Abstract
Gene expression systems that allow the regulation of bacterial genes during an infection are valuable molecular tools but are lacking for mycobacterial pathogens. We report the development of mycobacterial gene regulation systems that allow controlling gene expression in fast and slow-growing mycobacteria, including Mycobacterium tuberculosis, using anhydrotetracycline (ATc) as inducer. The systems are based on the Escherichia coli Tn10-derived tet regulatory system and consist of a strong tet operator (tetO)-containing mycobacterial promoter, expression cassettes for the repressor TetR and the chemical inducer ATc. These systems allow gene regulation over two orders of magnitude in Mycobacterium smegmatis and M.tuberculosis. TetR-controlled gene expression was inducer concentration-dependent and maximal with ATc concentrations at least 10- and 20-fold below the minimal inhibitory concentration for M.smegmatis and M.tuberculosis, respectively. Using the essential mycobacterial gene ftsZ, we showed that these expression systems can be used to construct conditional knockouts and to analyze the function of essential mycobacterial genes. Finally, we demonstrated that these systems allow gene regulation in M.tuberculosis within the macrophage phagosome.
INTRODUCTION
Mycobacterium tuberculosis, the etiologic agent of tuberculosis, infects about one-third of the world's population and kills two million people every year (1,2). Successful therapy of tuberculosis requires continuous treatment for at least 6 months with multiple drugs (2). Shortening the duration of chemotherapy selects for drug-resistant tuberculosis, which is difficult to treat and has a high mortality (3). The development of novel anti-mycobacterial drugs that would allow shorter chemotherapies and allow treatment of drug resistant tuberculosis is urgently needed.
Inducible gene expression systems are powerful tools for studying gene function and for the validation of drug targets in bacteria (4,5). Unfortunately, only a few regulated expression systems are available to control gene expression in mycobacteria (6–8). The most widely used system is controlled by two positive regulators (AmiC and AmiD) and one negative regulator (AmiA) and can be induced by the addition to the growth medium of short aliphatic amides, acetamide or butyramide (7–9). Acetamide-controlled expression systems have been used to analyze conditional mutants of Mycobacterium smegmatis (10,11) and M.tuberculosis (12) and for the overexpression of mycobacterial antigens in M.smegmatis (13). The currently available regulated expression systems are valuable for certain experiments that can be performed in liquid culture. None of the available systems, however, allows controlled regulation of mycobacterial gene expression during an infection.
Tet repressor (TetR) proteins regulate the expression of a family of tetracycline (Tc)-exporting proteins (14). In the absence of Tc, TetR tightly binds the tet operators (tetO) in the tetA promoter and suppresses transcription of tetA, which encodes the Tc exporter. Once Tc enters the cell it binds TetR and induces a conformational change that results in dissociation of TetR from tetO and thus induces expression of TetR-controlled genes. Induction of TetR occurs prior to inhibition of the ribosome by tetracyclines because affinity of TetR for these drugs is 103- to 105-fold higher than the affinity of the drugs for the ribosome (14). Tetracyclines can cross biological membranes by diffusion, enabling these inducers to penetrate most bacterial and eukaryotic cells. Efficient TetR-controlled expression systems have been developed for Gram-positive and Gram-negative bacteria and can be used to regulate the bacterial gene expression during infection of mice (15–18). Here, we describe the development of a TetR-controlled expression system that allows the efficient regulation of gene expression in mycobacteria in liquid culture and during infection of mammalian cells.
MATERIALS AND METHODS
Bacterial strains, plasmids, media and culture conditions
Table 1 lists the strains and plasmids used in this study. Mycobacteria were grown in Middlebrook 7H9 medium (Difco/VWR) with 0.2% glycerol and 0.05% Tween-80. For growth of M.tuberculosis H37Rv, the medium was supplemented with 0.5% BSA, 0.2% dextrose and 0.085% sodium chloride (ADN). For selection of recombinant mycobacteria, kanamycin and hygromycin were used at 30 and 50 μg/ml, respectively. Escherichia coli DH5α was grown in Luria–Bertani broth (LB) and kanamycin and hygromycin were used at 60 and 200 μg/ml, respectively.
Table 1.
Bacterial strains and plasmids used in this work
| Strains | Genotype | Source or reference |
|---|---|---|
| E.coli | ||
| DH5α | F- φ80dlacZΔM15 Δ(lacZYA-argF) U169 deoR recA1 endA1 hsdR17 (rK−mK+) phoA supE44 λ-thi-1 gyrA96 relA1 | Gibco, BRL |
| M.tuberculosis H37Rv | ATCC 25618 | |
| M.smegmatis mc2155 | (54) | |
| Plasmids | ||
| pMS2 | Hygr; pAL500 origin, ColE1 origin, contains multiple cloning site and 2 transcriptional terminators | (25) |
| pMGuvK | pMS2 derivative, Hygr, promoterless gfpuv, promoterless Kmr | This work and (25) |
| pMGK | pMS2 derivative, Hygr, promoterless gfp+, promoterless Kmr | This work and (24) |
| pWH520 | (27) | |
| pUV15 | pMS2 derivative, Hygr, Kmr, psmyc-gfp | This work and (25) |
| pUV15tetORs | pUV15 derivative, Hygr, Kmr, psmyc-tetR, pmyc1tetO-gfp+ | This work |
| pUV15tetORm | pUV15 derivative, Hygr, Kmr, pimyc-tetR, pmyc1tetO-gfp+ | This work |
| pME0L0 | pMS2 derivative, Hygr, promoterless lacZ | This work |
| pME0L1 | pMS2 derivative, Hygr, pmyc1tetOlacZ | This work |
| pME1sL1 | pMS2 derivative, Hygr, psmyc-tetR(B), pmyc1tetO-lacZ | This work |
| pME1mL1 | pMS2 derivative, Hygr, pimyc-tetR(B), pmyc1tetO-lacZ | This work |
| pMV306Km | Kmr, int, attP integrates at attB site on mycobacterial chromosome | (32) |
| pMC1s | pMV306Km derivative, Kmr, psmyc-tetR | This work |
Promoter trap library
Chromosomal DNA from M.tuberculosis H37Rv and M.smegmatis mc2155 was isolated as described previously (19) and partially digested with Sau3A. Digested DNA was analyzed using 5% polyacrylamide gels; the 50–500 bp fraction was purified (20) and ligated into dephosphorylated, BamH1-digested pMGuvK. Ligated DNA was electroporated into M.smegmatis and fluorescent colonies were selected on LB plates containing 50 μg/ml hygromycin. Plasmids were isolated and transformed into E.coli DH5α for amplification. Preparation of competent M.smegmatis and electroporation were as described previously (21).
Primer extension
Primer extension was as described previously (20) using a synthetic oligonucleotide gfp2 (5′-gcatcaccttcaccctctccactgac-3′) and 10 μg RNA from recombinant M.smegmatis. RNA was isolated as described previously (22).
Reporter gene constructs
Two gfp genes, gfpuv (23) and the more recently developed gfp+ (24), were used. The gfpuv gene was used as reporter in plasmid pMGuvK to isolate mycobacterial promoters in M.smegmatis on LB plates. The gfp+ gene was used in plasmid pMGK to quantify green fluorescent protein (GFP) activity of mycobacteria growing in liquid cultures. The lacZ gene was cloned from pSVlacZ (Promega) into pMS2 (25) leading to the plasmid pME0L0.
Reporter gene assays
GFP activity
M.smegmatis was transformed by electroporation, plated onto selective (50 μg/ml hygromycin) agar plates and incubated at 37°C. After 3 days of incubation, three colonies from each transformation were used to inoculate 3 ml liquid cultures and incubated for ∼60 h on a shaker (250 r.p.m.) at 37°C. The bacteria were then diluted 100-fold into fresh medium and grown as before for ∼15 h to reach the logarithmic growth phase. Optical density was measured, and bacteria were harvested by centrifugation and resuspended in phosphate-buffered saline (PBS) to a density of 2 OD580/0.1 ml. Then, 0.1 ml of the bacterial suspension was transferred into black 96-well plates, and fluorescence at 515 nm after excitation at 485 nm was measured in a fluorescence microplate reader (Molecular Devices). Experiments with M.tuberculosis were performed as described for M.smegmatis with the exceptions that plates were incubated for 18 days and liquid cultures were not shaken and incubated for 8–10 days, then diluted 1:10 and grown for 2–3 days to reach the logarithmic growth phase.
β-galactosidase activity
Bacteria were cultivated as for GFP measurements. An aliquot of 10 μl of a 330 μM solution of the fluorogenic β-galactosidase substrate C2FDG [Molecular Probes, (26)] was mixed with 0.1 ml of bacteria in a black 96-well plate. The plate was incubated in the dark for 3 h (M.smegmatis) or 1 h (M.tuberculosis) after which the cells were excited at 485 nm and fluorescence emission was measured at 515 nm in a fluorescence microplate reader (Molecular Devices). Incubation with the substrate was shorter for M.tuberculosis because higher absolute β-galactosidase activities were observed with this organism. For all GFP and β-galactosidase measurements, triplicate bacterial cultures were grown and measurements are representatives of at least three independent transformations. The fluorescence intensity was normalized to the cell density and expressed in relative fluorescence units (RFUs). Mycobacteria transformed with plasmids pMS2 containing no gfp/lacZ gene and pMGK or pME0L0 containing a promoterless gfp+ or lacZ gene were used to measure background fluorescence of the cells and background expression.
Construction of Pmyc1tetO and Ps/imyctetR
Tet operators were inserted into Psmyc using oligonucleotide-directed PCR mutagenesis (20). The PCR oligonucleotides were smyc-tet1 (5′-gagtttgtcctccctatcagtgatagataggctctgggagtacccgtctg-3′), smyc-tet2 (5′-ctgatagggaggacaaactctatcactgatagggagttctcccgctcgtcagagaccct-3′), gfp2 (5′-gcatcaccttcaccctctccact gac-3′) and poly3 (5′-gaactagttgattagctaagcagaagg-3′). A PCR product containing Pmyc1tetO, the tetO containing Psmyc derivative, was digested with XbaI and SphI and cloned into pUV15 to replace Psmyc. A Tn10-derived tetR gene was amplified by PCR from pWH520 (27) and cloned into pMS2 for extrachromosomal expression or into pMV306Km for integration onto the mycobacterial chromosome. The promoters Psmyc or Pimyc were used for the expression of TetR in mycobacteria. All constructs were verified by restriction analysis and DNA sequencing.
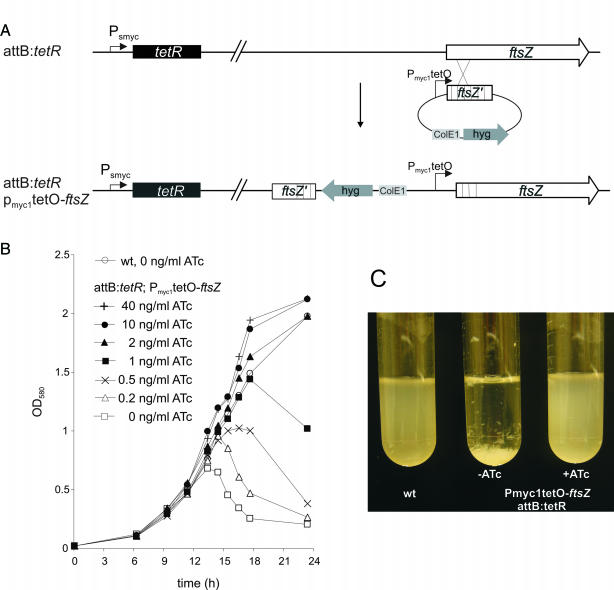
Construction of a conditional ftsZ knockout in M.smegmatis
The first 700 bp of M.smegmatis ftsZ were cloned downstream of Pmyc1tetO in a pMS2 derivative in which the pAL5000 origin of replication was deleted. The resulting plasmid, pKIftsZ, cannot replicate as an episome in mycobacteria. M.smegmatis that carried a Psmyc-tetR fusion in the attB site of the chromosome was electroporated with 1 μg of pKIftsZ and recombinants were selected on 7H11 agar plates that contained 50 μg/ml hygromycin and either no anhydrotetracycline (ATc) or 50 ng/ml ATc. Integration of the plasmid via homologous recombination was confirmed by PCR using primers that hybridized to Pmyc1tetO and to the 3′ region of ftsZ that was not part of pKIftsZ.
Macrophage infections
Murine bone marrow-derived macrophages were prepared as described previously (22,28) and seeded onto coverslips in 24-well plates. The cells were infected with live M.tuberculosis at a multiplicity of infection (MOI) of 5–10 bacteria to 1 macrophage. Four hours post infection, the macrophage monolayers were washed three times with warm PBS, followed by the addition of complete tissue culture medium containing 100 μg/ml amikacin to kill extracellular bacteria. Eight hours post infection the monolayers were washed again and medium was replaced. Twenty-four hours post infection, 100 ng/ml ATc were added where indicated and 72 h later the coverslips were analyzed by microscopy. Widefield images of infected cells were obtained using a Leica DMIRB fluorescence microscope equipped with a 40× 1.25 numerical aperture objective and a 1.5× magnification changer. Digital images were captured with a Photometrics CoolSnap HQ camera (Photometrics/Roper Scientific, Tucson, AZ) driven by MetaMorph image acquisition software (Universal Imaging, Downingtown, PA). Fluorescence images of all samples within one experiment were acquired under identical conditions, and the images are displayed at the same contrast so that all images are directly comparable. For the transmitted light images, phase contrast optics was used to allow visualization of the bacteria.
RESULTS
Isolation of strong mycobacterial promoters
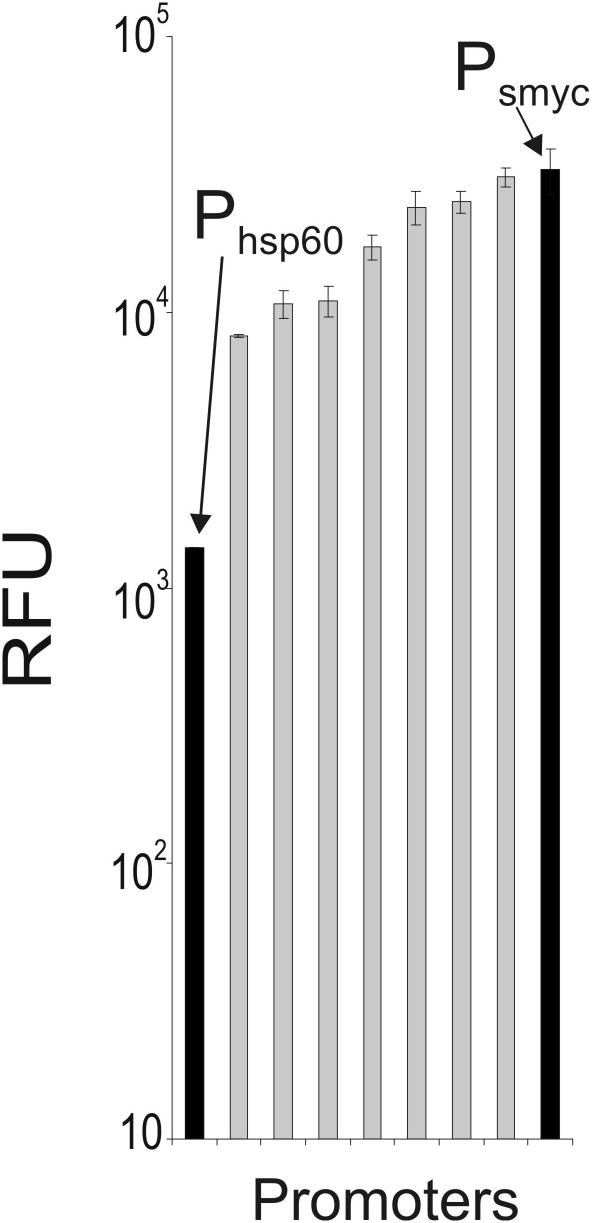
50–500 bp long fragments of chromosomal DNA from M.smegmatis and M.tuberculosis were prepared and ligated into an E.coli-mycobacteria shuttle vector upstream of a promoterless gfp gene. Plasmid ligations were directly transformed into M.smegmatis and colonies with GFP activity were identified on agar plates. The GFP activities of 77 fluorescent clones, 34 from the M.tuberculosis library and 43 from the M.smegmatis library, were measured in liquid culture and compared with the activity of GFP that was transcribed from the Mycobacterium bovis BCG hsp60 promoter. The hsp60 promoter has strong activity in M.smegmatis and M.tuberculosis and has been frequently used to overexpress proteins in mycobacteria (29–32). Clones isolated from the two DNA libraries had GFP activities that ranged from 5-fold less to 10-fold higher than that of hsp60-gfp containing bacteria. Plasmids isolated from the eight clones with the highest GFP activities in M.smegmatis were also analyzed in M.bovis BCG (Figure 1). These plasmids produced GFP activities in BCG that were similar to those detected in M.smegmatis. The plasmid that produced the highest GFP activity in M.smegmatis and M.bovis BCG, named pUV15, was chosen for further characterization.
Figure 1.
Isolation of DNA fragments with promoter activity in M.bovis BCG. M.bovis BCG was transformed with plasmids from the promoter library or control plasmid and GFP activity was measured in log phase cultures. The eight promoters with the highest activity in M.smegmatis (data not shown) were measured in M.bovis BCG. All measurements were carried out in triplicate. The fluorescence intensity was normalized to the cell density and expressed in RFUs. Data are averages and error bars represent standard deviations.
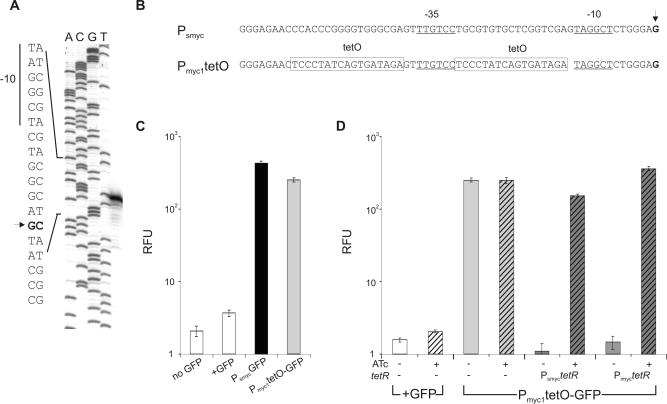
Insertion of tet operators into Psmyc generated a strong mycobacterial promoter that can be efficiently repressed by TetR
Sequencing of pUV15 revealed that it contains 290 bp of M.smegmatis DNA upstream of gfp. Of this fragment 189 bp are 100% identical to the upstream region of the putative rpsA gene of M.smegmatis. The 5′ end of the gfp mRNA encoded by pUV15 was identified by primer extension and located within the 189 bp fragment (Figure 2A). pUV15 produced no GFP activity in E.coli and the sequence elements that are located upstream of the putative transcriptional start point are only marginally similar to the E.coli −10 and −35 promoter consensus sequences. However, replacement of the putative −10 region TAGGCT with the sequence GGCTGG reduced GFP activity in M.smegmatis by 98%, suggesting that primer extension correctly identified the transcriptional start point within the cloned M.smegmatis DNA. This promoter was named Psmyc and is identical to that described by Kaps et al. (25). The 17 nt upstream and downstream of the putative −35 consensus sequence of Psmyc were replaced with tet operators (tetO) to construct a Psmyc derivative, Pmyc1tetO (Figure 2B), that might be susceptible to inhibition by the transcriptional repressor TetR. Pmyc1tetO had only 2-fold lower activity than Psmyc (Figure 2C). The effect of TetR on Pmyc1tetO activity was tested using an expression cassette, in which the strong promoter Psmyc was used to transcribe a derivative of the Tn10 tetR gene (27). Figure 2D demonstrates that the expression of TetR led to efficient, 200-fold repression of GFP activity in M.smegmatis.
Figure 2.
TetR-controlled gene expression in M.smegmatis. (A) Sequence of transcriptional start point of Psmyc determined by primer extension. Primer extension reactions were performed with RNA isolated during logarithmic growth. (B) Nucleotide sequence of Psmyc and Pmyc1tetO. Putative −10 and −35 promoter consensus sequences are underlined. Tet operator sequences are indicated by boxes. (C) Effect of incorporation of Tet operator sites on Psmyc activity. The bar labeled ‘no GFP’ represents the activity of bacteria transformed with a plasmid that does not contain gfp and shows background fluorescence of the bacteria; the bar labeled ‘+GFP’ contains the gfp reporter without Psmyc or Pmyc1tetO and shows background promoter activity of the plasmid; Psmyc-GFP shows GFP activity driven by Psmyc (black bar), and Pmyc1tetO-GFP shows GFP activity driven by Pmyc1tetO (gray bar). (D) Effect of ATc and tetR expression levels on activity of Pmyc1tetO. Crosshatched bars indicate the addition of 50 ng/ml ATc. Light gray bars show constitutive Pmyc1tetO activities. Dark gray bars indicate the expression of tetR by Psmyc (strong promoter see Figure 1) and by Pimyc (intermediate strength promoter). In (D), all fluorescence intensities were corrected for background fluorescence of the bacteria. All measurements were carried out in triplicate. The fluorescence intensity was normalized to the cell density and expressed in RFUs. Data are averages and error bars represent standard deviations.
Induction of Pmyc1tetO with ATc
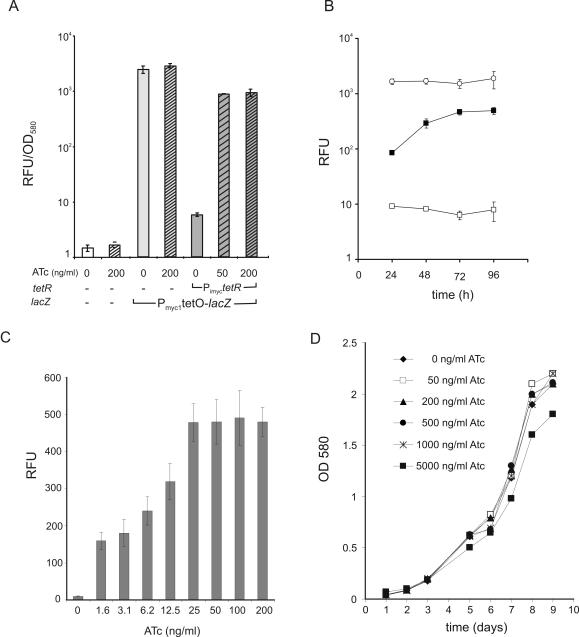
ATc, a tetracycline derivative with a very high affinity to Tn10 TetR and low toxicity (33), was used to measure induction of Pmyc1tetO. Addition of 50 ng/ml ATc to M.smegmatis transformed with Pmyc1tetO and Psmyc-tetR resulted in 150-fold induction of GFP (Figure 2D), but the GFP activity did not reach that observed in the absence of TetR. An expression cassette in which a weaker promoter, Pimyc (25), transcribed tetR was constructed and used to test whether reducing the intracellular amount of TetR could increase the efficiency of induction. Transformation of M.smegmatis with Pimyc-tetR produced amounts of TetR that were lower than those produced by Psmyc-tetR (data not shown) but sufficient to repress Pmyc1tetO by a factor of 170 (Figure 2D). Addition of 50 ng/ml ATc to M.smegmatis transformed with Pmyc1tetO and Pimyc-tetR resulted in complete induction of GFP (Figure 2D).
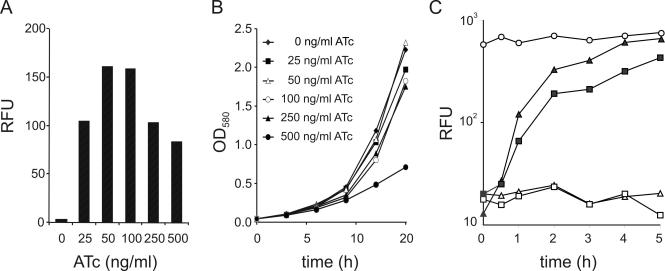
Induction of Pmyc1tetO by ATc in M.smegmatis is time- and dose-dependent and complete at a concentration that is 10-fold below the minimal inhibitory concentration of ATc
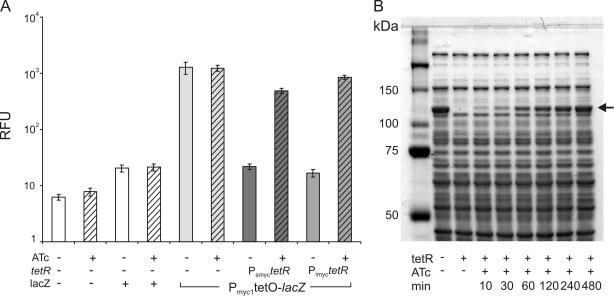
M.smegmatis was transformed with an episomal plasmid containing Pmyc1tetO-gfp and Pimyc-tetR and cultivated in the presence of different concentrations of ATc. Quantification of GFP activities after 15 h of cultivation demonstrated that concentrations of 50 and 100 ng/ml ATc gave maximal GFP activities (Figure 3A). Induction with 25 ng/ml ATc led to 1.6-fold lower GFP activities. Cultivation of M.smegmatis in the presence of 250 or 500 ng/ml ATc also led to reduced GFP activities. Only 500 ng/ml ATc caused significantly reduced growth of M.smegmatis (Figure 3B). These experiments demonstrated that induction is dose-dependent and complete at concentrations that are 10-fold below the minimal inhibitory concentration (MIC) of ATc. The kinetics of GFP induction was measured using Pimyc-tetR and Psmyc-tetR (Figure 3C). Maximal GFP activities in Pimyc-tetR containing bacteria were observed 4 h after the addition of ATc. Psmyc-tetR containing bacteria showed strong but incomplete and delayed induction. This induction was time-dependent and the kinetics of induction depended on the expression level of TetR. Analyses using a more sensitive reporter gene, lacZ, confirmed that regulation of almost two orders of magnitude was achieved with Pmyc1tetO (Figure 4A) and demonstrated that complete induction of Pmyc1tetO-lacZ resulted in high β-galactosidase protein levels (Figure 4B).
Figure 3.
Determination of the optimal inducer concentration, growth inhibition by ATc and kinetics of induction. (A) Inducer concentration. M.smegmatis cultures transformed with plasmids carrying Pmyc1tetO-gfp and Psmyc-tetR were grown into log phase (OD580 ∼ 0.5) and increasing amounts of ATc were added. Fifteen hours later, GFP activities were determined. (B) Growth curves in the presence of ATc. M.smegmatis was grown in 7H9 medium without and with different concentrations of ATc and optical densities of the cultures were recorded every 3 h. (C) Kinetics of induction. M.smegmatis transformed with plasmids containing Pmyc1tetO-gfp (circles), Pmyc1tetO-gfp + Psmyc-tetR (squares) and Pmyc1tetO-gfp + Pimyc-tetR (triangles) were grown into log phase (OD580 ∼ 0.5), then 50 ng/ml ATc was added (t = 0) to duplicate cultures of M.smegmatis transformed with Pmyc1tetO-gfp + Pimyc-tetR (filled squares) and Pmyc1tetO-gfp + Pimyc-tetR (filled triangles). GFP activity was determined 0.5, 1, 2, 3, 4 and 5 h after the addition of ATc.
Figure 4.
TetR controlled expression of lacZ in M.smegmatis. (A) TetR-controlled β-galactosidase activity. M.smegmatis was transformed as indicated underneath the bars. Bacteria were grown in 7H9 medium with 50 μg/ml hygromycin to an OD580 of ∼1.5. Cultures were diluted 1:100 into fresh medium without and with the addition of 50 ng/ml ATc (cross-hatched bars). After 15 h induction time, β-galactosidase activity was determined using the fluorescent substrate C2FDG. All fluorescence measurements were carried out in triplicate. The fluorescence intensity was normalized to the cell density and expressed in RFUs. Data are averages and error bars represent standard deviations. (B) Time course of TetR-controlled β-galactosidase expression. Lysates from M.smegmatis (18 μg protein) expressing lacZ with the Pmyc1tetO promoter were separated on a 9% SDS–PAGE gel. Lane 1, molecular weight marker; lane 2, constitutively expressed lacZ (no tetR); lane 3, repressed lacZ (+tetR); lanes 4–9, time course of induction of TetR-controlled lacZ by 50 ng/ml ATc. The arrow indicates the β-galactosidase protein band.
Pmyc1tetO allows construction of TetR-controlled, conditional knockouts in M.smegmatis
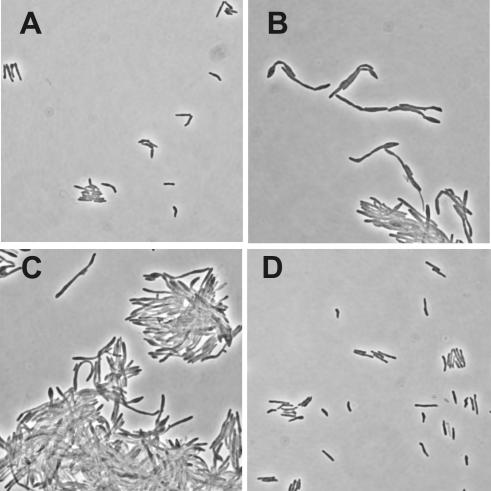
FtsZ is a bacterial tubulin homolog that is essential for cell division in most bacteria (34,35). A mycobacterial suicide plasmid was constructed, in which the first 700 bp of the M.smegmatis ftsZ gene were cloned downstream of Pmyc1tetO. Integration of this plasmid into the chromosome of M.smegmatis via homologous recombination replaced the native ftsZ promoter(s) with Pmyc1tetO (Figure 5A). M.smegmatis that contained tetR constitutively expressed by Psmyc in the chromosomal attB site (attB:tetR) was used to replace the ftsZ promoter with Pmyc1tetO. No recombinants were obtained with TetR expressing M.smegmatis unless ATc was included in the plates used for selection. Integration of Pmyc1tetO in front of ftsZ was confirmed by PCR (data not shown). The requirement of ATc for the growth of M.smegmatis attB:tetR Pmyc1tetO-ftsZ recombinants suggested that TetR efficiently repressed the expression of ftsZ and inhibited cell division. To test this hypothesis, M.smegmatis attB:tetR Pmyc1tetO-ftsZ was taken from ATc containing plates, grown in liquid culture in the presence of ATc until they reached the logarithmic growth phase, harvested and used to inoculate liquid media with different ATc concentrations. Removal of ATc led to the inhibition of growth and aggregation of M.smegmatis attB:tetR Pmyc1tetO-ftsZ but not of wt M.smegmatis (Figure 5B and C). Both phenotypes were delayed or reversed by the addition of ATc; low amounts of ATc delayed occurrence of the phenotypes and M.smegmatis attB:tetR Pmyc1tetO-ftsZ grew normally in the presence of 2–40 ng/ml ATc. Removal of ATc from M.smegmatis attB:tetR Pmyc1tetO-ftsZ cultures also resulted in the morphological changes that are expected to occur as a result of the inhibition of FtsZ (10): the bacteria became elongated, showed bud-like structures and formed large aggregates (Figure 6). Addition of 40 ng/ml ATc to the growth medium prevented these effects and the bacteria were indistinguishable from wt cells.
Figure 5.
TetR-controlled regulation of ftsZ expression in M.smegmatis. (A) Strategy for construction of the conditional ftsZ knockout. A suicide plasmid was used to replace the ftsZ upstream region with Pmyc1tetO in an M.smegmatis strain that had PsmyctetR integrated in the attB site. (B) Impact of ftsZ silencing on growth of M.smegmatis. Wt M.smegmatis and the conditional ftsZ knockout were grown in the presence of 40 ng/ml ATc into the mid log phase. The cultures were washed and diluted to an OD580 of 0.02 into fresh medium without and with different concentrations of ATc (t = 0). Growth in the presence and absence of ATc was followed by measuring optical densities of the cultures. (C) Photographs of wt M.smegmatis and the conditional ftsZ knockout cultures 20 h after growth without and with 40 ng/ml ATc.
Figure 6.
Morphology of the M.smegmatis conditional ftsZ knockout. Cultures grown in the presence or absence of ATc were examined by phase contrast microscopy with a 100× objective. (A) Wt M.smegmatis, (B, C and D) the conditional ftsZ knockout strain 9 h after removal of ATc (B), 12 h after removal of ATc (C) and in the presence of 40 ng/ml ATc (D).
TetR-controlled gene expression in M.tuberculosis
A replicative plasmid containing Pmyc1tetO-lacZ and Pimyc-tetR was transformed into M.tuberculosis H37Rv, and β-galactosidase activities were analyzed during in vitro growth. The β-galactosidase activities of M.tuberculosis containing Pmyc1tetO-lacZ and Pimyc-tetR were ∼400 times lower than the β-galactosidase activities of M.tuberculosis containing only Pmyc1tetO-lacZ and no tetR (Figure 7A). Addition of 50 or 200 ng/ml ATc led to a 150- and 160-fold increase in β-galactosidase activities after 96 h. Quantification of β-galactosidase activities 24, 48 and 72 h after the addition of ATc showed that full induction of β-galactosidase occurred 72 h after the addition of 50 ng/ml ATc (Figure 7B). Lowering the ATc concentration below 25 ng/ml resulted in a dose-dependent decrease in β-galactosidase expression levels (Figure 7C). Growth curves of M.tubercale H37Rv with different concentrations of ATc revealed that concentrations of up to 1 μg/ml did not decrease the growth rate (Figure 7D). These experiments demonstrated that the combination of Pmyc1tetO and Pimyc-tetR allows efficient gene repression and ATc concentration-dependent induction of gene expression in virulent M.tuberculosis. Induction can be achieved with ATc concentrations that are at least 20-fold below the MIC of the antibiotic.
Figure 7.
TetR controlled regulation of lacZ expression in M.tuberculosis H37Rv. (A) M.tuberculosis was transformed as indicated underneath the bars. Bacteria were grown at 37°C in 7H9 medium with 10% ADNaCl enrichment and 50 μg/ml hygromycin to an OD580 of ∼0.5. Cultures were then diluted 1:5 into fresh medium without and with addition of 50 or 200 ng/ml ATc (cross-hatched bars). After 96 h induction time, β-galactosidase activity was determined using the fluorescent substrate C2FDG. All fluorescence measurements were carried out in triplicate. The fluorescence intensity was normalized to the cell density and expressed in RFUs. Data are averages and error bars represent standard deviations. (B) Kinetics of β-galactosidase induction in H37Rv. H37Rv transformed with plasmids containing Pmyc1tetO-lacZ (circles), Pmyc1tetO-lacZ + Pimyc-tetR (squares) were grown into early log phase (OD580 ∼ 0.2), then 50 ng/ml ATc was added (t = 0) to cultures of H357Rv transformed with Pmyc1tetO-lacZ + Pimyc-tetR (filled squares). β-galactosidase activities were measured after 24, 48, 72 and 96 h induction time as described in (A). (C) Concentration-response curve of β-galactosidase induction in H37Rv. TetR-controlled lacZ was induced with different concentrations of ATc for 72 h, and β-galactosidase activities were determined as described in (A). (D). Growth curves in presence of ATc. M.tuberculosis was grown in enriched 7H9 medium without and with increasing concentrations of ATc and optical densities of the cultures were recorded every 24 h.
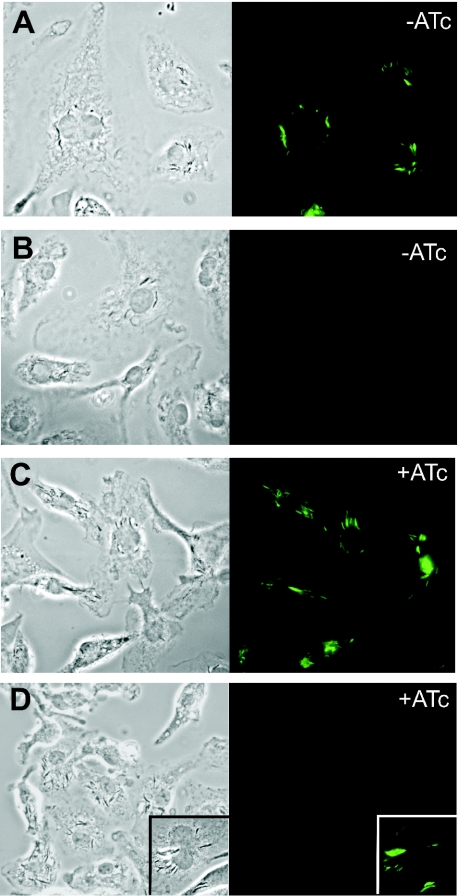
TetR-controlled gene expression in intraphagosomal M.tuberculosis
An important application of a TetR-controlled mycobacterial gene expression system is to regulate gene expression in mycobacteria that reside within macrophages or mice. We tested whether the TetR-controlled expression system allows the regulation of GFP expression within intraphagosomal M.tuberculosis. Bone marrow-derived murine macrophages were infected with M.tuberculosis carrying either constitutively expressed gfp (Pmyc1tetO-gfp) or TetR repressed gfp (Pmyc1tetO-gfp + Pimyc-tetR) on a replicative plasmid. Extracellular bacteria were killed using amikacin. ATc was added 24 h post infection. Treatment of infected macrophages with 100 ng/ml ATc for 72 h led to the induction of gfp in intracellular M.tuberculosis (Figure 8). This demonstrated that both promoters, Pmyc1tetO and Pimyc, are active in intracellular M.tuberculosis and that ATc gains access to the intraphagosomal bacteria to induce gene expression. Thus, the TetR-controlled expression system allows regulation of gene expression in intracellular M.tuberculosis.
Figure 8.
TetR-controlled GFP expression in intraphagosomal M.tuberculosis H37Rv. Murine bone marrow-derived macrophages grown on coverslips in 24 well plates were infected with live bacteria at an MOI of 5–10 bacteria: 1 macrophage. Four hours post infection, the macrophage monolayers were washed three times with warm PBS, followed by the addition of complete tissue culture medium containing 100 μg/ml amikacin to kill extracellular bacteria. The monolayers were washed again 8 h post infection. Twenty-four hours post infection, 100 ng/ml ATc were added to wells shown in (C) and 72 h later the coverslips were analyzed by microscopy as described in Materials and Methods. The left panels depict the cell monolayers in phase contrast and the right panels show the corresponding fluorescence image. Macrophages were infected with M.tuberculosis transformed with Pmyc1tetO-GFP (A); Pmyc1tetO-GFP + Pimyc-tetR (B and C). (D) Cells that are infected with Mtb lacking GFP and treated with ATc. Samples were prepared on a different day than those shown in (A–C), but the images were acquired under the exact same conditions. The relative brightness of control samples was similar on the two different days (insert: macrophages infected with Mtb constitutively expressing GFP).
DISCUSSION
The ideal bacterial expression system should be silent in the absence of inducer, quickly respond to non-toxic concentrations of inducer, allow regulation over a range of expression levels and, if used for the analysis of bacterial pathogens, permit gene regulation during infection of cells in tissue culture or animals. In an attempt to develop such a system for mycobacteria, we made use of the Tn10 encoded tet regulatory system of E.coli and adapted it for use in mycobacteria. A TetR responsive promoter was constructed by insertion of tet operators into regions of a strong mycobacterial promoter that are not essential for promoter activity. This promoter, Pmyc1tetO, mediated strong gene expression in M.smegmatis and M.tuberculosis and was efficiently repressed by TetR. However, some β-galactosidase activity was detected in the presence of Pmyc1tetO and TetR (Figure 4A). A similar level of β-galactosidase activity was measured in M.smegmatis using a vector that contained lacZ without Pmyc1tetO (Figure 4A). This background activity was therefore most likely due to an unidentified promoter within the plasmid that is not controlled by TetR. Pmyc1tetO independent β-galactosidase activity was also observed in M.tuberculosis. Elimination of this activity might further improve the expression system. However, the low level of gene expression that was detected in the presence of TetR did not interfere with effective silencing of ftsZ in M.smegmatis.
The efficient regulation of promoter activities by transcriptional repressors depends on several factors; one of them is the concentration of free repressor in the cell. Overexpression of a repressor can interfere with the efficient induction of promoter activity (17), as observed for induction of Pmyc1tetO in M.smegmatis and M.tuberculosis. In M.smegmatis, transcription of tetR by Pimyc allowed maximal induction, while Psmyc led to TetR levels that were incompletely induced. This effect was even more pronounced in M.tuberculosis. When tetR was expressed by the strong promoter Psmyc, induction of β-galactosidase expression with 200 ng/ml for 96 h was only 2-fold in M.tuberculosis (data not shown). In contrast, when tetR transcription was driven by Pimyc, a promoter of intermediate strength, induction was 160-fold under the same conditions. Thus, the use of promoters of different strength to drive transcription of tetR provides an additional mechanism to control expression levels of the regulated gene in the repressed and induced state.
The kinetics of induction of Pmyc1tetO with ATc was different in M.smegmatis and M.tuberculosis. The slower induction kinetics in M.tuberculosis may be explained by the increased resistance of this species to tetracycline. The MIC of tetracycline for M.smegmatis is 0.5 μg/ml (36) and identical to the concentration of ATc that inhibited growth of M.smegmatis in our experiments. In contrast, we did not detect any significant impact on the growth of M.tuberculosis after the addition of up to 5 μg/ml ATc. This is consistent with reports that tetracyclines have little activity against M.tuberculosis (37–39). A tetracycline resistance determinant, Tet(V), has been identified in M.smegmatis (40). M.tuberculosis contains two mycobacterial multidrug resistance efflux pumps, TapA and P55, that when expressed in M.smegmatis increase its MIC to tetracycline 4- to 8-fold (36,41). These efflux pumps may contribute to the increased tetracycline resistance of M.tuberculosis and explain the delayed kinetics of tetracycline-induced gene expression. It is also possible that M.tuberculosis is less permeable for tetracyclines than non-tuberculous mycobacteria. Nevertheless, access of ATc to TetR in M.tuberculosis was sufficient to allow full induction of TetR in response to 50 ng/ml ATc after 72 h (three generations).
A tetracycline-dependent, conditional ftsZ knockout was constructed in M.smegmatis by integrating Pmyc1tetO directly upstream of the start codon of ftsZ and simultaneously expressing TetR. Removal of ATc from this M.smegmatis strain resulted in phenotypes that are typical for inactivation of FtsZ: growth inhibition, filamentation, formation of bud-like structures and bacterial aggregation. Expression of ftsZ by Pmyc1tetO presumably led to normal FtsZ levels, because overexpression of FtsZ is toxic (42). Different genes most likely require different expression levels during normal growth. Pmyc1tetO is a strong promoter, and full induction may, for some genes, lead to growth inhibition. However, the use of different ATc concentrations (Figures 5B and 7C) showed that β-galactosidase and FtsZ expression levels were inducer concentration-dependent. This demonstrated that the TetR-controlled expression system can be used to fine-tune gene expression and suggests that promoter replacement is a generally applicable strategy to generate conditional knockouts in mycobacteria.
Gene expression systems that allow turning mycobacterial genes on and off during an infection will be valuable tools for the characterization of genes that are important for in vivo growth and persistence of M.tuberculosis. The system described here allows the control of M.tuberculosis gene expression within phagosomes of primary macrophages and may therefore also allow control of M.tuberculosis gene expression during an animal infection. The strategy that we used to construct Pmyc1tetO should be applicable to other mycobacterial promoters. Increasing knowledge of mycobacterial gene expression within the host (22,43–45) should allow the selection of promoters with robust in vivo activities and the construction of TetR-controlled promoters that can be induced during all phases of an M.tuberculosis infection. A TetR-controlled expression system for mycobacteria based on the tetZ resistance determinant from Corynebacterium glutamicum has also been constructed and has been successfully used to downregulate ftsZ via an antisense approach (M.C.J. Blokpoel personal communication). We used a Tn10-derived TetR for our studies because it and its close relatives are by far the best characterized TetRs. Mutants with altered specificities for operator binding (46,47), dimerization (48) and induction (49–51) as well as mutants with an altered response to ATc binding (52,53) provide an excellent basis for the rational improvement of tetracycline-inducible expression systems in mycobacteria.
Acknowledgments
The authors thank Carl Nathan, Michael Niederweis and Brian Robertson for helpful discussions and for critical review of this manuscript. We are grateful to Wolfgang Hillen for providing pWH520 and TetR antibodies. We thank Shuangping Shi for help with experiments. We are grateful to Tom Templeton and Lynda Pierini for help with microscopy. S.E. was supported by NIH grant ROI HL68525. D.S. was supported by an Ellison Medical Foundation New Scholar Award in Global Infectious Diseases. Funding to pay the Open Access publication charges for this article was provided by ROI HL68525 and the Ellison Medical Foundation.
REFERENCES
- 1.Barnes P.F., Cave M.D. Molecular epidemiology of tuberculosis. N. Engl. J. Med. 2003;349:1149–1156. doi: 10.1056/NEJMra021964. [DOI] [PubMed] [Google Scholar]
- 2.Frieden T.R., Sterling T.R., Munsiff S.S., Watt C.J., Dye C. Tuberculosis. Lancet. 2003;362:887–899. doi: 10.1016/S0140-6736(03)14333-4. [DOI] [PubMed] [Google Scholar]
- 3.Kremer L., Besra G.S. Re-emergence of tuberculosis: strategies and treatment. Expert Opin. Investig. Drugs. 2002;11:153–157. doi: 10.1517/13543784.11.2.153. [DOI] [PubMed] [Google Scholar]
- 4.DeVito J.A., Mills J.A., Liu V.G., Agarwal A., Sizemore C.F., Yao Z., Stoughton D.M., Cappiello M.G., Barbosa M.D., Foster L.A., et al. An array of target-specific screening strains for antibacterial discovery. Nat. Biotechnol. 2002;20:478–483. doi: 10.1038/nbt0502-478. [DOI] [PubMed] [Google Scholar]
- 5.Mnaimneh S., Davierwala A.P., Haynes J., Moffat J., Peng W.T., Zhang W., Yang X., Pootoolal J., Chua G., Lopez A., et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 6.Lim A., Boon C., Dick T. Inducibility of the Streptomyces traRts107-Ptra expression cassette in Mycobacterium smegmatis. Biol. Chem. 2000;381:517–519. doi: 10.1515/BC.2000.066. [DOI] [PubMed] [Google Scholar]
- 7.Mahenthiralingam E., Draper P., Davis E.O., Colston M.J. Cloning and sequencing of the gene which encodes the highly inducible acetamidase of Mycobacterium smegmatis. J. Gen. Microbiol. 1993;139:575–583. doi: 10.1099/00221287-139-3-575. [DOI] [PubMed] [Google Scholar]
- 8.Parish T., Mahenthiralingam E., Draper P., Davis E.O., Colston M.J. Regulation of the inducible acetamidase gene of Mycobacterium smegmatis. Microbiology. 1997;143:2267–2276. doi: 10.1099/00221287-143-7-2267. [DOI] [PubMed] [Google Scholar]
- 9.Roberts G., Muttucumaru D.G., Parish T. Control of the acetamidase gene of Mycobacterium smegmatis by multiple regulators. FEMS Microbiol. Lett. 2003;221:131–136. doi: 10.1016/S0378-1097(03)00177-0. [DOI] [PubMed] [Google Scholar]
- 10.Dziadek J., Rutherford S.A., Madiraju M.V., Atkinson M.A., Rajagopalan M. Conditional expression of Mycobacterium smegmatis ftsZ, an essential cell division gene. Microbiology. 2003;149:1593–1603. doi: 10.1099/mic.0.26023-0. [DOI] [PubMed] [Google Scholar]
- 11.Greendyke R., Rajagopalan M., Parish T., Madiraju M.V. Conditional expression of Mycobacterium smegmatis dnaA, an essential DNA replication gene. Microbiology. 2002;148:3887–3900. doi: 10.1099/00221287-148-12-3887. [DOI] [PubMed] [Google Scholar]
- 12.Gomez J.E., Bishai W.R. whmD is an essential mycobacterial gene required for proper septation and cell division. Proc. Natl Acad. Sci. USA. 2000;97:8554–8559. doi: 10.1073/pnas.140225297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Triccas J.A., Parish T., Britton W.J., Gicquel B. An inducible expression system permitting the efficient purification of a recombinant antigen from Mycobacterium smegmatis. FEMS Microbiol. Lett. 1998;167:151–156. doi: 10.1111/j.1574-6968.1998.tb13221.x. [DOI] [PubMed] [Google Scholar]
- 14.Berens C., Hillen W. Gene regulation by tetracyclines. Constraints of resistance regulation in bacteria shape TetR for application in eukaryotes. Eur. J. Biochem. 2003;270:3109–3121. doi: 10.1046/j.1432-1033.2003.03694.x. [DOI] [PubMed] [Google Scholar]
- 15.Geissendorfer M., Hillen W. Regulated expression of heterologous genes in Bacillus subtilis using the Tn10 encoded tet regulatory elements. Appl. Microbiol. Biotechnol. 1990;33:657–663. doi: 10.1007/BF00604933. [DOI] [PubMed] [Google Scholar]
- 16.Ji Y., Zhang B., Van S.F., Horn, Warren P., Woodnutt G., Burnham M.K., Rosenberg M. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science. 2001;293:2266–2269. doi: 10.1126/science.1063566. [DOI] [PubMed] [Google Scholar]
- 17.Lutz R., Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian F., Pan W. Construction of a tetR-integrated Salmonella enterica serovar Typhi CVD908 strain that tightly controls expression of the major merozoite surface protein of Plasmodium falciparum for applications in human Vaccine production. Infect. Immun. 2002;70:2029–2038. doi: 10.1128/IAI.70.4.2029-2038.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Soolingen D., Hermans P.W., de Haas P.E., Soll D.R., van Embden J.D. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J. Clin. Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J., Russell D.W. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 21.Hatful G.F., William R., Jacobs J. Molecular Genetics of Mycobacteria. Washington, DC: ASM Press; 2000. pp. 319–320. [Google Scholar]
- 22.Schnappinger D., Ehrt S., Voskuil M.I., Liu Y., Mangan J.A., Monahan I.M., Dolganov G., Efron B., Butcher P.D., Nathan C., et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crameri A., Whitehorn E.A., Tate E., Stemmer W.P. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 24.Scholz O., Thiel A., Hillen W., Niederweis M. Quantitative analysis of gene expression with an improved green fluorescent protein. p6. Eur. J. Biochem. 2000;267:1565–1570. doi: 10.1046/j.1432-1327.2000.01170.x. [DOI] [PubMed] [Google Scholar]
- 25.Kaps I., Ehrt S., Seeber S., Schnappinger D., Martin C., Riley L.W., Niederweis M. Energy transfer between fluorescent proteins using a co-expression system in Mycobacterium smegmatis. Gene. 2001;278:115–124. doi: 10.1016/s0378-1119(01)00712-0. [DOI] [PubMed] [Google Scholar]
- 26.Rowland B., Purkayastha A., Monserrat C., Casart Y., Takiff H., McDonough K.A. Fluorescence-based detection of lacZ reporter gene expression in intact and viable bacteria including Mycobacterium species. FEMS Microbiol. Lett. 1999;179:317–325. doi: 10.1111/j.1574-6968.1999.tb08744.x. [DOI] [PubMed] [Google Scholar]
- 27.Berens C., Altschmied L., Hillen W. The role of the N terminus in Tet repressor for tet operator binding determined by a mutational analysis. J. Biol. Chem. 1992;267:1945–1952. [PubMed] [Google Scholar]
- 28.Ehrt S., Schnappinger D., Bekiranov S., Drenkow J., Shi S., Gingeras T.R., Gaasterland T., Schoolnik G., Nathan C. Reprogramming of the macrophage transcriptome in response to interferon-gamma and Mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase. J. Exp. Med. 2001;194:1123–1140. doi: 10.1084/jem.194.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bastos R.G., Dellagostin O.A., Barletta R.G., Doster A.R., Nelson E., Osorio F.A. Construction and immunogenicity of recombinant Mycobacterium bovis BCG expressing GP5 and M protein of porcine reproductive respiratory syndrome virus. Vaccine. 2002;21:21–29. doi: 10.1016/s0264-410x(02)00443-7. [DOI] [PubMed] [Google Scholar]
- 30.Biet F., Kremer L., Wolowczuk I., Delacre M., Locht C. Mycobacterium bovis BCG producing interleukin-18 increases antigen-specific gamma interferon production in mice. Infect. Immun. 2002;70:6549–6557. doi: 10.1128/IAI.70.12.6549-6557.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.da Cruz F.W., McBride A.J., Conceicao F.R., Dale J.W., McFadden J., Dellagostin O.A. Expression of the B-cell and T-cell epitopes of the rabies virus nucleoprotein in Mycobacterium bovis BCG and induction of an humoral response in mice. Vaccine. 2001;20:731–736. doi: 10.1016/s0264-410x(01)00414-5. [DOI] [PubMed] [Google Scholar]
- 32.Stover C.K., de la Cruz V.F., Fuerst T.R., Burlein J.E., Benson L.A., Bennett L.T., Bansal G.P., Young J.F., Lee M.H., Hatfull G.F. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 33.Gossen M., Bujard H. Anhydrotetracycline, a novel effector for tetracycline controlled gene expression systems in eukaryotic cells. Nucleic Acids Res. 1993;21:4411–4412. doi: 10.1093/nar/21.18.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Errington J., Daniel R.A., Scheffers D.J. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 2003;67:52–65. doi: 10.1128/MMBR.67.1.52-65.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiss D.S. Bacterial cell division and the septal ring. Mol. Microbiol. 2004;54:588–597. doi: 10.1111/j.1365-2958.2004.04283.x. [DOI] [PubMed] [Google Scholar]
- 36.Silva P.E., Bigi F., de la Paz Santangelo M., Romano M.I., Martin C., Cataldi A., Ainsa J.A. Characterization of P55, a multidrug efflux pump in Mycobacterium bovis and Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2001;45:800–804. doi: 10.1128/AAC.45.3.800-804.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hobby G.L., Lenert T.F. Antituberculous activity of tetracycline and related compounds. Am. Rev. Tuberc. 1955;72:367–372. doi: 10.1164/artpd.1955.72.3.367. [DOI] [PubMed] [Google Scholar]
- 38.Kamala T., Herbert D., Venkatesan P., Paramasivan C.N. Minimal inhibitory concentrations of lomefloxacin and minocycline against drug-sensitive and drug-resistant isolates of M.tuberculosis compared on L-J and 7H11 media. Int. J. Lepr. Other Mycobact. Dis. 1997;65:375–378. [PubMed] [Google Scholar]
- 39.Shaila M.S., Gopinathan K.P., Ramakrishnan T. Protein synthesis in Mycobacterium tuberculosis H37Rv and the effect of streptomycin in streptomycin-susceptible and -resistant strains. Antimicrob. Agents Chemother. 1973;4:205–213. doi: 10.1128/aac.4.3.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Rossi E., Blokpoel M.C., Cantoni R., Branzoni M., Riccardi G., Young D.B., De Smet K.A., Ciferri O. Molecular cloning and functional analysis of a novel tetracycline resistance determinant, tet(V), from Mycobacterium smegmatis. Antimicrob. Agents Chemother. 1998;42:1931–1937. doi: 10.1128/aac.42.8.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ainsa J.A., Blokpoel M.C., Otal I., Young D.B., De Smet K.A., Martin C. Molecular cloning and characterization of Tap, a putative multidrug efflux pump present in Mycobacterium fortuitum and Mycobacterium tuberculosis. J. Bacteriol. 1998;180:5836–5843. doi: 10.1128/jb.180.22.5836-5843.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dziadek J., Madiraju M.V., Rutherford S.A., Atkinson M.A., Rajagopalan M. Physiological consequences associated with overproduction of Mycobacterium tuberculosis FtsZ in mycobacterial hosts. Microbiology. 2002;148:961–971. doi: 10.1099/00221287-148-4-961. [DOI] [PubMed] [Google Scholar]
- 43.Shi L., Jung Y.J., Tyagi S., Gennaro M.L., North R.J. Expression of Th1-mediated immunity in mouse lungs induces a Mycobacterium tuberculosis transcription pattern characteristic of nonreplicating persistence. Proc. Natl Acad. Sci. USA. 2003;100:241–246. doi: 10.1073/pnas.0136863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Talaat A.M., Lyons R., Howard S.T., Johnston S.A. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl Acad. Sci. USA. 2004;101:4602–4607. doi: 10.1073/pnas.0306023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Timm J., Post F.A., Bekker L.G., Walther G.B., Wainwright H.C., Manganelli R., Chan W.T., Tsenova L., Gold B., Smith I., et al. Differential expression of iron-, carbon-, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc. Natl Acad. Sci. USA. 2003;100:14321–14326. doi: 10.1073/pnas.2436197100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Helbl V., Hillen W. Stepwise selection of TetR variants recognizing tet operator 4C with high affinity and specificity. J. Mol. Biol. 1998;276:313–318. doi: 10.1006/jmbi.1997.1540. [DOI] [PubMed] [Google Scholar]
- 47.Helbl V., Tiebel B., Hillen W. Stepwise selection of TetR variants recognizing tet operator 6C with high affinity and specificity. J. Mol. Biol. 1998;276:319–324. doi: 10.1006/jmbi.1997.1539. [DOI] [PubMed] [Google Scholar]
- 48.Schnappinger D., Schubert P., Pfleiderer K., Hillen W. Determinants of protein-protein recognition by four helix bundles: changing the dimerization specificity of Tet repressor. EMBO J. 1998;17:535–543. doi: 10.1093/emboj/17.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henssler E.M., Scholz O., Lochner S., Gmeiner P., Hillen W. Structure-based design of Tet repressor to optimize a new inducer specificity. Biochemistry. 2004;43:9512–9518. doi: 10.1021/bi049682j. [DOI] [PubMed] [Google Scholar]
- 50.Krueger C., Schmidt A., Danke C., Hillen W., Berens C. Transactivator mutants with altered effector specificity allow selective regulation of two genes by tetracycline variants. Gene. 2004;331:125–131. doi: 10.1016/j.gene.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 51.Scholz O., Kostner M., Reich M., Gastiger S., Hillen W. Teaching TetR to recognize a new inducer. J. Mol. Biol. 2003;329:217–227. doi: 10.1016/s0022-2836(03)00427-3. [DOI] [PubMed] [Google Scholar]
- 52.Kamionka A., Bogdanska-Urbaniak J., Scholz O., Hillen W. Two mutations in the tetracycline repressor change the inducer anhydrotetracycline to a corepressor. Nucleic Acids Res. 2004;32:842–847. doi: 10.1093/nar/gkh200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scholz O., Henssler E.M., Bail J., Schubert P., Bogdanska-Urbaniak J., Sopp S., Reich M., Wisshak S., Kostner M., Bertram R., et al. Activity reversal of Tet repressor caused by single amino acid exchanges. Mol. Microbiol. 2004;53:777–789. doi: 10.1111/j.1365-2958.2004.04159.x. [DOI] [PubMed] [Google Scholar]
- 54.Snapper S.B., Melton R.E., Mustafa S., Kieser T., Jacobs W.R., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 1990;4:1991–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]