Abstract
HIV-1 replication can be efficiently inhibited by intracellular expression of an siRNA targeting the viral RNA. However, HIV-1 escape variants emerged after prolonged culturing. These RNAi-resistant viruses contain nucleotide substitutions or deletions in or near the targeted sequence. We observed an inverse correlation between the level of resistance and the stability of the siRNA/target-RNA duplex. However, two escape variants showed a higher level of resistance than expected based on the duplex stability. We demonstrate that these mutations induce alternative folding of the RNA such that the target sequence is occluded from binding to the siRNA, resulting in reduced RNAi efficiency. HIV-1 can thus escape from RNAi-mediated inhibition not only through nucleotide substitutions or deletions in the siRNA target sequence, but also through mutations that alter the local RNA secondary structure. The results highlight the enormous genetic flexibility of HIV-1 and provide detailed molecular insight into the sequence specificity of RNAi and the impact of target RNA secondary structure.
INTRODUCTION
Double-stranded RNA (dsRNA) can induce RNA interference (RNAi) in cells, resulting in sequence-specific degradation of the homologous single-stranded RNA (1,2). RNAi is an evolutionarily conserved process that may provide the host with a mechanism directed against transposable elements (3) and infecting viruses (4–6). The dsRNA trigger is processed by a ribonuclease (Dicer) into the effector molecules, ∼22 nt double-stranded RNAs termed short interfering RNAs (siRNAs) (7,8). One strand of the siRNA duplex is incorporated into the RNA-induced silencing complex (RISC), which subsequently binds and cleaves complementary RNA sequences (9,10). The efficiency of target RNA cleavage is affected by the stability of the siRNA/target-RNA duplex, which depends on the sequence complementarity between the siRNA and its target RNA (11), the nucleotide composition of the duplex and the precise position of nucleotide mismatches (12,13). Moreover, it has been suggested that RNAi efficiency is affected by the accessibility of the target RNA, which may be influenced by protein binding (12) and the formation of RNA secondary structure (14–18).
Introduction of siRNAs into cells has proven to be a powerful tool to suppress gene expression. Transfection of synthetic siRNAs into cells results in transient inhibition of the targeted gene (19). Long-term gene suppression can be achieved by the introduction of vectors that stably express short hairpin RNAs (shRNAs) that are processed into siRNAs by Dicer (20,21).
RNAi may be a powerful new method for intracellular immunization against human immunodeficiency virus type 1 (HIV-1). It has been demonstrated in short-term assays that HIV-1 replication can be inhibited by synthetic siRNAs targeting either viral RNA sequences or cellular mRNAs encoding protein cofactors that support HIV-1 replication (22–27). Recently, we demonstrated long-term inhibition of HIV-1 replication in human T cells that stably express siRNAs directed against the viral Nef gene (28). However, viral escape variants that were no longer inhibited by siRNA-Nef emerged. The siRNA-Nef target sequence in these RNAi-resistant viruses was either partially or completely deleted, or modified by nucleotide substitutions, which demonstrates the exquisite sequence-specificity of the RNAi mechanism. In this study, we analyzed the sequence changes in nine escape viruses and performed experiments to study the resistance mechanism. We observed a strong correlation between the stability of the siRNA/target-RNA duplex and the level of RNAi resistance. In addition, two viruses were found to escape from RNAi through mutations that induce an alternative secondary structure of the target RNA. These results demonstrate that occlusion of an siRNA-target sequence by RNA secondary structure reduces RNAi efficiency. Moreover, our results highlight the extreme versatility of HIV-1 and its evolutionary capacity to escape from RNAi-mediated antiviral therapy.
MATERIALS AND METHODS
Cells and viruses
SupT1 T-cells transduced with pRetro-SUPER expressing siRNA-Nef were cultured in RPMI 1640 medium containing 10% fetal calf serum (FCS), 100 U/ml penicillin and 100 U/ml streptomycin at 37°C and 5% CO2. SupT1 cells (2.5 × 105 cells in 1 ml medium) were infected with wild-type or mutant HIV-1 LAI (1 ng of CA-p24), and viral replication was monitored by determining the CA-p24 level in the supernatant by ELISA.
C33A cervix carcinoma cells were grown as a monolayer in DMEM supplemented with 10% FCS and minimal essential medium nonessential amino acids at 37°C and 5% CO2. C33A cells were transfected by the calcium phosphate method. Briefly, cells were grown in 3 ml of culture medium in 10 cm2 wells to 60% confluency. A mix of 5 μg wild-type or mutant HIV-1 LAI DNA in 110 μl water, 125 μl of 50 mM HEPES (pH 7.1), 250 mM NaCl, 1.5 mM Na2HPO4 and 15 μl of 2 M CaCl2, was incubated at room temperature for 20 min and added to the culture medium. The culture medium was changed after 16 h and viruses were harvested 3 days post-transfection.
DNA constructs
The full-length molecular HIV-1 clone LAI (29) was used to produce wild-type and mutant viruses. Nucleotide numbers presented here refer to the position on the genomic HIV-1 RNA transcript, with +1 being the capped G residue. The mutant proviral DNA sequences were PCR-amplified from cellular DNA with the 5′ Env primer tTA1-AD (+8269 to +8289) and the 3′ U5 primer CN1 (+9253 to +9283). The PCR fragments were digested with XhoI and BspEI, and cloned into the plasmid Blue-3′LTR (30). The XhoI–BglI fragments (1709 bp) of these plasmids were cloned into the wild-type LAI clone, resulting in the full-length mutant clones R1–R9.
The firefly luciferase expression vector pGL3 control (Promega) was used to construct the wild-type and mutant reporter-Nef target plasmids (pGL3-Nef). An approximately 250 bp Nef fragment (+8448 to +8698) was PCR amplified from the full-length molecular clones with the primers EW1 (5′-ACGTCTAGAATTCTGAGACGAGCTGAGCCAGCA-3′) and EW3 (5′-GACTCTAGACTGCAGGAGTGAATTAGCCCTTCCA-3′). The PCR product was digested with XbaI and cloned into the XbaI site located downstream of the luciferase gene in pGL3 control. The forward orientation of the insert was checked by sequence analysis.
To construct the m1–m4 mutants, base changes were introduced into pGL3-Nef by mutagenesis PCR (31). Mutagenic (m) primers EWmut1 (5′-ACAGCAGCTACCAATCCTGCTTGTGC -3′, mismatching nucleotide underlined; m1), EWmut2 (5′-CACAAGTAGGAATACAGCAGCTACCAACCTGCTTGTGC-3′; m2), EWmut3 (5′-CACAAGTAGTAATACAGCAG-3′; m3 and m4), and the general primers EW1 (Primer 1), EW2 (5′-TGAGGCCCGGTACCTGAGGTGTGACT-3′; primer 2), and EW3 (Primer 3) were used with the wild-type pGL3-Nef (m1, m2 and m4) or the R8 pGL3-Nef (m3) template. Briefly, PCR reactions were performed with primer M plus primer 3, and with primer 1 plus primer 2. The PCR products were purified, mixed and PCR amplified with primers 1 and 3 as described previously (31). The PCR fragments were digested with XbaI and cloned in the corresponding site of pGL3 control. All mutations were verified by sequence analysis.
The pRetro-SUPER-shNef vector, which expresses siRNA-Nef under control of the H1 RNA polymerase III promoter was digested with EcoRI and XhoI, and the 314 bp expression cassette was ligated into the EcoRI/XhoI sites of pBluescriptII (KS+) (Stratagene) to produce pBS-siRNA-Nef. Plasmid pRL-CMV (Promega) expresses renilla luciferase under control of the CMV promoter.
Luciferase assay
C33A cells were grown in 1 ml culture medium in 2 cm2 wells to 60% confluence and transfected by the calcium phosphate method. A 100 ng wild-type or mutant pGL3-Nef was mixed with 0.5 ng pRL-CMV, 0.5–500 ng pBS-siRNA-Nef and completed with pBluescriptII to 1 μg of DNA in 15 μl water. The DNA was mixed with 25 μl of 2× HBS and 10 μl of 0.6 M CaCl2, incubated at room temperature for 20 min and added to the culture medium. The culture medium was refreshed after 16 h. After another 24 h, the cells were lysed in 150 μl of Passive Lysis Buffer (PLB) (Promega) by shaking for 20 min at room temperature. The cell lysate was centrifuged and 10 μl of the supernatant was used to measure firefly and renilla luciferase activities with the Dual-luciferase Reporter Assay System (Promega). The renilla luciferase expression in transfected cells allowed us to correct for variation in transfection efficiency.
In silico RNA analysis
The stability of the duplex of siRNA-Nef (UGUGCUUCUAGCCAGGCAC) and wild-type or mutant Nef target sequences (+8448 to +8698) was predicted with the Hybridization Mfold program (32,33) at http://www.bioinfo.rpi.edu/applications/mfold. The RNA structure of the wild-type and mutant target sequences (+8524 to +8579) was predicted with the RNA Mfold program (32,33). Similar results were obtained with larger RNA fragments (+8448 to +8698; data not shown).
RNA structure probing
Wild-type LAI and R8 proviral plasmids were used as template for PCR amplification with primers EWr1 (5′-AATTTAATACGACTCACTATAGGGGTGGGAGCAGCATCTCGAG-3′; T7 RNA-polymerase promoter underlined) and EWr2 (5′-TGAATTAGCCCTTCCAGTCC-3′). The resulting PCR product contains a T7 RNA-polymerase promoter upstream of the HIV-1 nucleotides (+8474 to +8694). DNA products were purified with a PCR purification kit (Qiagen). RNA transcripts were produced by in vitro transcription with the Megashortscript T7 transcription kit (Ambion), and transcripts were purified on a NucAway™ spin column (Ambion). RNA concentrations were determined by spectrophotometry.
Wild-type and R8 RNA (20 pmol) were denatured in 60 μl water at 85°C for 3 min followed by snap cooling on ice. After addition of 20 μl 4× MO buffer (final concentration: 125 mM KAc, 2.5 mM MgAc, 25 mM HEPES, pH 7.0) and incubation for 30 min at 37°C, the transcripts were incubated with 5 mM lead(II) acetate at room temperature. Samples (15 μl) were taken at 0, 5, 15, 25 min, and cleavage was stopped by adding 3 μl of 1 M EDTA. RNA products were purified over a NucAway™ spin column (Ambion). Oligonucleotide EWr2 (5′-TGAATTAGCCCTTCCAGTCC-3′; +8675 to +8694) was 5′ end labeled with the kinaseMax kit (Ambion) in the presence of 1 μl of [γ-32P]ATP (0.37 MBq/μl, Amersham Biosciences). Three picomoles of 32P-labeled oligonucleotide EWr2 was annealed to 3 pmol of the lead(II)-treated RNA by incubation at 85°C for 3 min followed by slow cooling to 60°C. The primer was extended at 60°C for 1 h using the Thermoscript reverse transcriptase (Invitrogen). After adding 20 μl gel-loading buffer II (Ambion), the samples were heated to 95°C and 10 μl was analyzed on a denaturing 6% acrylamide gel. A sequence ladder of the wild-type Nef region was produced with the 32P-labeled oligonucleotide EWr2, the pGL3-Nef plasmid as template and the thermo sequenase cycle sequencing kit (USB).
Electrophoretic mobility shift assay (EMSA)
The siRNA-Nef antisense oligonucleotide UGUGCUUCUAGCCAGGCAC (Eurogentec) was 5′ end labeled with the kinaseMax kit (Ambion) in the presence of 1 μl of [γ-32P]ATP (0.37 MBq/μl, Amersham Biosciences). Wild-type and R8 RNA were denatured in 30 μl water at 85°C for 3 min followed by snap cooling on ice. After addition of 10 μl 4× MO buffer (final concentration: 125 mM KAc, 2.5 mM MgAc, 25 mM HEPES, pH 7.0), the RNA was renatured at 37°C for 30 min. The transcripts were diluted in 1× MO buffer to a final concentration varying from 0 to 7.5 μM in MO buffer. The 5′-labeled oligonucleotide was added (2.6 nM) and the samples (20 μl) were incubated for 30 min at room temperature. After adding 4 μl non-denaturing loading buffer (50% glycerol with bromophenol blue), the sample was analyzed on a non-denaturing 4% acrylamide gel. Electrophoresis was performed at 150 V at room temperature, and the gel was subsequently dried. Quantification of the free and bound oligonucleotide was performed with a Phosphor Imager (Molecular Dynamics).
RESULTS
Selection of RNAi-resistant HIV-1 variants
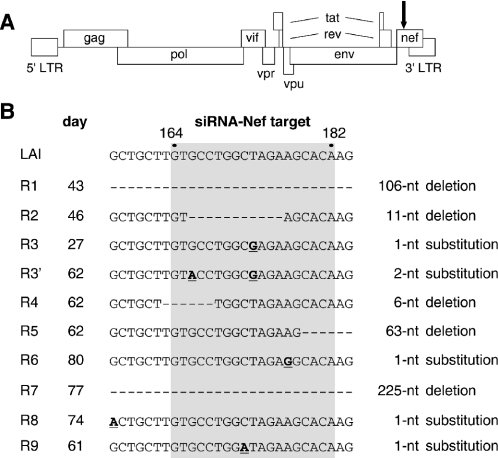
We previously demonstrated potent inhibition of HIV-1 replication in the SupT1 T-cell line by stable expression of an siRNA directed against the viral Nef gene (28). This target sequence is located near the 3′ end of the viral genome (Figure 1A), and is present in both the unspliced genomic RNA and all spliced subgenomic RNAs. This replication block is apparently not absolute as HIV-1 escape variants appeared after several weeks of culture. We previously reported the acquisition of either a nucleotide substitution or a deletion in the Nef target sequence in such escape viruses. We now selected additional escape variants and analyzed a total of nine cultures by RT–PCR of the Nef segment and subsequent sequence analysis (Figure 1B). A single nucleotide substitution was observed within the 19 nt target sequence in three escape viruses (R3, R6, R9). The R3 virus had acquired an additional nucleotide substitution at a later time (sample R3′). This finding suggests that the single R3 mutation did not provide complete RNAi-resistance. Partial or complete deletion of the Nef target was observed in five cultures (R1, R2, R4, R5, R7). These changes obviously affect the Nef open reading frame, resulting in the synthesis of a Nef protein with an internal deletion or a truncated protein due to a frameshift mutation. Since the Nef protein is not essential for HIV-1 replication in T-cell lines, these mutations will have no major impact on viral replication in our culture system. Most surprisingly, the R8 escape virus has no mutation in the siRNA-Nef target sequence, but does instead have a single nucleotide substitution 7 nt upstream of the target sequence.
Figure 1.
HIV-1 escape variants that resist siRNA-Nef inhibition. (A) Schematic representation of the HIV-1 LAI proviral genome. The position of the siRNA-Nef target sequence is indicated with an arrow. This sequence is present in both the unspliced and all spliced forms of HIV-1 RNA. (B) HIV-1 LAI variants resistant to siRNA-Nef were selected in nine independent cultures. The Nef target sequence (gray box indicates nucleotides 164–182 of the Nef gene; nucleotides 8553–8571 in the LAI RNA genome) and flanking sequences are shown for the wild-type (LAI) and the evolved RNAi-resistant viruses (R1–R9). The day at which the escape variants were sequenced is indicated. Deletions are shown as dashes, substitutions are underlined and in bold. In the R1 virus, nucleotides 125–230 of the Nef gene are deleted. In the R5 virus, nucleotides 179–241 are deleted. In the R7 virus, we observed deletion of nucleotides 44–268 and a T269A substitution.
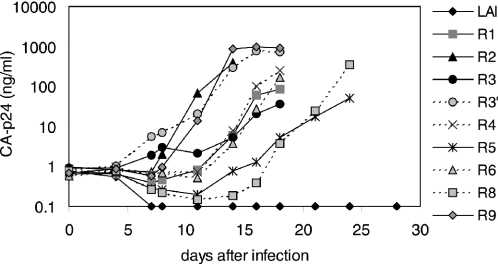
To verify that the observed mutations in or near the target sequence mediated the RNAi-resistant phenotype, we introduced the mutant Nef sequences in the HIV-1 LAI molecular clone. SupT1 cells that stably express siRNA-Nef were infected with wild-type or mutant HIV-1 virus. Wild-type HIV-1 is potently inhibited by the siRNA-Nef and did not initiate a spreading infection (Figure 2). In contrast, all mutant viruses replicate, demonstrating that the observed mutations in the Nef gene confer resistance against siRNA-Nef. The different escape variants do seem to replicate with different efficiencies. For instance, R3′ is more fit than the R3 virus, suggesting that the additional mutation in the target sequence provides a higher level of resistance against siRNA-Nef. However, we decided not to focus in more detail on the differences in replication fitness because it is a complex phenotype made up of several factors (level of RNAi-resistance, impact of Nef protein modification, potential removal of RNA replication signals, etc.).
Figure 2.
Nef mutations provide resistance against siRNA-Nef. The Nef mutations shown in Figure 1B were cloned in the HIV-1 LAI molecular clone. Virus stocks were produced in transiently transfected C33A cells and used to infect SupT1 cells stably expressing siRNA-Nef. Virus replication was monitored by determining the level of CA-p24 in the culture supernatant. Similar results were obtained in independent infection experiments. The R7 variant was excluded from further analysis since it contained a complete deletion of the siRNA-Nef target sequence very similar to the R1 variant. The wild-type LAI virus that does not replicate on these SupT1-siRNA-Nef cells replicates efficiently on control SupT1 cells [results not shown, see also (28)].
The siRNA/target-RNA duplex stability influences the level of RNAi resistance
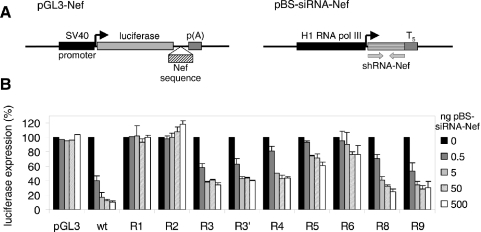
To accurately quantify the level of RNAi resistance, we made reporter gene constructs in which the wild-type or mutant Nef target sequence was placed downstream of the luciferase reporter gene (Figure 3A, pGL3-Nef). These constructs were co-transfected with an increasing amount of an siRNA-Nef expressing plasmid (pBS-siRNA-Nef) into the human cervix carcinoma cell line C33A, and luciferase production was measured after 48 h (Figure 3B). Reporter gene expression of the construct with the wild-type Nef sequence was significantly reduced by co-transfection with 0.5 ng pBS-siRNA-Nef (40% residual expression), and nearly complete inhibition (∼10% residual expression) was obtained with higher amounts of siRNA-Nef. Expression of the reporter gene construct in which the target sequence was completely deleted (R1) was not inhibited by siRNA-Nef, demonstrating complete resistance. The constructs with a partial deletion of the target sequence showed complete (R2, 11 nt deletion) or partial resistance against siRNA-Nef (R4 and R5, with a deletion of 5 and 4 nt in the target sequence, respectively). Expression of the latter constructs was inhibited marginally at low amounts of pBS-siRNA-Nef, and high siRNA-Nef levels resulted in partial inhibition (40–60% residual expression). The constructs with a single nucleotide substitution showed either partial resistance (R3 and R9) or nearly complete resistance (R6). The acquisition of a second mutation in the R3 virus (R3′) only marginally increased the level of resistance in this assay system. The construct with the mutation upstream of the target sequence (R8) also demonstrates partial resistance against siRNA-Nef in this assay.
Figure 3.
Quantification of the level of RNAi resistance. (A) A 250 bp Nef fragment (nucleotides 8448–8698 in the LAI RNA genome) that encompasses the target and flanking regions was cloned downstream of the firefly luciferase gene in the pGL3 reporter plasmid. These reporter gene constructs were cotransfected into C33A cells with the siRNA-Nef expressing plasmid pBS-siRNA-Nef. (B) Luciferase expression observed after transfection of the reporter constructs with increasing amounts of pBS-siRNA-Nef. The level of expression observed in the absence of siRNA-Nef was set at 100% for each reporter construct. This level did not vary significantly for the different constructs. The mean values obtained in three independent experiments are shown (±SE).
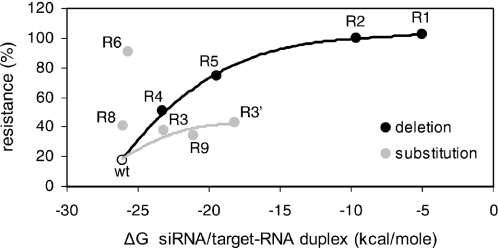
Since RNAi is dependent on the sequence-specific base-pairing of siRNA to the target RNA sequence, reduced binding due to mismatches may explain the resistance-phenotype of all escape variants, except for the R8 variant with a nucleotide substitution outside the target sequence. We therefore calculated the predicted thermodynamic stability (ΔG) of the siRNA/target-RNA interaction for the wild-type and mutant targets and plotted this value against the measured level of resistance (Figure 4). The siRNA-Nef forms a perfectly base-paired duplex with the wild-type target (ΔG = −26.1 kcal/mol), consistent with efficient inhibition by siRNA-Nef (18% resistance). Both the substitution and deletion mutants show reduced siRNA/target-RNA duplex stability that correlates with increased resistance against siRNA-Nef (Figure 4). However, two escape variants (R6 and R8) do not follow this general pattern and show an exceptionally high level of resistance. The nucleotide substitution in R8 is located upstream of the target sequence. The siRNA/target-RNA duplex stability is thus not affected for this mutant, yet it is fairly resistant against RNAi. The R6 mutant contains an A-to-G substitution in the target sequence that has only a minor effect on the siRNA/target-RNA duplex because an U–A base pair is replaced by an U–G base pair. Whereas this substitution only marginally affects the stability of the duplex (ΔG = −25.7 kcal/mol), the R6 mutant shows nearly complete resistance.
Figure 4.
RNAi-resistance due to reduced stability of the siRNA/target-RNA duplex. The thermodynamic stability (ΔG) of the siRNA-Nef/target-RNA duplex was calculated with the Hybridization Mfold program and plotted against the level of resistance as observed in Figure 3B with 5 ng of pBS-siRNA-Nef. The high level of resistance of the mutants R8 and R6 does not correlate with their predicted duplex stability.
Changes in target RNA structure can cause RNAi resistance
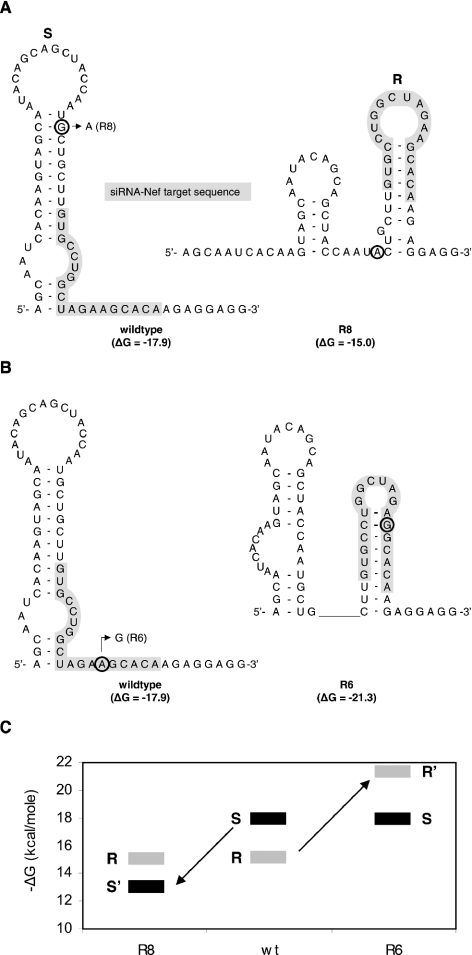
Binding of siRNA-Nef to its target sequence may also be affected by the accessibility of the target sequence, and thus by local RNA structure. Since RNAi resistance of the R6 and R8 mutants could not be explained by an altered stability of the siRNA/target-RNA duplex, we examined the effect of these mutations on the local RNA structure. We first used the Mfold program (32,33) to predict the secondary structure of the wild-type, R6 and R8 Nef target RNA. The energetically most favorable RNA structure of the wild-type Nef region (Figure 5A; ΔG = −17.9 kcal/mol) is an extended hairpin (S hairpin) that partially overlaps with the siRNA target sequence (marked in gray), but the 3′ half of the target sequence is predicted to be single-stranded. The G-to-A substitution upstream of the target sequence in R8 destabilizes this hairpin structure (ΔG = −13.0 kcal/mol), and the RNA is likely to fold an alternative, more stable conformation with two smaller hairpins (Figure 5A; ΔG = −15.0 kcal/mol). In this alternative conformation, the second hairpin (R hairpin) encompasses the complete target sequence of which both the 5′ and 3′ ends are occluded by base-pairing. This new hairpin configuration of the target sequence may prevent the binding of siRNA-Nef.
Figure 5.
Local target RNA structure can cause resistance against RNAi. (A) The predicted RNA structures (RNA Mfold program) of the wild-type and R8 Nef sequences. The target sequence is highlighted by a gray box. The G-to-A substitution 7 nt upstream of the target sequence in R8 is encircled. This mutation disrupts the preferred sensitive (S) hairpin (ΔG = −17.9 → ΔG = −13.0 kcal/mol), resulting in folding of a more stable alternative structure with the resistant (R) hairpin and a small upstream stem loop (ΔG = −15.0 kcal/mol). (B) RNA structures of the wild-type and R6 Nef sequences. The A-to-G substitution at nucleotide 14 of the target sequence in R6 is indicated. This mutation does not affect the S hairpin, but profoundly stabilizes the alternative R hairpin through formation of two additional base pairs (ΔG = −17.9 → ΔG = −21.3 kcal/mol). (C) The thermodynamic stability (ΔG) of the RNAi-sensitive (S) and the RNAi-resistant (R) structures are indicated for the wild-type (wt) and mutant (R6 and R8) Nef sequences. In wild-type RNA, the S structure (ΔG = −17.9 kcal/mol) is more stable than the R structure (ΔG = −15.1 kcal/mol). In mutant R8, the sensitive structure is destabilized (S′; ΔG = −13.0 kcal/mol) and energetically less favorable than the resistant structure (ΔG = −15.0 kcal/mol). Stabilization of the R hairpin in mutant R6 makes this conformation (R′; ΔG = −21.3 kcal/mol) more favorable than the sensitive conformation (S; ΔG = −17.9 kcal/mol).
This scenario is confirmed by inspection of the R6 escape variant. In fact, the R6 mutation in the target sequence does not reduce the stability of the wild-type S hairpin, but it profoundly stabilizes the alternative R hairpin (Figure 5B; ΔG = −21.3 kcal/mol). The A-to-G substitution in the target sequence allows the formation of two additional base pairs in this hairpin, thereby reducing the loop size from 10 to 6 nt. Thus, the exceptionally high level of resistance of R6 is probably due to occlusion of the target sequence in a stable hairpin structure, although a contribution of the slightly reduced stability of the siRNA/target-RNA duplex due to the formation of the G–U base pair cannot be excluded. In Figure 5C, the stability of the RNAi-sensitive (S) and the RNAi-resistant (R) conformations for the wild-type (wt), R6 and R8 RNAs are plotted. The wild-type can fold both RNA structures, but the S conformation is favored because it is more stable than the R conformation. The S conformation is destabilized (S′) in mutant R8, which thus becomes energetically less favorable than the R conformation. Stabilization of the R hairpin in mutant R6 makes this alternative conformation (R′) more favorable than the wild-type S structure.
The S and R RNA structure equilibrium dictates the level of RNAi resistance
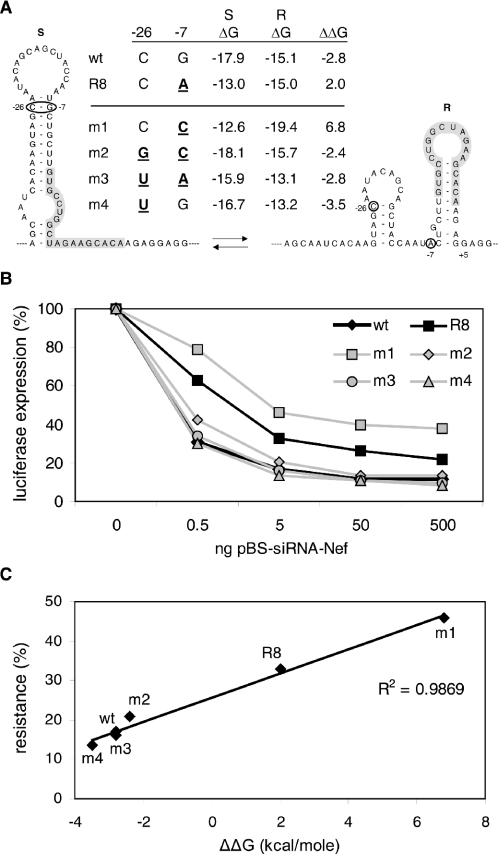
We designed additional mutants to confirm that the presence or absence of the wild-type S hairpin determines RNAi sensitivity. All new mutants affect the −26/−7 base pair of the S hairpin that was opened in the R8 escape variant, and these nucleotide changes are thus positioned outside the actual Nef target sequence (Figure 6A). The RNA Mfold program was used to estimate the thermodynamic stability of the S and R conformations and RNAi sensitivity was determined in the luciferase-assay system (Figure 6B). The G-to-C substitution at position −7 in mutant m1 resembles the R8 mutation and similarly opens the S hairpin. Like the R8 mutant, this m1 mutant shows partial resistance against siRNA-Nef (Figure 6B). However, the m1 mutation also stabilizes the alternative R conformation (m1: ΔGR = −19.4 kcal/mol) by the formation of two additional base pairs in the R hairpin (−7/+5 and −8/+6). This explains the higher level of resistance of m1 in comparison with R8 in which only hairpin S is destabilized. This resistant phenotype of m1 is reversed almost completely in the double mutant m2 by the introduction of a compensatory base change at position −26 (C-to-G), which restores base-pairing and preferential formation of the S hairpin. However, the m2 mutant is slightly more resistant than the wild-type. This can be explained by the relatively higher stability of the R hairpin in mutant m2 due to the −7 change. The −26 mutation in m2 also affects the stability of the small upstream hairpin of the R conformation. The ΔG values plotted in Figure 6A do include all these effects.
Figure 6.
Stability of the S hairpin determines the level of resistance. (A) Mutant pGL3-Nef reporter constructs with nucleotide substitutions upstream of the siRNA-Nef target sequence were constructed (mutations at positions −7 and/or −26 are in bold and underlined). The stability (ΔG) of the S and R structures was predicted with the Mfold program. The ΔΔG, providing a measure of the S and R structure equilibrium, was calculated (ΔΔG= ΔGS − ΔGR). (B) Luciferase expression after transfection of the reporter constructs with increasing amounts of pBS-siRNA-Nef. The level of expression in the absence of siRNA-Nef was set at 100% for each reporter construct. (C) The ΔΔG of the structures is plotted against the level of resistance as observed in Figure 6B with 5 ng of pBS-siRNA-Nef.
In the double mutant m3, we restored base-pairing in the original escape variant R8 (which has a G-to-A at position −7) by introducing a compensatory mutation (C-to-U at position −26). Accordingly, this compensation reversed the RNAi-resistant phenotype of R8. The corresponding single mutant m4, which contains only the C-to-U mutation at position −26, maintains base-pairing capacity in the S hairpin with a U–G base pair. Indeed, this mutant remains RNAi sensitive. The −26 mutation in m3 and m4 also affects the stability of the smaller hairpin in the R conformation, which is included in the ΔGR calculations. These combined results strongly suggest that it is not the identity of the −26/−7 base pair per se, but rather the stabilization of the S hairpin structure that is critical for RNAi sensitivity.
We calculated the differences in ΔG of the S and R conformations (ΔΔG), which reflects the S–R structure equilibrium, incorporating all mutational effects on the two conformations (Figure 6A). A negative ΔΔG value indicates that the RNA preferentially folds the S hairpin structure, whereas a positive value reflects a preference for the R hairpin conformation. Preference for the S hairpin correlates with RNAi sensitivity and mutations that shift the equilibrium towards the R hairpin conformation result in increased resistance against siRNA-Nef (Figure 6C).
RNA structure probing
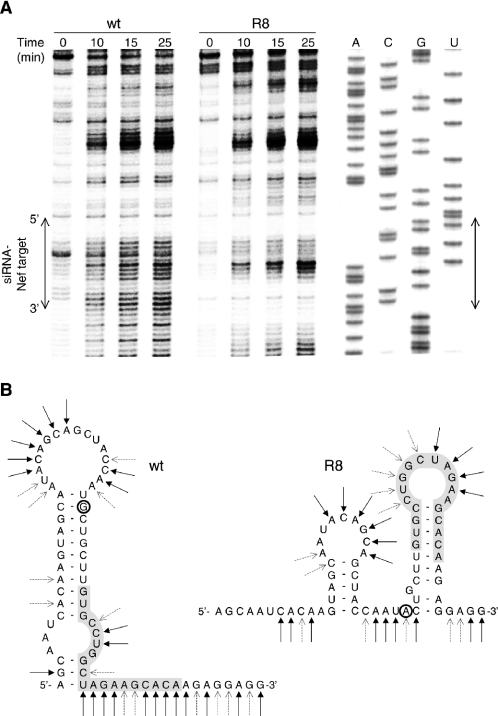
To demonstrate that the R8 mutation does indeed affect the local RNA structure, we performed RNA structure probing experiments on the Nef region using lead(II), which is known to preferentially cleave single-stranded RNA (Figure 7A). The cleavage pattern of the wild-type RNA is consistent with the proposed RNA secondary structure model (Figure 7B, left). For instance, we observed high reactivity in the loop and single-stranded regions and low reactivity in the base-paired stem. The 3′ end of the siRNA-Nef target sequence in the wild-type RNA is highly sensitive to lead, indicating that this region is single stranded. The cleavage pattern of the R8 mutant is different and confirms the proposed alternative folding (Figure 7B, right). In contrast to the wild-type RNA, the 3′ end of the siRNA-Nef target sequence is resistant to lead-induced cleavage, indicating that this region is base-paired as in hairpin R. The R8 RNA shows increased lead-sensitivity near the center of the target sequence, which corresponds with the exposed loop of the R hairpin. Domains further upstream and downstream on the wild-type and R8 RNA show a very similar lead-cleavage pattern, indicating that the change in RNA secondary structure occurs locally. These findings confirm that the R8 mutation results in alternative RNA folding that affects the presentation of the target sequence.
Figure 7.
RNA structure probing of the wild-type (wt) and R8 Nef sequence. (A) Wild-type and R8 RNA were treated with 5 mM lead(II) acetate for different times as indicated above the lanes. Sites of lead-induced cleavage were detected by primer extension on the RNA. The wild-type sequence was analyzed in parallel to assess cleavage positions, and the position of the siRNA-Nef target sequence is indicated with an arrow. (B) The lead-induced cleavage sites are represented on the secondary structure models of wild-type and R8 RNA. Strong and weak cleavages are indicated by solid and dotted arrows, respectively.
Stable target RNA structure blocks siRNA binding
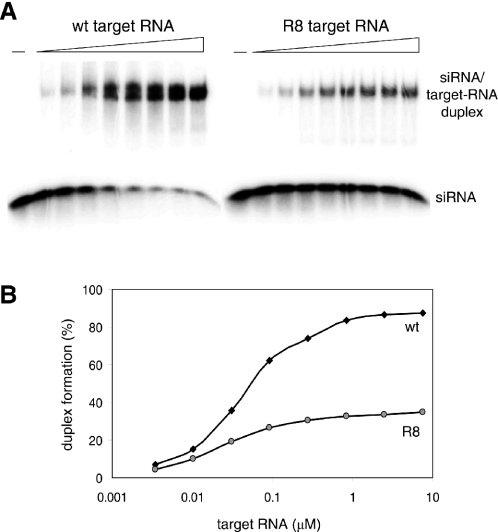
To demonstrate directly that the alternative structure formed by R8 RNA does occlude the Nef target sequence from binding to the complementary siRNA-Nef strand, we performed binding assays. A 19 nt RNA oligonucleotide corresponding to the antisense strand of the siRNA-Nef was radioactively labeled, incubated at room temperature with increasing amounts of wild-type and R8 target RNA, and analyzed in an electrophoretic mobility shift assay (Figure 8A). Binding of this siRNA-Nef oligonucleotide to the target RNA results in a profound shift on a non-denaturing gel. The free siRNA-Nef and the siRNA-Nef/target-RNA duplex bands were quantitated to calculate the percentage of binding (Figure 8B). The siRNA-Nef bound less efficiently to R8 RNA than to wild-type RNA. Whereas almost all siRNA is shifted at high levels of wild-type RNA, the same amount of R8 RNA is not able to shift more than 39% of the siRNA-Nef. Because RNA structures should be temperature sensitive, we also performed assays at elevated temperatures. Indeed, the binding defect of the R8 mutant was diminished under such conditions (results not shown). Furthermore, kinetic experiments with an excess of siRNA-Nef showed that binding to wild-type RNA is significantly faster than to R8 RNA (3- to 4-fold difference at room temperature, results not shown). Since these two RNA templates do not differ in the actual target sequence, these results indicate that the alternative structure of R8 does occlude the target sequence from interacting with siRNA-Nef.
Figure 8.
Target RNA structure can inhibit siRNA binding. (A) Radioactively labeled oligonucleotide simulating the antisense strand of siRNA-Nef was incubated with an increasing amount of wild-type or R8 RNA. The siRNA/target-RNA duplex formation was analyzed by EMSA. (B) Free and bound siRNA oligonucleotide was quantified to calculate the level of duplex formation (bound siRNA/free + bound siRNA).
DISCUSSION
HIV-1 replication can be inhibited efficiently through RNAi by targeting the viral Nef sequence. However, RNAi-resistant viruses emerged that contained nucleotide substitutions or deletions in or near the siRNA-Nef target sequence. We recloned these mutant Nef sequences in the HIV-1 genome and demonstrate that these mutations do indeed confer resistance against siRNA-Nef. We used a reporter–target gene construct to accurately quantitate the level of resistance. For most mutants, we observed an inverse correlation between the level of resistance and the stability of the siRNA/target-RNA duplex. Two mutants did not follow this trend and demonstrated an unexpected high level of resistance. We demonstrate that these mutations induce alternative folding of the RNA that occludes the target sequence from binding to siRNA-Nef. These results demonstrate that the efficiency of RNAi-mediated inhibition depends on the efficiency of siRNA-binding to the target RNA. This interaction can be diminished by nucleotide substitutions or deletions in the target sequence that cause mismatches with the siRNA, or by mutations that induce an RNA structure in which the target sequence is occluded.
It has previously been reported that a mismatch between the target and siRNA can affect RNAi to a variable degree, depending on the position of the mismatch in the duplex (13,34). Kinetic analyses suggested that different regions of the siRNA play distinct roles regarding target recognition, cleavage and product release (35). The 5′ end of the siRNA contributes to initial target RNA binding, and mutations in either the central or the 3′ region of the target affect RNAi efficiency most dramatically (13,34). Most nucleotide substitutions that we observed in the RNAi-resistant HIV-1 variants are located in the center of the Nef target sequence, consistent with this notion. Mutations in the 5′ region of the target RNA have a milder effect, which may explain the small difference in resistance of the early R3 and late R3′ variants. The partial deletion mutants R4 and R5 differ significantly in their levels of resistance against siRNA-Nef (respectively 50% and 75% with 5 ng of siRNA-Nef-expressing plasmid). Focusing on the siRNA-Nef target sequence, R4 has a 5 nt deletion at the 5′ side and R5 a 4 nt deletion at the 3′ side. The fact that R5 is more resistant than R4 is also consistent with the notion that mismatches in the 3′ region in the target RNA are less well tolerated by the RNAi machinery.
Several studies have suggested an inhibitory effect of target RNA structure on RNAi efficiency (14–18). However, these studies mainly compared the efficiency of different siRNA and target sequences, such that only correlations with target RNA structure could be suggested. Furthermore, these studies did not experimentally verify the proposed RNA structures. We assayed the efficiency of a single target sequence when present in a different structural context. We confirmed these alternative RNA secondary structures in probing experiments. We demonstrate that occlusion of the target sequence by RNA structure inhibits binding of the siRNA, and thus reduces RNAi efficiency. Interestingly, the wild-type RNAi-sensitive hairpin S occludes only the 5′ side of the target sequence, whereas both the 5′ and 3′ sides are base-paired in the alternative RNAi-resistant structure. This result indicates that an accessible 3′ target end is sufficient for target recognition, which is consistent with the idea that this 3′ region is mainly responsible for target recognition and binding (35).
It has previously been suggested that RNA hairpins are poor targets for RNAi because the corresponding antisense siRNAs will also have the ability to fold a stem–loop structure, which may hamper their activity (14). Although the 19 nt siRNA-Nef can indeed form a hairpin (6 bp stem, 6 nt loop: ΔG=−5.1 kcal/mol), we know that this siRNA-Nef is very effective in silencing of the wild-type virus, indicating that this siRNA structure is not inhibitory.
Gene therapy to impose HIV-1 specific RNAi could become a realistic approach to potently inhibit virus replication (36–38). The siRNA directed against the viral Nef gene demonstrated a potent and sequence-specific antiviral effect. However, due to the high mutation rate of HIV-1, the virus can escape from RNAi-mediated inhibition (28,39). The fact that RNAi-resistant HIV-1 variants can emerge not only through deletions or substitutions in the siRNA target sequence, but also through mutations that alter the local RNA structure, demonstrates the extreme flexibility of the HIV-1 virus. Some escape viruses have lost the ability to encode a functional Nef protein. Whereas complete inactivation of the accessory Nef function has a relatively minor impact on the HIV-1 replication fitness in vitro, it significantly attenuates replication in vivo (40,41). It is therefore less likely that HIV-1 will acquire resistance in vivo by deletion or frameshifting of the Nef open reading frame. Nevertheless, the virus may still evolve resistance by accumulating silent point mutations. It is therefore important to identify additional targets in highly conserved sequences in essential HIV-1 genes. There may be other constraints in the viral genome, e.g. overlapping open reading frames, which could be used to design potent siRNAs from which escape is very difficult. Furthermore, targeting of a sequence located in or near a functionally essential secondary RNA structure motif could be advantageous. Escape mutations (even silent codon changes) may disrupt such replication signals and are thus less likely to emerge. Additionally, the transcripts of cellular cofactors can be targeted, but the knockdown of such functions should not affect viability of the host cell. Ideally, HIV-1 should be targeted by multiple siRNAs against essential HIV-1 sequences (42). Similar to current antiviral drug combination therapies, this should prevent the emergence of escape mutants that are resistant to RNAi-mediated inhibition.
Acknowledgments
This research was sponsored by The Netherlands Organisation for Health Research and Development (ZonMw; VICI grant) and The Netherlands Organisation for Scientific Research (NWO-CW; TOP grant). The Open Access publication charges for this article were waived by Oxford University Press.
REFERENCES
- 1.Fire A., Xu S., Montgomery M.K., Kostas S.A., Driver S.E., Mello C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Hammond S.M., Bernstein E., Beach D., Hannon G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 3.Ketting R.F., Haverkamp T.H., van Luenen H.G., Plasterk R.H. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 4.Vance V., Vaucheret H. RNA silencing in plants—defense and counterdefense. Science. 2001;292:2277–2280. doi: 10.1126/science.1061334. [DOI] [PubMed] [Google Scholar]
- 5.Waterhouse P.M., Wang M.B., Lough T. Gene silencing as an adaptive defence against viruses. Nature. 2001;411:834–842. doi: 10.1038/35081168. [DOI] [PubMed] [Google Scholar]
- 6.Bennasser Y., Le S.Y., Yeung M.L., Jeang K.T. HIV-1 encoded candidate micro-RNAs and their cellular targets. Retrovirology. 2004;1:43. doi: 10.1186/1742-4690-1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zamore P.D., Tuschl T., Sharp P.A., Bartel D.P. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 9.Nykanen A., Haley B., Zamore P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- 10.Martinez J., Patkaniowska A., Urlaub H., Luhrmann R., Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 11.Zeng Y., Cullen B.R. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holen T., Amarzguioui M., Wiiger M.T., Babaie E., Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res. 2002;30:1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pusch O., Boden D., Silbermann R., Lee F., Tucker L., Ramratnam B. Nucleotide sequence homology requirements of HIV-1-specific short hairpin RNA. Nucleic Acids Res. 2003;31:6444–6449. doi: 10.1093/nar/gkg876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo K.Q., Chang D.C. The gene-silencing efficiency of siRNA is strongly dependent on the local structure of mRNA at the targeted region. Biochem. Biophys. Res. Commun. 2004;318:303–310. doi: 10.1016/j.bbrc.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 15.Yoshinari K., Miyagishi M., Taira K. Effects on RNAi of the tight structure, sequence and position of the targeted region. Nucleic Acids Res. 2004;32:691–699. doi: 10.1093/nar/gkh221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vickers T.A., Koo S., Bennett C.F., Crooke S.T., Dean N.M., Baker B.F. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J. Biol. Chem. 2003;278:7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- 17.Kretschmer-Kazemi F.R., Sczakiel G. The activity of siRNA in mammalian cells is related to structural target accessibility: a comparison with antisense oligonucleotides. Nucleic Acids Res. 2003;31:4417–4424. doi: 10.1093/nar/gkg649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bohula E.A., Salisbury A.J., Sohail M., Playford M.P., Riedemann J., Southern E.M., Macaulay V.M. The efficacy of small interfering RNAs targeted to the type 1 insulin-like growth factor receptor (IGF1R) is influenced by secondary structure in the IGF1R transcript. J. Biol. Chem. 2003;278:15991–15997. doi: 10.1074/jbc.M300714200. [DOI] [PubMed] [Google Scholar]
- 19.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 20.Brummelkamp T.R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 21.Paul C.P., Good P.D., Winer I., Engelke D.R. Effective expression of small interfering RNA in human cells. Nat. Biotechnol. 2002;20:505–508. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- 22.Coburn G.A., Cullen B.R. Potent and specific inhibition of human immunodeficiency virus type 1 replication by RNA interference. J. Virol. 2002;76:9225–9231. doi: 10.1128/JVI.76.18.9225-9231.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacque J.M., Triques K., Stevenson M. Modulation of HIV-1 replication by RNA interference. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee N.S., Dohjima T., Bauer G., Li H., Li M.J., Ehsani A., Salvaterra P., Rossi J. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- 25.Novina C.D., Murray M.F., Dykxhoorn D.M., Beresford P.J., Riess J., Lee S.K., Collman R.G., Lieberman J., Shankar P., Sharp P.A. siRNA-directed inhibition of HIV-1 infection. Nature Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- 26.Qin X.F., An D.S., Chen I.S.Y., Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl Acad. Sci. USA. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Omoto S., Ito M., Tsutsumi Y., Ichikawa Y., Okuyama H., Andi B.E., Saksena N.K., Fuji Y. HIV-1 nef suppression by virally encoded microRNA. Retrovirology. 2004;1:44. doi: 10.1186/1742-4690-1-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das A.T., Brummelkamp T.R., Westerhout E.M., Vink M., Madiredjo M., Bernards R., Berkhout B. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J. Virol. 2004;78:2601–2605. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peden K., Emerman M., Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 30.Klaver B., Berkhout B. Comparison of 5′ and 3′ long terminal repeat promoter function in human immunodeficiency virus. J. Virol. 1994;68:3830–3840. doi: 10.1128/jvi.68.6.3830-3840.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mikaelian I., Sergeant A. A general and fast method to generate multiple site directed mutations. Nucleic Acids Res. 1992;20:376. doi: 10.1093/nar/20.2.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathews D.H., Sabina J., Zuker M., Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 33.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amarzguioui M., Holen T., Babaie E., Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haley B., Zamore P.D. Kinetic analysis of the RNAi enzyme complex. Nature Struct. Mol. Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 36.Haasnoot P.C., Cupac D., Berkhout B. Inhibition of virus replication by RNA interference. J. Biomed. Sci. 2003;10:607–616. doi: 10.1159/000073526. [DOI] [PubMed] [Google Scholar]
- 37.Silva J.M., Hammond S.M., Hannon G.J. RNA interference: a promising approach to antiviral therapy? Trends Mol. Med. 2002;8:505–508. doi: 10.1016/s1471-4914(02)02421-8. [DOI] [PubMed] [Google Scholar]
- 38.Kitabwalla M., Ruprecht R.M. RNA interference—a new weapon against HIV and beyond. N. Engl. J. Med. 2002;347:1364–1367. doi: 10.1056/NEJMcibr022294. [DOI] [PubMed] [Google Scholar]
- 39.Boden D., Pusch O., Lee F., Tucker L., Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J. Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deacon N.J., Tsykin A., Solomon A., Smith K., Ludford-Menting M., Hooker D.J., McPhee D.A., Greenway A.L., Ellett A., Chatfield C., et al. Genomic structure of an attenuated quasi species of HIV-1 from blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 41.de Ronde A., Klaver B., Keulen W., Smit L., Goudsmit J. Natural HIV-1 NEF accelerates virus replication in primary human lymphocytes. Virology. 1992;188:391–395. doi: 10.1016/0042-6822(92)90772-h. [DOI] [PubMed] [Google Scholar]
- 42.Berkhout B. RNA interference as an antiviral approach: targeting HIV-1. Curr. Opin. Mol. Ther. 2004;6:141–145. [PubMed] [Google Scholar]