Abstract
Since its initial description, the yeast two-hybrid (Y2H) system has been widely used for the detection and analysis of protein–protein interactions. Mating-based strategies have been developed permitting its application for automated proteomic interaction mapping projects using both exhaustive and high-throughput strategies. More recently, a number of prokaryotic two-hybrid (P2H) systems have been developed but, despite the many advantages such Escherichia coli-based systems have over the Y2H system, they have not yet been widely implemented for proteomic interaction mapping. This may be largely due to the fact that high-throughput strategies employing bacterial transformation are not as amenable to automation as Y2H mating-based strategies. Here, we describe the construction of novel conjugative P2H system vectors. These vectors carry a mobilization element of the IncPα group plasmid RP4 and can therefore be mobilized with high efficiency from an E.coli donor strain encoding all of the required transport functions in trans. We demonstrate how these vectors permit the exploitation of bacterial conjugation for technically simplified and automated proteomic interaction mapping strategies in E.coli, analogous to the mating-based strategies developed for the Y2H system.
INTRODUCTION
Protein–protein interactions play important functional roles in virtually every cellular process. They mediate many disease states and biological processes important for the pathogenesis of bacterial and viral infections (1), and the specificity of these interactions makes them ideal targets for novel therapeutic agents (2–4). The identification of protein interactions can also be a critical step in the determination of unknown protein functions, and as many of the predicted genes in the ever growing list of completed genome sequences are of unknown function, the construction of protein interaction maps can aid the functional annotation of genome sequences and further our understanding of complex biological processes. The detection and analysis of such molecular interactions has thus become the latest challenge facing molecular biologists in the post-genomic era. High-throughput, easy to handle, and preferably automatable, technologies are required to facilitate the full exploitation of the enormous volume of genomic sequence data now available. As a result, a wide variety of biochemical and genetic-based assays have been developed for the detection of protein–protein interactions, and the yeast two-hybrid (Y2H) system (5) currently represents one of the most powerful genetic-based approaches for the detection of protein–protein interactions in vivo.
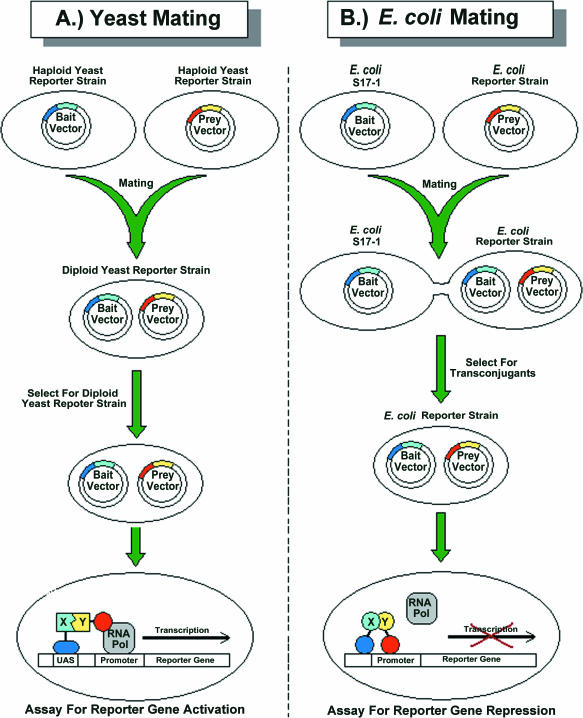
The Y2H system employs hybrid proteins, generally termed Bait and Prey hybrid proteins, for the detection of protein–protein interactions via reporter gene activation resulting in a readily detectable phenotype (see Figure 1A). The Y2H system is highly sensitive, often detecting interactions undetected by other techniques such as co-immunoprecipitation, and since its initial description, has spawned the development of a whole repertoire of similar genetic-based systems for the detection of a wide range of biomolecular interactions [reviewed in (6–9)]. The Y2H system itself has been widely applied not only to detect novel interacting partners for proteins of interest by performing library screens (10,11) but also to study interactions between defined protein pairs to identify minimal domains (12) and critical residues (13) required for interaction. The system has been adapted, through the use of yeast mating types (14), for high-throughput and automated protein interaction mapping strategies (see Figure 2A). It has been applied for the construction of protein interaction maps for functional groups of proteins (14–16), small proteomes (17,18) and more recently has been applied for the analysis of more complex proteomes (19–24).
Figure 1.
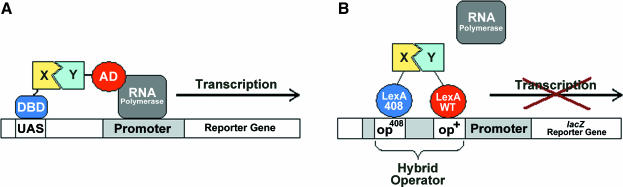
Schematic representation depicting the detection of protein–protein interactions in two-hybrid systems. (A) The Y2H system: X represents a given protein fused to a specific DNA binding domain (DBD). In library screens this protein is termed the ‘Bait’. Y represents a given protein, or a pool of proteins encoded by a DNA library fused to a transcriptional activation domain (AD). This fusion protein is often termed the ‘Prey’. If X and Y interact, the AD is brought to the vicinity of the DNA bound DBD and transcription is activated from the adjacent promoter resulting in a clearly detectable phenotype. (B) The LexA repressor-based P2H system: X represents a given Bait protein fused to a mutant LexA DBD (LexA408) and Y represents a given Prey protein, or library of Prey proteins, fused to a wild-type LexA DBD (LexAWT). Interactions between X and Y produce a heterodimeric LexA repressor capable of binding to a hybrid (op408/op+) operator site and repressing reporter gene expression.
Figure 2.
Mating-based protein–protein interaction mapping. (A) Application of yeast mating for protein–protein interaction mapping: The Bait vector carrying a cloned gene of interest is transformed into a haploid yeast reporter strain of one mating type (MAT a). This is then mated either with individual Prey clones, or with pools of Prey clones, maintained in a haploid yeast reporter strain of opposite mating type (MAT α). Diploid yeast reporter strains carrying both a Bait and a Prey vector are selected and assayed for reporter gene activation to detect protein–protein interactions. (B) Application of bacterial mating for protein–protein interaction mapping using the LexA repressor-based P2H system. This is analogous to yeast-mating strategies. A mobilizable Bait vector carrying a cloned gene of interest is transformed into E.coli strain S17-1. This is then mated either with individual Prey clones, or with pools of Prey clones, maintained in the E.coli reporter strain for the P2H system. Transconjugant E.coli reporter strain cells carrying both a Bait and a Prey vector are selected for and then assayed for reporter gene repression to detect protein–protein interactions.
In recent years, there has been a growing interest in the development of prokaryotic two-hybrid (P2H) systems as these present many advantages over Y2H systems and represent an experimental alternative to the yeast-based systems. A large number of P2H systems have been described previously (25–39) [reviewed in (40,41)], but despite their many advantages, P2H systems still have not been widely implemented for large-scale or proteomic scale protein interacting mapping in the same way as the Y2H system. Such applications of the P2H systems described to date, with the exception of the system developed by Joung and co-workers (31) that permits the exploitation of phage transduction, require the use of bacterial transformation to introduce plasmids into appropriate Escherichia coli reporter strains to test for interactions (42). While transformation-based strategies are possible, they are technically more involved and therefore less conducive to automation than the technically simplistic mating-based strategies developed for Y2H systems. It is probably for this very reason that the Y2H system has remained the preferred approach for large-scale protein interaction mapping.
Here, we describe the construction of novel Bait vectors for use with the P2H system developed by Dmitrova et al. (39) and based on the E.coli LexA repessor protein. The LexA protein comprises an N-terminal DNA-binding domain (DBD) and a C-terminal dimerization domain and only binds to operator sequences, comprising two inverted repeat half sites (op+/op+), as a dimer. It has been demonstrated that the natural dimerization domain of this repressor protein can be replaced with heterologous homodimerizing domains, and the resulting chimeric protein can still function as an efficient repressor (43,44). In order to permit the detection of heterologous protein–protein interactions, Dmitrova et al. (39) generated a mutant LexA DBD, the LexA408 DBD, with an altered binding specificity for a mutant operator sequence, op408. One protein of an interacting pair, X, is fused C-terminally to this LexA408 DBD, while the other, Y, is fused C-terminally to a wild-type LexA DBD (LexAWT DBD). The hybrid proteins are then co-expressed, from separate plasmids, in a specially constructed E.coli reporter strain, SU202, carrying a chromosomally integrated reporter gene. This reporter gene comprises a lacZ gene placed under the transcriptional control of a modified sulA promoter bearing hybrid op408/op+ LexA operator sequences. Only a heterologous interaction between X and Y generates a heterodimeric protein, with one LexAWT DBD and one LexA408 DBD, capable of binding the hybrid op408/op+ operator to repress the expression of the lacZ reporter gene (see Figure 1B).
The new Bait vectors that we have constructed carry a region of the conjugative plasmid RP4, called a mob site (mobilization element). These vectors can therefore be transferred by highly efficient bacterial conjugation from a donor E.coli strain called S17-1, bearing a chromosomally integrated RP4 plasmid encoding all of the required plasmid mobilization functions (45), into a recipient reporter E.coli strain already harbouring the Prey vector for the system. The resulting transconjugant E.coli reporter strain carries both the Prey and Bait vectors for the system and can be assayed to detect protein–protein interactions. This bacterial mating therefore achieves the same result as the mating of yeast cells (see Figure 2B). Using the model interaction of the eukaryotic Jun and Fos leucine zippers, we demonstrate how selected Bait clones can be mated with pools of Prey clones for technically simplified screening of libraries to identify interacting partners with an efficiency unattainable using bacterial transformation procedures or yeast mating. We also demonstrate how each clone in a Bait library could be mated with each individual clone in an arrayed Prey library in a matrix format using a technically simplistic and automatable protocol (see Figure 3). Exhaustive screening in this fashion would reveal all possible interactions between proteins in the Prey and Bait libraries. The strategies we describe here are analogous to those currently implemented for high-throughput proteomic interaction mapping using Y2H mating-based approaches (15,19,21,22,46–48) and should therefore permit the application of this LexA P2H system, or any of the previously described P2H systems modified to incorporate such a mobilizable vector, for similar proteomic interaction mapping projects.
Figure 3.
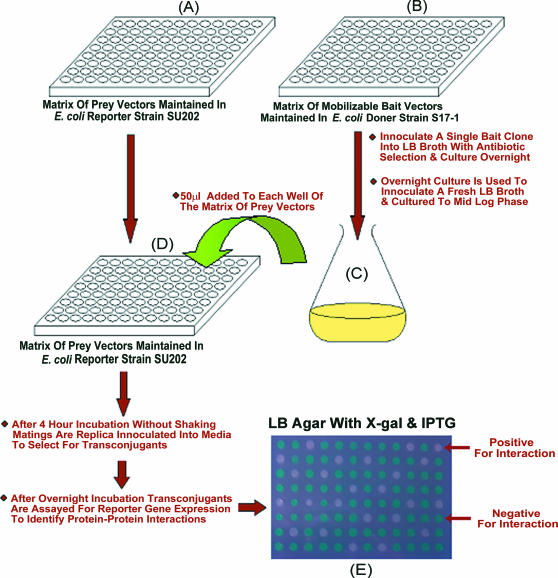
Exhaustive protein interaction mapping in E.coli using a P2H system and bacterial conjugation. In order to carry out exhaustive protein interaction mapping, matrices of Prey and Bait clones would be constructed by cloning PCR-amplified open reading frames (ORFs) into the Prey and Bait vectors for the P2H system. The cloning of amplified ORFs would initially be carried out using standard E.coli strains and once constructed Prey vectors would be transformed into the P2H systems reporter strain SU202 to form a matrix of recipient Prey clones (A), and Bait vectors would be transformed into an E.coli strain, such as S17-1, to form a matrix of donor Bait (B) clones, respectively. Each donor Bait clone would then mated with each Prey clone by simply culturing the Bait clone in bulk in LB broth (C) and adding an aliquot to each well of a freshly grown plate of recipient Prey clones (D). Following a 4 h incubation, matings would be replica inoculated into fresh plates to select for transconjugants, incubated overnight and finally a sample of each transconjugant would be spotted onto the surface of selective media to screen for protein–protein interactions. Above transconjugants exhibiting the jun and fos interaction, and thus harbouring plasmids pPC810 and pMS604, were detected by spotting onto LB IPTG X-gal indicator plates (E) and were identified as white culture spots and after 24 h incubation at 37°C. Transconjugants not exhibiting the jun and fos interaction, and thus harbouring plasmids pPC810 and pPC605, appeared as blue culture spots.
MATERIALS AND METHODS
The plasmids and strains utilized in this work are listed in Table 1 and a more detailed description of some of these is given below.
Table 1.
Bacterial strains and plasmids used in this study
| E.coli | Relevant genotype | Reference | ||
|---|---|---|---|---|
| XL1-Blue | recA1, hsdR17(rk−, mK+), supE44, lac, [F′, proAB+lacIq, lacZΔM15::Tn10(TetR)] | Stratagene | ||
| SU202 | lexA71::Tn5(Def)sulA211 ΔlacU169, F′[lacIqlacZΔM15::Tn9] | (39) | ||
| S17-1 | thi pro res− mod+ SmR TpRrecA1 RP-4-2[Tc::Mu; Km::Tn7] | (45) | ||
| Plasmids | Origin | Resistance | Features | Reference |
| pMS604 | ColE1 | Tetr | lexAWT-fos construct | (44) |
| pPC605 | ColE1 | Tetr | lexAWT-MCS construct | This study |
| pDP804 | p15a | Ampr | lexA408-jun construct | (39) |
| pJQ200ks | p15a | Gmr | mob sacB | (49) |
| pJQ200-NS | p15a | Gmr | mob sacB | This study |
| pPC810 | p15a | Gmr | mob sacB lexA408-jun construct | This study |
| pPC811 | p15a | Gmr | mob sacB lexA408-MCS construct | This study |
Bacterial strains and plasmids
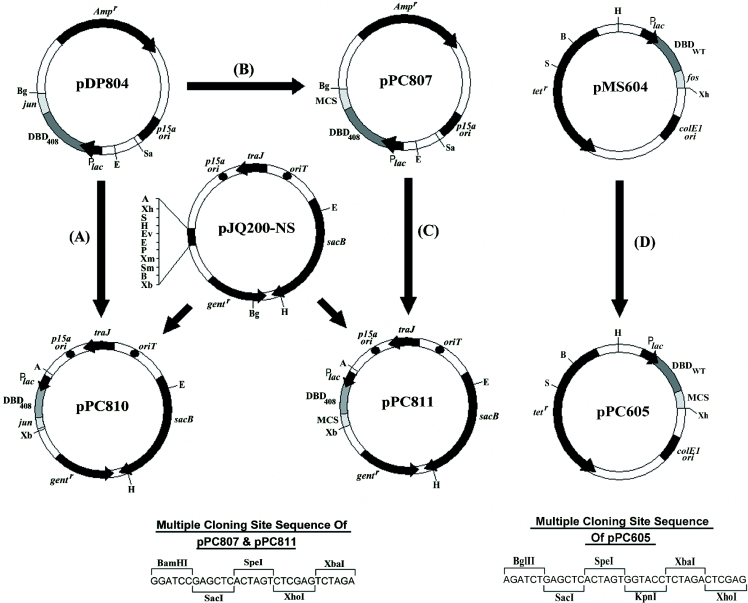
Plasmids pMS604 and pDP804 are designated in this work as Prey and Bait vectors, respectively (see Figure 4). Plasmid pMS604 (44) expresses a chimeric protein consisting of a wild-type LexA DBD fused to the Fos leucine zipper (LexAWT-Fos). Plasmid pDP804 (39) expresses a chimeric protein consisting of a mutated LexA DBD, called LexA408, fused to the Jun leucine zipper (LexA408-Jun). The hybrid proteins on both of these plasmids are expressed from UV5lac promoters and thus expression can be induced using isopropyl-β-d-thiogalactopyranoside (IPTG).
Figure 4.
Schematic representation depicting the construction of novel mobilizable Bait vectors for use with a LexA repressor-based P2H system (see Materials and Methods for full construction details). Plasmids pDP804, pPC807, pPC810 and pPC811 all carry a LexA408 DBD coding sequence (DBD408) while plasmids pMS604 and pPC605 carry a LexAWT DBD coding sequence (DBDWT). Restriction sites are given as follows: BglII (Bg), EcoRI (E), SacII (Sa), ApaI (A), XhoI (Xh), SalI (S), HindIII (H), EcoRV (Ev), PstI (P), XmaI (Xm), SmaI (Sm), BamHI (B), XbaI (Xb).
Plasmid pJQ200-sk (49) carries an RP4 mob site and can thus be mobilized with high efficiency from an E.coli strain, such as S17-1 described below. Plasmid pJQ200-NS (see Figure 4) was derived from plasmid pJQ200-sk. The NotI–SacII–SacI restriction sites of the multiple cloning site (MCS) of pJQ200-sk were eliminated by restricting the vector sequentially with NotI and then with SacI, filling in the resulting cohesive ends with Klenow and then re-ligating the blunt ends. This was carried out to permit the incorporation of these restriction sites as unique sites within a newly designed MCS. The pJQ200-sk plasmid and pJQ200-NS also carry a sacB gene that is lethal to E.coli cells grown in the presence of 5% sucrose. E.coli strains cured of any plasmid carrying this gene can be easily selected by streaking colonies onto Luria–Bertani (LB) agar lacking antibiotic selection for the plasmid and containing 5% sucrose.
The E.coli strain S17-1 (45) was used as a donor strain for bacterial matings. This strain harbours a chromosomally integrated IncPα group plasmid RP4 encoding all of the transfer functions necessary for plasmid mobilization. These products can function in trans mediating the mobilization of any plasmid carrying a recognizable RP4 mobilizable element called a mob site.
The E.coli strain SU202 (39) was the reporter strain used for the detection of heterologous protein–protein interactions. The strain is both kanamycin resistant (due to the chromosomally inserted Tn5) and chloramphenicol resistant (due to the Tn9 inserted on the F′ factor), but kanamycin was preferentially used for transconjugant selection. This is because the F factor does not carry a mutation in the traD gene and thus was found to be transferable during bacterial matings complicating transconjugant selection. The strain also carries a chromosomally integrated reporter gene consisting of a lacZ gene placed under the transcriptional control of a modified sulA promoter bearing a hybrid op408/op+ LexA operator sequence.
E.coli strains were routinely cultured on LB medium (50). Transformation was by the method of Inoue et al. (51) and E.coli XL1-Blue was used for routine transformation and plasmid preparation. Antibiotics were used at the following concentrations: 20 μg kanamycin ml−1, 10 μg tetracycline ml−1, 10 μg gentamicin ml−1 and 50 μg streptomycin ml−1. IPTG was used at a concentration of 1 mM for the induction of hybrid protein expression, and β-galactosidase activity was assayed by plating onto either LB agar containing IPTG and the chromogenic substrate X-gal at a concentration of 50 μg ml−1 or onto MacConkey agar containing 10% lactose and IPTG (available from Oxoid).
Molecular biology techniques were performed using standard protocols (50). Plasmid DNA was isolated from cultures by the alkaline lysis method (52). PCRs were carried out using the temperature gradient block in a Thermo Hybaid PCR Express thermocycler. Restricted DNA fragments for sub-cloning were cut from ethidium bromide-stained gels as required and purified using the Perfectprep gel clean up kit as directed by the manufacturers (Eppendorf).
Construction of the mobilizable Bait vector pPC810 expressing a LexA408-Jun hybrid protein
The region of plasmid pDP804 carrying the entire lexA408-jun expression construct, including the Plac UV5 promoter region, was sub-cloned onto the mobilizable plasmid pJQ200-NS. Plasmid pDP804 was linearized by digestion with EcoRI, treated with Klenow to fill in the cohesive ends generated and subsequently digested with BglII. The 524 bp blunt-BglII lexA408-jun fragment was then gel purified. Plasmid pJQ200-NS was linearized by digestion with XhoI, the cohesive ends were filled by Klenow treatment and the vector was subsequently digested with BamHI. The blunt-BamHI pJQ200-NS plasmid was then ligated with the gel-purified lexA408-jun blunt-BglII fragment and the resulting vector was named pPC810 (see Figure 4A).
Construction of the mobilizable Bait vector pPC811 facilitating genetic fusions with LexA408
A region of pDP804 extending from its unique SacII site to the end of the LexA408 DBD coding sequence was amplified using the following primers: LexA-F1, 5′-CCGCGGCCCTCTCACTTCCTTGTTAAGTATCTTCC-3′ and LexA-R1, 5′-AGATCTAAGCTTCTAGACTCGAGACTAGTGAGCTCGGATCCTGGTTCACCGGCAGCCACACGACCTACC-3′. The reverse primer was designed to incorporate a MCS, terminating in a BglII site, C-terminally to the lexA408 sequence (underlined bases correspond to the added MCS sequence). The resulting 717 bp PCR product was cloned into the pCR2.1 vector of a TA cloning kit (Invitrogen) and verified by sequencing. The fragment was sub-cloned as a SacII–BglII fragment back into the original pDP804 plasmid replacing the original lexA408-jun construct with a lexA408-MCS construct and the resulting plasmid was named pPC807 (see Figure 4B). The lexA408-MCS construct was then sub-cloned from plasmid pPC807 onto pJQ200-NS as follows. Plasmid pPC807 was linearized by digestion with EcoRI, treated with Klenow to fill in the cohesive ends generated and subsequently digested with XbaI. The resulting 426 bp blunt-XbaI lexA408-MCS fragment was then gel purified. Plasmid pJQ200-NS was linearized by digestion with XhoI, the cohesive ends were filled by Klenow treatment and the vector was subsequently digested with XbaI. The blunt-XbaI pJQ200-NS plasmid was then ligated to the gel-purified lexA408-MCS blunt-XbaI fragment and the resulting vector was named pPC811 (see Figure 4C).
Construction of a Prey vector pPC605 facilitating genetic fusions with LexAWT
A region of pMS604 extending from the unique HindIII site to the end of the LexAWT DBD coding sequence was amplified using the following primers; LexAW-F1: 5′-CTGTGATAAACTACCGCATTAAAGCTTATCGAT-3′ and LexAW-R1:5′-ctcgagtctagaggtaccactagtgagctcagatct TGGTTCACCGGCAGCCACACGACCTACC-3′. The reverse primer was designed to incorporate a MCS, terminating in an XhoI site, C-terminally to the lexAWT sequence (underlined bases correspond to the added MCS sequence). The resulting 465 bp PCR product was cloned into the pCR2.1 vector of a TA cloning kit (Invitrogen) and sequenced. The fragment was then sub-cloned as a HindIII–XhoI fragment back into the original pMS604 plasmid replacing the original lexAWT-fos construct with a lexAWT-MCS construct and the resulting plasmid was named pPC605 (see Figure 4D).
Performing E.coli surface matings for the detection of protein–protein interactions
Bacterial conjugation was carried out according to the following protocol. Donor S17-1 strains, carrying either pPC810 or pPC811, and recipient SU202 strains, carrying either pMS604 or pPC605, were inoculated into LB broth with appropriate antibiotic selection and incubated overnight at 37°C. Overnight donor and recipient cultures were used to inoculate fresh LB broth cultures without antibiotic selection and grown to mid-log phase (OD600 0.3–0.5). Donor and recipient cultures were then mixed in a 4:1 ratio (based on culture OD600) in a 1.5 ml microfuge tube (usually 800 μl of donor culture with 200 μl of recipient culture) and the cells were pelleted by centrifugation. The pellet was resuspended in 40 μl of LB and spotted onto 0.45 μm filter paper placed on an LB agar plate. After a 4 h incubation at 37°C, the filter paper was immersed in 2 ml of LB broth, agitated, the resulting cell suspension diluted and plated on selective media. Total recipient and total transconjugant counts were determined by plating on LB agar supplemented with kanamycin, tetracycline and LB agar supplemented with kanamycin, tetracycline and gentamycin, respectively. The transfer frequency (Tf) was then calculated as the number of transconjugants per recipient according to the equation:
For the detection of protein–protein interactions, 1 ml of the recovered post-mating cell suspension was inoculated into 9 ml of LB broth with appropriate antibiotics for selection of transconjugants and incubated, without shaking, overnight at 37°C. This was to permit outgrowth of transconjugants prior to the induction of hybrid protein expression, which was subsequently achieved by plating on selective media incorporating IPTG and X-gal to screen for protein–protein interactions.
Performing liquid mating in an automated 96 well format for detection of protein–protein interactions
Recipient SU202 strains carrying either pMS604 or pPC605 were inoculated into LB broth with appropriate antibiotic selection and incubated overnight at 37°C. These overnight cultures were then distributed in 150 μl aliquots into separate wells of a 96 well plate to prepare a matrix of recipient Prey clones that was stored at 4°C. The Prey matrix of recipient clones was replica inoculated into a 96 well plate with 50 μl of fresh LB broth in each well. The plate was then sealed using sterile pierceable adhesive aluminium foil and incubated overnight at 37°C on a microtitre plate shaker. A donor E.coli S17-1 strain carrying either pPC810 or pPC811 was inoculated into 5 ml of LB with appropriate antibiotic selection, and incubated overnight with shaking at 37°C. The overnight donor culture was then used to inoculate a fresh LB broth and incubated at 37°C until mid-log phase (OD600 0.3–0.5). Matings were prepared by transferring 50 μl of the donor culture to the wells of the Prey matrix of recipient clones. The 96 well mating plate was incubated at 37°C without shaking for 4 h, and then a 1 μl aliquot from each of the wells was transferred to the corresponding well in a fresh transconjugant selection plate. The transconjugant selection plate contained 150 μl of fresh LB media, with appropriate antibiotics for transconjugant selection (kanamycin, tetracycline and gentamycin for the selection of SU202 harbouring both a Prey and a Bait vector), in each well. The initial transconjugant selection plate was covered with Breatheasy sealing film (available from Greiner Ltd or Genetix Ltd) and then incubated overnight at 37°C with shaking. This initial transconjugant selection plate was used to replica inoculate a second fresh transconjugant selection plate to further purify transconjugants by transferring a 1 μl aliquot from each well of the initial transconjugant selection plate to its corresponding position in the second transconjugant selection plate. The media used in this round of selection also incorporated 1 mM IPTG to induce the expression of hybrid genes from the Prey and Bait vectors. Following incubation at 37°C overnight, a sample of culture from each well was spotted, using a autoclavable polypropylene 96 well replication tool (available from Sigma Aldrich Ltd), onto the surface of indicator plates, which could be either LB agar incorporating appropriate antibiotic selection, IPTG and X-gal or MacConkey agar incorporating 10% lactose and IPTG to test for protein–protein interactions. Indicator plates were incubated at 37°C overnight and the following day the positions in the matrix in which the protein–protein interactions occurred were identified by inspection of culture spot colour (on LB agar incorporating IPTG and Xgal: white spots for protein–protein interactions and blue if there were no interactions; on MacConkey agar: white/pink for protein–protein interactions or magenta if there were no interactions). It was found to be important that cultures be grown in the presence of IPTG prior to spotting onto the final indicator media. In the LexA-based system, positive protein–protein interactions between hybrid proteins result in repression of a constitutively expressed lacZ reporter system, and SU202 reporter strain cells are therefore phenotypically Lac positive in the absence of hybrid gene expression. Culturing in IPTG allows degradation of any pre-existing β-galactosidase enzyme prior to spotting onto indicator plates to evaluate reporter gene expression and identify interactions. All of the above liquid transfer steps, mating setup and transconjugant selections, were carried out by the RoboAmp automated liquid handling system (available from MWG-Biotech Milton Keynes) but can be carried out manually using autoclavable 96 pin replication tools available from Genetix Ltd.
RESULTS AND DISCUSSION
Construction of novel mobilizable P2H system vectors: exploitation of bacterial conjugation for P2H system screening
In order to detect protein–protein interactions using the P2H systems described to date, both the Prey and Bait vectors for the systems have to be introduced into an appropriate E.coli reporter strain by bacterial transformation. The resulting transformants are then assayed for reporter gene activation/repression or a reconstituted enzyme activity in order to detect protein–protein interactions. We proposed that bacterial conjugation could be exploited as a technically simplified and more efficient means of introducing plasmids in combination to test for protein–protein interactions. The IncPα plasmid RP4 is a self-transmissible antibiotic resistance plasmid. It has been demonstrated that E.coli plasmids carrying a cis-acting mobilization (Mob) region can be transferred between E.coli strains, and into many other gram-negative bacteria, using the RP4-specific mobilization system with transfer efficiencies approaching unity when matings are carried out under optimal conditions (45,53). These mobilizable plasmids have mainly been used as suicide vectors for introducing transposons and cloned genes into non-E.coli hosts for mutagenesis and expression studies (49,54,55).
To permit the exploitation of the RP4 plasmid mobilization system for P2H system screening procedures, we have constructed two novel mobilizable Bait vectors for use with the LexA repressor-based P2H system developed by Dmitrova and co-workers (39). A fragment of pDP804, carrying the entire lexA408-jun expression construct, was cloned into plasmid pJQ200-NS to form plasmid pPC810. We also constructed a second plasmid, pPC811, in which the jun sequence was replaced with a MCS by cloning a PCR-amplified lexA408-MCS expression construct into plasmid pJQ200-NS. Plasmid pJQ200-NS was derived from pJQ200-sk and therefore carries an RP4 mobilizable element, the mob site. This site permits the efficient mobilization of this plasmid, and therefore the new Bait plasmids pPC810 and pPC811, from a donor strain of E.coli such as S17-1. This strain of E.coli carries a chromosomally integrated RP4 encoding all of the required transfer functions and its use ensures that once a plasmid has been mobilized to a recipient strain it cannot be further mobilized. An alternative strategy for mobilizing plasmids would be to use a helper plasmid, such as pRK600 (56). The helper plasmid also carries all of the transfer functions required to mobilize plasmids and can be used to mobilize plasmids out of any E.coli strain. However, in addition to mobilizing the desired plasmid, it is also transmissable. We preferred to use the S17-1 strain as a donor because it avoided any complications arising as a result of the helper plasmid entering the recipient.
The Bait plasmids pPC810 and pPC811 can be mobilized with high frequency into SU202 reporter cells already harbouring a Prey vector
Having constructed the novel mobilizable Bait vectors pPC810 and pPC811, E.coli surface matings were carried out to demonstrate that these plasmids could be mobilized into the E.coli SU202 reporter strain, already harbouring a Prey vector, to detect protein–protein interactions. Matings were prepared to mobilize pPC810 into SU202 harbouring either the pMS604 Prey plasmid (expressing a LexAWT-Fos hybrid protein) or the pPC605 plasmid (in which the fos sequence had been replaced by a MCS sequence and therefore only expressed a LexAWT DBD). The pPC811 plasmid was mated in the same manner to verify all four possible combinations of the Bait and Prey vectors. Surface matings were undertaken by mixing mid-log phase donor and recipient cultures in a ratio of 4:1 based on the culture OD600 values. It was expected that the use of excess donor in this way would maximize the transfer frequencies ensuring that every recipient cell would receive a copy of a Bait vector. Matings were prepared in duplicate with one mating of each duplicate being removed and plated on selective media after 4 h incubation at 37°C, while the second mating was incubated overnight. Very high transfer efficiencies of 3–5 × 10−1 were achieved for the 4 h matings. For the duplicate matings that had been incubated overnight, there appeared to be a significant reduction in the transfer efficiencies obtained compared with those observed for the 4 h matings. The explanation for this apparent drop in transfer frequency is that transconjugants initially grow considerably slower than either the recipient or donor strains, requiring a period of outgrowth to permit plasmid establishment after mating. From the observed transfer frequencies for the 4 h matings, it was obvious that not all recipient cells received a copy of a Bait plasmid. It is therefore likely that when matings are incubated for prolonged periods, the recipient cells outgrow the transconjugants and, as transfer frequency is calculated using total transconjugant and total recipient counts, there is therefore an apparent drop in the calculated transfer frequency. From this experiment, it was concluded that a 4 h incubation was sufficient for carrying out highly efficient bacterial matings.
Having demonstrated that the novel Bait vectors could be mobilized into reporter SU202 cells already harbouring Prey vectors, we had to demonstrate that the resulting transconjugants displayed a β-galactosidase phenotype consistent with the combination of Prey and Bait plasmids they possessed. We found that when recovered mating cell suspensions were immediately plated on media selecting for both transconjugants and screening for protein–protein interactions simultaneously, growth and colour development, particularly of interaction negative and therefore Lac+ clones, required prolonged incubation periods of 2 days. We found that better results were obtained when selection of transconjugants and induction of hybrid protein expression for the detection of interactions was carried out sequentially. Recovered mating cell suspensions were therefore subjected to an outgrowth period in transconjugant selective LB broth lacking IPTG prior to plating on selective media incorporating IPTG to induce hybrid protein expression and X-gal to permit detection of protein–protein interactions. All of the transconjugants displayed the expected phenotype, i.e. those harbouring a combination of pPC810 with pMS604 and thus co-expressing both a LexA408-Jun and LexAWT-Fos hybrid proteins resulted in white colonies indicative of protein–protein interactions, while all other vector combinations gave blue colonies indicating no protein–protein interactions.
Bacterial conjugation can be used for highly efficient library screening
It has been reported that when performed under optimal conditions, the frequency of transfer of RP4 mobilizable vectors can approach unity (53). In our mating experiments, where matings were carried out using log phase cultures of both donors and recipients, transfer frequencies of ∼3–5 × 10−1 were routinely observed which equivocates to one in every two to three recipient cells successfully receiving a copy of a Bait vector. This means that the bacterial mating should permit the screening of complex libraries of pooled Prey clones to identify interacting partners for a defined Bait protein with an efficiency far greater than that attainable using bacterial transformation. Such mating strategies are also technically simpler to carry out, as cells do not have to be made competent and transformed.
In order to simulate a library screen, libraries of Prey clones were prepared. The OD600 of two overnight cultures, SU202-pMS604 and SU202-pPC605, were measured and then the cells were pelleted and resuspended in saline solution to an OD600 of 0.4. A set of serial dilutions of the SU202-pMS604 culture, down to 10−3, was prepared using the SU202-pPC605 culture as the diluent. The resulting 10−3 dilution therefore represents a library of SU202 Prey clones, in which an interacting partner, LexAWT-Fos encoded by plasmid pMS604, occurs at a known frequency of 1:1000 among non-interacting species represented by clones harbouring the pPC605 plasmid and thus only expressing the LexAWT DBD. This library was screened by carrying out a surface mating with an S17-1 donor strain carrying plasmid pPC810. Recovered matings were initially plated to determine the transfer frequency for the mating and an optimal dilution factor that would give ∼500 transconjugants per plate. A 1 ml aliquot of the recovered mating was also inoculated into 9 ml of broth excluding IPTG to permit outgrowth of transconjugants. After the overnight outgrowth step, the transconjugant culture was diluted and a volume equivalent to 10 000 transconjugants plated over 20 plates of selective media to yield a plating density of ∼500 colonies per plate and to screen for protein–protein interactions. White clones were recovered at a 5-fold higher frequency than expected. This was due to proliferation of transconjugants during the overnight outgrowth step evident from the higher than expected total transconjugant counts observed on plates. It was always observed that when plated on solid selective media to screen for protein–protein interactions, white clones, expressing interacting pairs of hybrid proteins, grew more rapidly than blue clones expressing non-interacting pairs of hybrid protein. This is likely to be due to the fact that in these clones high-level expression of β-galactosidase is repressed as a result of protein–protein interactions and they therefore express one less high molecular weight protein and have a growth advantage over Lac+ clones in which no interactions occur. The overnight outgrowth step was carried out using broth excluding IPTG and one might therefore expect that, in the absence of hybrid protein expression, all transconjugants would proliferate in an unbiased manner and that the frequency of white to blue clones would therefore be unaffected by such proliferation. However, even in the absence of IPTG, there is hybrid gene expression (data not shown) and thus there would be some positive selection for clones harbouring interacting pairs of proteins during the outgrowth step resulting in the culture becoming enriched for such clones. The implication of the growth advantage exhibited by transconjugants harbouring positive protein–protein interactions is, as observed in the above experiment, that a disproportionate number of positive white colonies may be recovered when performing library screens due to amplification of a single transconjugant harbouring a single interacting protein pair. In addition, the extent of the growth advantage would probably depend on the strength of a particular protein–protein interaction, and therefore if a particular Bait protein used in a library screen had multiple interacting partners, there may be further enrichment for the strongest interacting pair. However, these effects could be very simply overcome by reducing the duration of the outgrowth step thus minimizing transconjugant amplification, and the above experiment clearly demonstrated that E.coli mating-based strategies using our novel mobilizable Bait vectors could be used to carry out efficient library screening for the identification of protein–protein interactions.
The sacB gene facilitates recovery and identification of Prey plasmids isolated during library screening and verification of protein–protein interactions
When clones exhibiting positive protein–protein interactions are obtained during library screening using Y2H systems, the identity of the cloned DNA on the Prey vector has to be determined. This often requires rescuing the Prey vector, via transformation into E.coli, and then sequencing the cloned DNA. The interaction is then verified again by re-introducing the recovered Prey vector into a fresh yeast reporter strain along with the Bait vector. Similar steps have to be taken when P2H systems are used for carrying out library screens using transformation-based protocols. We proposed that the sacB gene present on our conjugative P2H system Bait vectors would facilitate the simplified rescue of Prey vectors after library screening and verification of identified protein–protein interactions.
To verify that the white clones retrieved in our library screen did in fact contain the pMS604 Prey plasmid, expressing the LexAWT-Fos protein, 12 random white clones were picked and cured of the pPC810 plasmid by exploiting the presence of the sacB gene on this plasmid. Three blue clones were also cured in the same way as controls. Clones were picked and streaked on media selecting only for the Prey vector and containing 5% sucrose to induce loss of the Bait vector. Loss of the Bait vector was verified on the basis of gentamycin sensitivity. Plasmid DNA was isolated from each of the clones and the identity of the plasmid in each clone verified by carrying out a BamHI–BglII double digest that permits the distinction of pMS604 from pPC605. All of the white clones were verified as having the pMS604 Prey plasmid and all of the blue clones were verified as having the pPC605 plasmid.
While curing in the manner described above provides a simple and fast way of obtaining the Prey vector in isolation for identification either by restriction analysis or by sequencing, it also permits verification of protein–protein interactions. If the observed Lac− phenotype of a transconjugant clone is due to protein–protein interactions between hybrid proteins encoded by the Prey and Bait vectors, and not due to a mutational event in the E.coli cell, then curing the clone of the Bait vector should result in a reversion to a Lac+ phenotype. This was demonstrated by streaking all of the cured clones on selective LB agar incorporating IPTG and X-gal and observing that all clones generated blue colonies. If further validation is required, cured clones can subsequently be re-mated to re-introduce the original Bait vector.
Bacterial conjugation can be used for exhaustive automated protein interaction mapping
In addition to permitting highly efficient and technically simplified screening of libraries of pooled Prey clones, the mobilizable Bait vectors should also permit the implementation of the LexA-based P2H system for automated exhaustive protein interaction mapping strategies analogous to those developed for Y2H systems. Libraries of Prey and Bait clones can be constructed, the Prey constructs being transformed into and maintained in the E.coli SU202 reporter strain and Bait constructs being transformed into and maintained in E.coli S17-1. Each of these clone libraries can be maintained in a matrix format, either in 96 well or 384 well formats, where the identity of each clone at each position in the matrices is known. Each clone in the Bait library matrix can then be mated with each clone in the Prey library in a matrix and the resulting transconjugants tested for reporter gene expression to detect protein–protein interactions (see Figure 3). This exhaustive screening in a matrix fashion reveals all possible interactions between proteins in the Prey and Bait libraries. To demonstrate the feasibility of this strategy, we prepared a matrix of Prey clones by distributing cultures of SU202 harbouring either pMS604 or pPC605 into individual wells of a 96 well plate. This master matrix plate was used to replica inoculate fresh 96 well plates for use in mating experiments. These simply involved the addition of logarithmic S17-1-pPC810 culture to each well of the Prey matrix to setup liquid matings. Following two rounds of transconjugant selection and purification, transconjugants were spotted onto the surface of indicator media, to screen for protein–protein interactions. These were identified according to the colour of colony spots after overnight incubation, and every occurrence within the matrix of SU202 clones harbouring the pMS604 plasmid (and thus expressing the LexA-Fos hybrid) was successfully identified (see photo insert in Figure 3).
In the above matrix experiment, liquid matings were employed with all of the steps being carried out using a RoboAmp liquid handling system. However, bacterial matings could in fact be carried out using a range of protocols including those developed for carrying out yeast matings. For example, in the protocol described by Cagney et al. (46) and employed by Uetz et al. (21), Bait clones were first spotted onto the surface of solid media and, having been allowed to dry, Preys were then spotted on top of the Baits and the plates incubated to allow mating. It is envisaged that carrying out bacterial matings in such a manner would further increase the efficiency and speed with which interaction mapping could be undertaken.
CONCLUSIONS
The Y2H system has been widely implemented for the detection and analysis of protein–protein interactions. Through the exploitation of yeast mating types, technically simple and automatable strategies have been developed, permitting its application to high-throughput and exhaustive proteomic interaction mapping. It has therefore become an important component technology in the field of functional genomics helping to further our understanding of complex biological processes, annotate genome sequences and facilitating the identification of novel therapeutic targets.
P2H systems present many advantages over yeast-based technologies, which largely derive from the ease with which E.coli can be genetically manipulated, the lack of cellular compartmentalization, its faster growth rate and the higher transformation efficiencies that are attainable permitting rapid and more efficient screening of complex libraries. The systems permit the investigation of prokaryotic protein–protein interactions in a prokaryotic genetic background, but they can also be used for the analysis of eukaryotic proteins. This may be particularly desirable in circumstances where homologous yeast proteins interfere with an interaction by interacting with and sequestering one of the interacting partners leading to false negatives, or by acting as a bridge between two proteins leading to false positives. The absence of such homologous eukaryotic proteins in E.coli may result in the observation of less false positives and negatives. Transcriptional activation in E.coli differs fundamentally from the mechanism in yeast and therefore false positives that occur with the Y2H system as a result of reporter gene activation by non-specific acidic activator sequences may not occur in E.coli-based P2H systems. E.coli-based systems can also permit analysis of eukaryotic proteins that are toxic when expressed in yeast because they interfere with the function of yeast homologues. However, it is important to note that while P2H systems have many advantages, they do suffer from many of the same problems encountered by other hybrid technologies. Whenever fusion proteins are used for the detection of protein–protein interactions, there is the possibility that sites required for interaction may be occluded by the fused moieties. In addition, P2H systems may not be suitable for the analysis of all eukaryotic proteins due to problems associated with expression, stability and incorrect folding of eukaryotic proteins in bacterial cells. P2H systems should therefore be considered as a complementary technology to the Y2H system.
Despite their many proposed advantages, P2H systems have not yet found the same widespread application to large-scale protein interaction mapping as Y2H systems. One of the key advantages of E.coli-based systems is the higher transformation efficiencies attainable which promise more efficient library screening for identifying protein–protein interactions. However, although efficient, bacterial transformation is not particularly amenable to automated strategies. In an era where high-throughput automatable techniques are becoming ever more important in order to permit the full exploitation of the vast amounts of genomic sequence data, the technical simplicity of Y2H mating-based strategies has meant that the Y2H system has remained the preferred approach. We have constructed novel mobilizable P2H system Bait vectors for the LexA-based P2H system originally developed by Dmitrova and co-workers (39). We have developed E.coli mating-based strategies employing these vectors, demonstrating how they can be used for carrying out library screens to identify protein–protein interactions with an efficiency greater than that attainable with either bacterial transformation or yeast-mating strategies. Construction of similar mobilizable vectors for a P2H system permitting direct selection for protein–protein interactions (28,29,35) should permit even more efficient library screening than that possible with systems that employ genetic screening for interactions rather than selection where plating densities are restricted (30,33,39). The added benefit of the sacB gene on our Bait vectors permits simple recovery of Prey vectors for the identification of cloned insert DNA and the verification of identified protein–protein interactions. We have also demonstrated how the technical simplicity of E.coli mating-based P2H strategies permits their automation and facilitates exhaustive protein interaction mapping using matrices of Bait and Prey clones.
The P2H system developed by Joung et al. (31) permits the use of highly efficient phage transduction to combine Bait and Prey vectors to test for protein–protein interactions and could potentially be automated to facilitate high-throughput protein interaction mapping, although no such application has yet been reported. However, the fact that the strategies we have described here are analogous to those that have been developed for implementing Y2H mating-based strategies should give our conjugative LexA-based P2H system the added benefit of permitting its implementation for high-throughput proteomic interaction mapping using similar automated platforms and protocols to those already developed for carrying out automated Y2H mating-based strategies. In addition, while the vectors we have constructed are for use with the LexA-based P2H system, the work reported here demonstrates how through the simple incorporation of a mob element similar mobilizable vectors could be constructed for any of the previously described P2H systems broadening their applicability by permitting their implementation for high-throughput protein interaction mapping in a similar manner. Such applications of P2H systems will permit further evaluation of the potential strengths and weaknesses of each of these systems. The high efficiency of bacterial mobilization could be further exploited to facilitate the screening of highly complex peptide aptamer libraries to identify peptides capable of specifically disrupting designated protein–protein interactions in a similar manner to that described by Colas et al. (2). Ultimately, the use of mobilizable vectors and E.coli mating-based strategies may finally allow P2H systems and all of the proposed advantages of E.coli-based P2H systems to be fully exploited helping to increase the speed and efficiency with which protein interaction mapping can be undertaken for the determination of protein functions and the identification of novel therapeutic targets.
Acknowledgments
This publication has emanated from research that was conducted with the financial support of Enterprise Ireland through the Research Innovation Fund (RIF). We would like to thank all colleagues who provided strains and plasmids used in this study and the National Centre for Sensors Research at Dublin City University for its additional support. Funding to pay the Open Access publication charges for this article was provided by Enterprise Ireland through the Research Innovation Fund (RIF).
REFERENCES
- 1.Karimova G., Ladant D., Ullmann A. Two-hybrid systems and their usage in infection biology. Int. J. Med. Microbiol. 2002;292:17–25. doi: 10.1078/1438-4221-00182. [DOI] [PubMed] [Google Scholar]
- 2.Colas P., Cohen B., Jessen T., Grishina I., McCoy J., Brent R. Genetic selection of peptide aptamers that recognize and inhibit cyclin-dependent kinase 2. Nature. 1996;380:548–550. doi: 10.1038/380548a0. [DOI] [PubMed] [Google Scholar]
- 3.Vidal M., Endoh H. Prospects for drug screening using the reverse two-hybrid system. Trends Biotechnol. 1999;17:374–381. doi: 10.1016/s0167-7799(99)01338-4. [DOI] [PubMed] [Google Scholar]
- 4.Park S.H., Raines R.T. Genetic selection for dissociative inhibitors of designated protein–protein interactions. Nat. Biotechnol. 2000;18:847–851. doi: 10.1038/78451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fields S., Song O.-K. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 6.Vidal M., Legrain P. Yeast forward and reverse ‘n’-hybrid systems. Nucleic Acids Res. 1999;27:919–929. doi: 10.1093/nar/27.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brent R., Finley R.L., Jr Understanding gene and allele function with two-hybrid methods. Annu. Rev. Genet. 1997;31:663–704. doi: 10.1146/annurev.genet.31.1.663. [DOI] [PubMed] [Google Scholar]
- 8.Frederickson R.M. Macromolecular matchmaking: advances in two-hybrid and related technologies. Curr. Opin. Biotechnol. 1998;9:90–96. doi: 10.1016/s0958-1669(98)80090-6. [DOI] [PubMed] [Google Scholar]
- 9.Drees B.L. Progress and variations in two-hybrid and three-hybrid technologies. Curr. Opin. Chem. Biol. 1999;3:64–70. doi: 10.1016/s1367-5931(99)80012-x. [DOI] [PubMed] [Google Scholar]
- 10.Chien C., Bartel P., Sternglanz R., Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl Acad. Sci. USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen J.B., Walberg M.W., Edwards M.C., Elledge S.J. Finding prospective partners in the library: the two-hybrid system and phage display find a match. Trends Biochem. Sci. 1995;20:511–516. doi: 10.1016/s0968-0004(00)89119-7. [DOI] [PubMed] [Google Scholar]
- 12.Holt K.H., Olson L., Moye-Rowley W.S., Pessin J.E. Phosphatidylinositol 3-kinase activation is mediated by high-affinity interactions between distinct domains within the p110 and p85 subunits. Mol. Cell. Biol. 1994;14:42–49. doi: 10.1128/mcb.14.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li B., Fields S. Identification of mutations in p53 that affect its binding to SV40 large T antigen by using the yeast two-hybrid system. FASEB J. 1993;7:957–963. doi: 10.1096/fasebj.7.10.8344494. [DOI] [PubMed] [Google Scholar]
- 14.Finley R., Jr, Brent R. Interaction mating reveals binary and ternary connections between Drosophila cell cycle regulators. Proc. Natl Acad. Sci. USA. 1994;91:12980–12984. doi: 10.1073/pnas.91.26.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fromont-Racine M., Rain J.C., Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nature Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 16.Flores A., Briand J.-F., Gadal O., Andrau J.-C., Rubbi L., Van Mullem V., Boschiero C., Goussot M., Marck C., Carles C., et al. A protein–protein interaction map of yeast RNA polymerase III. Proc. Natl Acad. Sci. USA. 1999;96:7815–7820. doi: 10.1073/pnas.96.14.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartel P.L., Roecklein J.A., SenGupta D., Fields S. A protein linkage map of Escherichia coli bacteriophage T7. Nature Genet. 1996;12:72–77. doi: 10.1038/ng0196-72. [DOI] [PubMed] [Google Scholar]
- 18.Flajolet M., Rotondo G., Daviet L., Bergametti F., Inchauspe G., Tiollais P., Transy C., Legrain P. A genomic approach of the hepatitis C virus generates a protein interaction map. Gene. 2000;242:369–379. doi: 10.1016/s0378-1119(99)00511-9. [DOI] [PubMed] [Google Scholar]
- 19.Ito T., Tashiro K., Muta S., Ozawa R., Chiba T., Nishizawa M., Yamamoto K., Kuhara S., Sakaki Y. From the cover: Toward a protein–protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc. Natl Acad. Sci. USA. 2000;97:1143–1147. doi: 10.1073/pnas.97.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giot L., Bader J.S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 21.Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 22.Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rain J.C., Selig L., De Reuse H., Battaglia V., Reverdy C., Simon S., Lenzen G., Petel F., Wojcik J., Schachter V., et al. The protein–protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 24.Ito T., Ota K., Kubota H., Yamaguchi Y., Chiba T., Sakuraba K., Yoshida M. Roles for the two-hybrid system in exploration of the yeast protein interactome. Mol. Cell Proteomics. 2002;1:561–566. doi: 10.1074/mcp.r200005-mcp200. [DOI] [PubMed] [Google Scholar]
- 25.Hu J.C., O'Shea E.K., Kim P.S., Sauer R.T. Sequence requirements for coiled-coils: analysis with lambda repressor-GCN4 leucine zipper fusions. Science. 1990;250:1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- 26.Bunker C.A., Kingston R.E. Identification of a cDNA for SSRP1, an HMG-box protein, by interaction with the c-Myc oncoprotein in a novel bacterial expression screen. Nucleic Acids Res. 1995;23:269–276. doi: 10.1093/nar/23.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cairns M.T., Green A.J., White P.M., Johnston P.G., Brenner S. A novel bacterial vector system for monitoring protein–protein interactions in the cAMP-dependent protein kinase complex. Gene. 1997;185:5–9. doi: 10.1016/s0378-1119(96)00601-4. [DOI] [PubMed] [Google Scholar]
- 28.Karimova G., Pidoux J., Ullmann A., Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl Acad. Sci. USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pelletier J.N., Campbell-Valois F.-X., Michnick S.W. Oligomerization domain-directed reassembly of active dihydrofolate reductase from rationally designed fragments. Proc. Natl Acad. Sci. USA. 1998;95:12141–12146. doi: 10.1073/pnas.95.21.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kornacker M.G., Remsburg B., Menzel R. Gene activation by the AraC protein can be inhibited by DNA looping between AraC and a LexA repressor that interacts with AraC: possible applications as a two-hybrid system. Mol. Microbiol. 1998;30:615–624. doi: 10.1046/j.1365-2958.1998.01096.x. [DOI] [PubMed] [Google Scholar]
- 31.Joung J.K., Ramm E.I., Pabo C.O. A bacterial two-hybrid selection system for studying protein–DNA and protein–protein interactions. Proc. Natl Acad. Sci. USA. 2000;97:7382–7387. doi: 10.1073/pnas.110149297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hays L.B., Chen Y.S., Hu J.C. Two-hybrid system for characterization of protein–protein interactions in E.coli. BioTechniques. 2000;29:288–290. doi: 10.2144/00292st04. 292, 294passim. [DOI] [PubMed] [Google Scholar]
- 33.Di Lallo G., Castagnoli L., Ghelardini P., Paolozzi L. A two-hybrid system based on chimeric operator recognition for studying protein homo/heterodimerization in Escherichia coli. Microbiology. 2001;147:1651–1656. doi: 10.1099/00221287-147-6-1651. [DOI] [PubMed] [Google Scholar]
- 34.Dove S.L., Hochschild A. Conversion of the omega subunit of Escherichia coli RNA polymerase into a transcriptional activator or an activation target. Genes Dev. 1998;12:745–754. doi: 10.1101/gad.12.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dove S.L., Joung J.K., Hochschild A. Activation of prokaryotic transcription through arbitrary protein–protein contacts. Nature. 1997;386:627–630. doi: 10.1038/386627a0. [DOI] [PubMed] [Google Scholar]
- 36.Hu C.-D., Chinenov Y., Kerppola T.K. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 37.Ozawa T., Takeuchi T.M., Kaihara A., Sato M., Umezawa Y. Protein splicing-based reconstitution of split green fluorescent protein for monitoring protein–protein interactions in bacteria: improved sensitivity and reduced screening time. Anal. Chem. 2001;73:5866–5874. doi: 10.1021/ac010717k. [DOI] [PubMed] [Google Scholar]
- 38.Ding Z., Zhao Z., Jakubowski S.J., Krishnamohan A., Margolin W., Christie P.J. A novel cytology-based, two-hybrid screen for bacteria applied to protein–protein interaction studies of a type IV secretion system. J. Bacteriol. 2002;184:5572–5582. doi: 10.1128/JB.184.20.5572-5582.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dmitrova M., Younes-Cauet G., Oertel-Buchheit P., Porte D., Schnarr M., Granger-Schnarr M. A new LexA-based genetic system for monitoring and analyzing protein heterodimerization in Escherichia coli. Mol. Gen. Genet. 1998;257:205–212. doi: 10.1007/s004380050640. [DOI] [PubMed] [Google Scholar]
- 40.Ladant D., Karimova G. Genetic systems for analyzing protein–protein interactions in bacteria. Res. Microbiol. 2000;151:711–720. doi: 10.1016/s0923-2508(00)01136-0. [DOI] [PubMed] [Google Scholar]
- 41.Hu J.C., Kornacker M.G., Hochschild A. Escherichia coli one- and two-hybrid systems for the analysis and identification of protein–protein interactions. Methods. 2000;20:80–94. doi: 10.1006/meth.1999.0908. [DOI] [PubMed] [Google Scholar]
- 42.Malek J.A., Wierzbowski J.M., Tao W., Bosak S.A., Saranga D.J., Doucette-Stamm L., Smith D.R., McEwan P.J., McKernan K.J. Protein interaction mapping on a functional shotgun sequence of Rickettsia sibirica. Nucleic Acids Res. 2004;32:1059–1064. doi: 10.1093/nar/gkh254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt-Dorr T., Oertel-Buchheit P., Pernelle C., Bracco L., Schnarr M., Granger-Schnarr M. Construction, purification, and characterization of a hybrid protein comprising the DNA binding domain of the LexA repressor and the Jun leucine zipper: a circular dichroism and mutagenesis study. Biochemistry. 1991;30:9657–9664. doi: 10.1021/bi00104a013. [DOI] [PubMed] [Google Scholar]
- 44.Porte D., Oertel-Buchheit P., Granger-Schnarr M., Schnarr M. Fos leucine zipper variants with increased association capacity. J. Biol. Chem. 1995;270:22721–22730. doi: 10.1074/jbc.270.39.22721. [DOI] [PubMed] [Google Scholar]
- 45.Simon R., Priefer U., Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology. 1983;1:784–791. [Google Scholar]
- 46.Cagney G., Uetz P., Fields S. High-throughput screening for protein–protein interactions using two-hybrid assay. Methods Enzymol. 2000;328:3–14. doi: 10.1016/s0076-6879(00)28386-9. [DOI] [PubMed] [Google Scholar]
- 47.Zhong J., Zhang H., Stanyon C.A., Tromp G., Finley R.L., Jr A strategy for constructing large protein interaction maps using the yeast two-hybrid system:regulated expression arrays and two-phase mating. Genome Res. 2003;13:2691–2699. doi: 10.1101/gr.1134603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolonin M.G., Zhong J., Finley R.L. Interaction mating methods in two-hybrid systems. Methods Enzymol. 2000;328:26–46. doi: 10.1016/s0076-6879(00)28388-2. [DOI] [PubMed] [Google Scholar]
- 49.Quandt J., Hynes M.F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 51.Inoue H., Nojima H., Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 52.Birnboim H.C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Connell M.P., Hynes M.F., Puehler A. Incompatibility between a Rhizobium Sym plasmid and a Ri plasmid of Agrobacterium. Plasmid. 1987;18:156–163. doi: 10.1016/0147-619x(87)90043-6. [DOI] [PubMed] [Google Scholar]
- 54.Simon R., Quandt J., Klipp W. New derivatives of transposon Tn5 suitable for mobilization of replicons, generation of operon fusions and induction of genes in gram-negative bacteria. Gene. 1989;80:161–169. doi: 10.1016/0378-1119(89)90262-x. [DOI] [PubMed] [Google Scholar]
- 55.Kovach M.E., Elzer P.H., Hill D.S., Robertson G.T., Farris M.A., Roop R.M., II, Peterson K.M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 56.Finan T.M., Kunkel B., De Vos G.F., Signer E.R. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]