUSSION
In vitro selection technology is a powerful method to search for target sites of ribozymes and antisense nucleic acids. We previously showed that selection using a biotinylated DNA probe was useful for long target-RNA sequences, and was able to elucidate the location of RNA–RNA interactions (33). Aptamers targeted to HCV IRES domain II specifically bound to the apical loop of domain II via a loop–loop interaction (33). In this report, we confirmed that the selection strategy was suitable for even longer RNA targets, such as domain III–IV. Using in vitro selection, the consensus complementary sequence against the apical loop of domain IIId was detected (Figure 2). It is surprising to note that the RNAs obtained using affinity selection selectively recognized domain IIId, though there were several stem–loop structures in domain III–IV. This convergence suggests that the apical loop of domain IIId has the greatest ability to interact with exogenous RNA molecules. Tallet-Lopez and co-workers carried out selection against domain IIId using short hairpin RNAs, but failed to identify any targeting RNA structures. They clarified suitable antisense oligonucleotides targeted to this domain (13).
From a structural and functional point of view, it is interesting to note that aptamers focused on the apical loop of domain IIId. This structure, which was solved by NMR (18,19), contains a helical stem with non-Watson–Crick base pairs and an apical loop (Figure 1C). The apical loop, 5′-UUGGGU-3′, was more disordered than any other region of the domain. Lukavsky and colleagues used ketoxal-probing analysis to show that the GGG triplet of the loop interacts directly with the 40S ribosomal subunit (18). Jubin and co-workers reported that the GGG triplet functions essentially for IRES-dependent translation and RNA folding (24). In this report, we showed that aptamer 3-07 has a high affinity for the apical loop of domain IIId. The G base that is next to the 5′ end of the consensus sequence might interact with C270 of the stem of the domain. However, the CCC of the consensus sequence was critical for the binding (Table 3). NMR data previously showed that base U264 of domain IIId stacks with the base pair G263–C270 (18); this stacking conformation might result in mispairing between U264 and aptamer 3-07. Furthermore, the structure of G267–C270 is unique, whereas U269 bulges out and the direction of the sugar backbone is inverted (18,19). Therefore, the A base at the 5′ end of the aptamer consensus sequence might be able to pair with U269 and stabilize the RNA–RNA interaction. RNase T1 digestion of the domain IIId GGG triplet was clearly protected by aptamer 3-07 (Figure 6). This indicates that aptamer 3-07 exclusively interacts with the apical loop of domain IIId.
The aptamers obtained in this study bound to domain IIId under similar KD values (Table 2). However, there was a significant difference for IRES-dependent translation inhibition in vitro (Figure 3). It is probably due to the difference of target molecules. We used domain IIId in the binding experiment and whole IRES sequence in the translation inhibition assay, respectively. Aptamer 3-07 reduced luciferase activity by 90%. We think its strong inhibitory activity is due to its distinctive structure. From the results of RNase A mapping analysis, the consensus sequence of aptamer 3-07, ACCCA, forms a folded internal loop (Figure 4B). The interaction between the apical loop of domain IIId and the internal loop of aptamer 3-07 might be effective in inhibition of translation. Aldaz-Caroll and co-workers suggested that such apical loop–internal loop interaction is stable for RNA–RNA complex formation (32).
Aptamer 3-07 strongly inhibited the IRES-dependent translation of firefly-luciferase mRNA in vitro. Mutation results indicate that the existence of the consensus sequence and the whole structure of 3-07 are necessary for maximum inhibition. Both 3-07(GGG) and 3-07(GGAAAG) mutants almost lost their inhibition ability, but very weak inhibition was detected in 3-07(GGAAAG) mutant (Figure 5B). Binding data also showed similar tendency (Table 3). On the other hand, we could detect more inhibition ability on both mutants in vivo assay (Figure 7). We cannot precisely explain this inconsistency of in vitro and in vivo. But when we compare two mutants, 3-07(GGAAAG) is effective than 3-07(GGG). This finding suggests that first the consensus is important and second the structure is important for inhibition.
We isolated RNA aptamers capable of binding to the HCV IRES domain IIId. Our selection system showed the character of RNA–RNA interaction of structural RNA. This system is capable of applying to other targets and allowing elucidation of natural or unnatural RNA–RNA interaction. One of the obtained aptamers inhibited IRES-dependent translation both in vitro and in vivo. Combinations with advanced gene delivery and expression systems will provide more effective function of aptamer 3-07, leading as anti-HCV drug.
Acknowledgments
We thank Prof. K. Shimotohno for providing plasmid p5′IL-U3′X, F. Nishikawa for helpful discussions and for synthesizing biotinylated RNA, Dr J. Hwang for useful advice on Biacore-apparatus handling and Dr P.K.R. Kumar for helpful discussions. K.K. and K.F. were supported by New Energy and Industrial Technology Development Organization (NEDO) fellowships. Funding to pay the Open Access publication charges for this article was provided by National Institute of Advanced Industrial Science and Technology.
REFERENCES
- 1.Rosenberg S. Recent advances in the molecular biology of hepatitis C virus. J. Mol. Biol. 2001;313:451–464. doi: 10.1006/jmbi.2001.5055. [DOI] [PubMed] [Google Scholar]
- 2.Kato N., Hijikata M., Ootsuyama Y., Nakagawa M., Ohkoshi S., Sugimura T., Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl Acad. Sci. USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsukiyama-Kohara K., Iizuka N., Kohara M., Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang C., Sarnow P., Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pestova T.V., Shatsky I.N., Flether S.P., Jackson R.J., Hellen C.U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sizova D.V., Kolupaeva V.G., Pestova T.V., Shatsky I.N., Hellen C.U. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolupaeva V.G., Pestova T.V., Hellen C.U. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J. Virol. 2000;74:6242–6250. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kieft J.S., Zhou K., Jubin R., Doudna J.A. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanecak R., Brown-Driver V., Fox M.C., Azad R.F., Furusako S., Nozaki C., Ford C., Sasmor H., Anderson K.P. Antisense oligonucleotide inhibition of hepatitis C virus gene expression in transformed hepatocytes. J. Virol. 1996;70:5203–5212. doi: 10.1128/jvi.70.8.5203-5212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ali N., Pruijn G.J., Kenan D.J., Keene J.D., Siddiqui A. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. J. Biol. Chem. 2000;275:27531–27540. doi: 10.1074/jbc.M001487200. [DOI] [PubMed] [Google Scholar]
- 11.Anwar A., Ali N., Tanveer R., Siddiqui A. Demonstration of functional requirement of polypyrimidine tract-binding protein by SELEX RNA during hepatitis C virus internal ribosome entry site-mediated translation initiation. J. Biol. Chem. 2000;275:34231–34235. doi: 10.1074/jbc.M006343200. [DOI] [PubMed] [Google Scholar]
- 12.Macejak D.G., Jensen K.L., Pavco P.A., Phipps K.M., Heinz B.A., Colacino J.M., Blatt L.M. Enhanced antiviral effect in cell culture of type I interferon and ribozymes targeting HCV RNA. J. Viral. Hepat. 2001;8:400–405. doi: 10.1046/j.1365-2893.2001.00321.x. [DOI] [PubMed] [Google Scholar]
- 13.Tallet-Lopez B., Aldaz-Carroll L., Chabas S., Dausse E., Staedel C., Toulme J.-J. Antisense oligonucleotides targeted to the domain IIId of the hepatitis C virus IRES compete with 40S ribosomal subunit binding and prevent in vitro translation. Nucleic Acids Res. 2003;31:734–742. doi: 10.1093/nar/gkg139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ray P.S., Das S. Inhibition of hepatitis C virus IRES-mediated translation by small RNAs analogous to stem–loop structures of the 5′-untranslated region. Nucleic Acids Res. 2004;32:1678–1687. doi: 10.1093/nar/gkh328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nulf C.J., Corey D. Intracellular inhibition of hepatitis C virus (HCV) internal ribosomal entry site (IRES)-dependent translation by peptide nucleic acids (PNAs) and locked nucleic acids (LNAs) Nucleic Acids Res. 2004;32:3792–3798. doi: 10.1093/nar/gkh706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spahn C.M., Kieft J.S., Grassucci R.A., Penczek P.A., Zhou K., Doudna J.A., Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 17.Gallego J., Varani G. The hepatitis C virus internal ribosome-entry site: a new target for antiviral research. Biochem. Soc. Trans. 2002;30:140–145. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 18.Lukavsky P.J., Otto G.A., Lancaster A.M., Sarnow P., Puglisi J.D. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nature Struct. Biol. 2000;7:1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- 19.Klinck R., Westhof E., Walker S., Afshar M., Collier A., Aboul-Ela F. A potential RNA drug target in the hepatitis C virus internal ribosomal entry site. RNA. 2000;6:1423–1431. doi: 10.1017/s1355838200000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collier A.J., Gallego J., Klinck R., Cole P.T., Harris S.J., Harrison G.P., Aboul-ela F., Varani G., Walker S. A conserved RNA structure within the HCV IRES eIF3-binding site. Nature Struct. Biol. 2002;9:375–380. doi: 10.1038/nsb785. [DOI] [PubMed] [Google Scholar]
- 21.Kieft J.S., Zhou K., Grech A., Jubin R., Doudna J.A. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nature Struct. Biol. 2002;9:370–374. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- 22.Psaridi L., Georgopoulou U., Varaklioti A., Mavromara P. Mutational analysis of a conserved tetraloop in the 5′ untranslated region of hepatitis C virus identifies a novel RNA element essential for the internal ribosome entry site function. FEBS Lett. 1999;453:49–53. doi: 10.1016/s0014-5793(99)00662-6. [DOI] [PubMed] [Google Scholar]
- 23.Kieft J.S., Zhou K., Jubin R., Murray M.G., Lau J.Y., Doudna J.A. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 24.Jubin R., Vantuno N.E., Kieft J.S., Murray M.G., Doudna J.A., Lau J.Y., Baroudy B.M. Hepatitis C virus internal ribosome entry site (IRES) stem loop IIId contains a phylogenetically conserved GGG triplet essential for translation and IRES folding. J. Virol. 2000;74:10430–10437. doi: 10.1128/jvi.74.22.10430-10437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 26.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 27.Gold L., Polisky B., Uhlenbeck O., Yarus M. Diversity of oligonucleotide functions. Annu. Rev. Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 28.Scarabino D., Crisari A., Lorenzini S., Williams K., Tocchini-Valentini G.P. tRNA prefers to kiss. EMBO J. 1999;18:4571–4578. doi: 10.1093/emboj/18.16.4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duconge F., Toulme J.-J. In vitro selection identifies key determinants for loop–loop interactions: RNA aptamers selective for the TAR RNA element of HIV-1. RNA. 1999;5:1605–1614. doi: 10.1017/s1355838299991318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lodmell J.S., Ehresmann C., Ehresmann B., Marquet R. Convergence of natural and artificial evolution on an RNA loop–loop interaction: the HIV-1 dimerization initiation site. RNA. 2000;6:1267–1276. doi: 10.1017/s1355838200000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tok J.B.-H., Cho J., Rando R.R. RNA aptamers that specifically bind to a 16S ribosomal RNA decoding region construct. Nucleic Acids Res. 2000;28:2902–2910. doi: 10.1093/nar/28.15.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aldaz-Carroll L., Tallet B., Dausse E., Yurchenko L., Toulme J.-J. Apical loop–internal loop interactions: a new RNA–RNA recognition motif identified through in vitro selection against RNA hairpins of the hepatitis C virus mRNA. Biochemistry. 2002;41:5883–5893. doi: 10.1021/bi0121508. [DOI] [PubMed] [Google Scholar]
- 33.Kikuchi K., Umehara T., Fukuda K., Hwang J., Kuno A., Hasegawa T., Nishikawa S. RNA aptamers targeted to domain II of hepatitis C virus IRES that bind to its apical loop region. J. Biochem. (Tokyo) 2003;133:263–270. doi: 10.1093/jb/mvg036. [DOI] [PubMed] [Google Scholar]
- 34.Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:202–287. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]
- 35.Honda M., Beard M.R., Ping L.H., Lemon S.M. A phylogenetically conserved stem–loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 1999;73:1165–1174. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]