Abstract
A new experimental technique for genome-wide detection of integration sites of polymorphic retroelements (REs) is described. The technique allows one to reveal the absence of a retroelement in an individual genome provided that this retroelement is present in at least one of several other genomes under comparison. Since quite a number of genomes are compared simultaneously, the search for polymorphic REs insertions is very efficient. The technique includes two whole-genome selective PCR amplifications of sequences flanking REs: one for a particular genome and another one for a mixture of ten different genomes. A subsequent subtractive hybridization of the obtained amplicons with DNA of a particular genome as driver results in isolation of polymorphic insertions. The technique was successfully applied for identification of 41 new polymorphic human AluYa5/Ya8 insertions. Among them, 18 individual Alu elements first sequenced in this work were not found in the available human genome databases. This result suggests that significant part of polymorphic REs were not identified during genome sequencing and remain to be detected and characterized. The proposed method does not depend on preliminary knowledge of evolutionary history of retroelements and can be applied for identification of insertion/deletion polymorphic markers in genomes of different species.
INTRODUCTION
Retroelements (REs) are known to comprise a significant portion of the human genome and might have a serious impact on genome functioning and evolution (1,2). Of special interest for human genome studies are recently integrated and polymorphic REs.
Polymorphic REs offer considerable advantages over other types of polymorphisms. In particular, they are stable, and although it can be assumed that preexisting REs might be excised from the genome, no experimental evidence is presently known in favor of this assumption. Therefore, the presence of an RE represents identity by descent, whereas the ancestral state of the RE insertion polymorphism is known to be the absence of the RE. Thus, identification of young, recently integrated and polymorphic REs is of considerable interest for deeper understanding of primate genome evolution and relationships of various human populations, as well as for the development of new powerful tools for gene mapping.
Several groups of authors described quite a number of polymorphic and human-specific Alu, L1 and HERV-K integrations (3–11). However, no comprehensive experimental genome-wide search for polymorphic RE insertions was done so far, though bioinformatic screening of genomic databases for Alu and L1 was performed (4,6,10,12,13). A rationale for the authors' approach was that small subsets of both Alu and L1 families amplified within the genome during recent evolutionary time (4–5 Mya) may be not fixed and may therefore display considerable polymorphism in human population. Indeed, Alu retroposition activity in human was increased some time after the divergence of the human and chimpanzee lineages, mostly due to the two most recently formed human Alu subfamilies, Alu Ya5 and Alu Yb8. The latest estimates of these subfamilies' ages assign their amplification to a period between 2.5 and 3.5 Mya (4,6,14). Therefore, a computer search for recently integrated REs in the human genomic DNA sequence databases could provide candidate polymorphic retroelement integrations. This technique was applied to the identification of recently integrated Alu family members (4). As a result, 2640 and 1852 representatives of the youngest Ya5 and Yb8 groups of Alu, respectively, were identified in the human genome draft sequence. An experimental analysis of 475 of these elements revealed that although over 99% of them were restricted to the human genome, only 106 investigated Alu inserts were polymorphic in the genomes of various human populations. Similar analysis was also performed for 262 representatives of the evolutionarily young L1Hs Ta family and authors found that 115 of them were polymorphic in humans (9,10): 29% and 69% of the loci that contained inserts of Ta-0 and the evolutionarily youngest Ta-1 subfamilies, respectively, were found to be polymorphic. Although RE polymorphisms are widely used for studies of human genome variability, most recently published data demonstrate that many polymorphic RE integrations are lacking from the available human genome databases (15,16). Apparently, the computer searches should be accompanied by experimental checking of which of the REs identified are indeed polymorphic. In addition, many polymorphic insertions can be absent from the available human genome sequence since not only it contains gaps but it also represents only a few haplotypes taken for sequencing just by chance. Moreover, the computer search is impossible for non-sequenced genomes, in particular those of primates other than human. Unfortunately, known experimental techniques did not permit to make genome-wide analyses, although some approaches were successfully applied to detecting polymorphic retroelements (15–18).
In this report, we describe a general approach to experimental identification of polymorphic REs in the human genome without any preliminary knowledge of the genome sequences. Using this approach, we identified 41 new polymorphic Alu insertions, 18 of which were not found in the published versions of the human genome sequence, therefore being undetectable by computer search.
MATERIALS AND METHODS
DNA samples
Human DNA samples were isolated from peripheral blood lymphocytes by standard phenol/chloroform extraction and ethanol precipitation. DNA sampling from East European and Asian populations has been described elsewhere (19). The samples kindly provided by Dr E. Khusnutdinova (Institute of Biochemistry and Genetics, Ufa Research Center, Russian Academy of Sciences, Ufa, 450054 Bashkortostan, Russia) represented DNAs of 11 individuals of various ethnic origin including Chuvash, Tabasaran, Tatar, Mari, Belorussian, Nogay, Lezgin, Uzbek, Kazakh and Russian. A mixture of their DNAs was used as tracer DNA, whereas driver DNA was prepared from an Andi (Northeast branch of the North Caucasian family) male individual DNA sample.
Three genomic DNA samples isolated from placentas were used for optimization of PCR conditions.
Preparation of driver DNA
Andi genomic DNA (500 ng) was digested with 20 U of RsaI endonuclease (Fermentas) in 50 μl of 1× restriction buffer Y+Tango (Fermentas) at 37°C for 2 h. The digested DNA was then purified by phenol/chloroform extraction, ethanol precipitated, dissolved in 10 μl of sterile water and ligated to an excess of the adapter (15 μM) at 16°C overnight using T4 DNA ligase (Promega). To form the adapter, 150 μM A1A2 and a12 oligonucleotides (Table 1) were annealed in TM (10 mM Tris–HCl, pH 7.8, 10 mM MgCl2) buffer. The ligation was terminated by incubation at 65°C for 15 min. The ligates were then separated from free adapter molecules by passing through a QIAquick PCR Purification Kit (Qiagen). A fraction of Alu Ya5 and Ya8 5′ flanking sequences was amplified using the selective PCR suppression technique (8,20). A1 and T1 primers (Table 1) were used in the first round of PCR. T1 primer corresponds to a fragment of the Alu sequence containing three mutations characteristic of the Alu Y subfamily, as well as two mutations distinguishing Alu Ya branch. The PCR mixture contained 10 ng of the ligate in 25 μl of 1× PCR Buffer for Advantage 2 (BD Biosciences, Clontech) containing 200 μM each of dNTPs, 0.4 μM each of primers and 0.5 μl of 50× Advantage 2 polymerase mix (BD Biosciences, Clontech). The PCR was carried out in a thermal cycler (OmniGene Hybaid) as follows: 72°C for 4 min (to extend 3′ ends of the DNA duplexes) and 23 cycles at 95°C for 25 s, 65°C for 25 s, 72°C for 80 s. The PCR products were 1000-fold diluted and re-amplified with A2 and T2 primers (Table 1) in the second PCR round (16 cycles at 94°C for 20 s, 68°C for 20 s, 72°C for 90 s). T2 primer was designed against the utmost 5′ end of Alu repeats. The PCR products obtained were purified by phenol/chloroform extraction, ethanol precipitated and dissolved in 100 μl of sterile water.
Table 1.
Oligonucleotides/primers used for the library construction
| A1A2 | TGTAGCGTGAAGACGACAGAAAGGGCGTGGTGCGGAGGGCGGT |
| A1 | TGTAGCGTGAAGACGACAGAA |
| A2 | AGGGCGTGGTGCGGAGGGCGGT |
| a12 | ACCGCCCTCC |
| A3A4 | AGCAGCGAACTCAGTACAACAAGTCGACGCGTGCCCGGGCTGGT |
| A3 | AGCAGCGAACTCAGTACAACA |
| A4 | AGTCGACGCGTGCCCGGGCTGGT |
| a34 | ACCAGCCC |
| A5A6 | GTAATACGACTCACTGGAGGGCGAGCGTGGTCGCCGCCGAGGTG |
| A5 | GTAATACGACTCACTGGAGGGC |
| A6 | GAGCGTGGTCGCCGCCGAGGTG |
| a56 | CACCTCGGC |
| T1 | TCACCGTTTTAGCCGGGA |
| T2 | GTGAGCCACCGCGCCCGG |
To remove A2-originated termini (residual adapter fragments), 3.75 μg of the purified PCR product was digested with 20 U of RsaI endonuclease (Fermentas) in 200 μl of 1× restriction buffer Y+Tango (Fermentas) at 37°C for 2 h. This last stage is crucial for avoiding cross-hybridization.
Preparation of tracer DNA
Chuvash, Tabasaran, Tatar, Mari, Belorussian, Nogay, Lezgin, Uzbek, Kazakh and Russian human genomic DNAs (100 ng each) were mixed and digested with 40 U of RsaI endonuclease (Fermentas) in 100 μl of 1× restriction buffer Y+Tango (Fermentas) at 37°C for 2 h. The digested DNA was then purified by phenol/chloroform extraction, ethanol precipitated, dissolved in 10 μl of sterile water, ligated to an excess of A1A2/a12 adapter, and two-round PCR amplified as described for driver DNA preparation. Prior to the PCR reaction, 60 pmol of oligonucleotide T2 was 5′ phosphorylated in 10 μl of 1× PNK Buffer A (Fermentas), containing 1 mM ATP and 10 U of T4 Polynucleotide Kinase (Fermentas). The PCR product was then purified by phenol/chloroform extraction, ethanol precipitated and dissolved in 100 μl of sterile water.
The purified PCR product (900 ng) was digested with RsaI endonuclease as described for driver DNA to remove A2-originated termini, and purified by passing through a QIAquick PCR Purification Kit. To remove protruding dA at 3′ ends added by AdvanTaq DNA Polymerase, the restricted DNA fragments were treated with Klenow fragment of DNA polymerase I (Fermentas) for 15 min at room temperature in 10 μl of 1× Klenow buffer (Fermentas) and additionally incubated for 15 min in the presence of 125 μM each of dNTPs. Klenow fragment was inactivated by heating at 70°C for 10 min. A 125 ng aliquots of the resulting DNA were ligated to adapters A3A4/a34 (1.7 μM) or A5A6/a56 to form the tracer A and tracer B portions, respectively.
Subtractive hybridization
Tracer A and B samples (16 ng each) were separately mixed with 900 ng of the RsaI digested driver DNA. DNA samples were purified by phenol/chloroform extraction, precipitated with ethanol and dissolved in 2.5 μl of hybridization buffer (0.5 M NaCl, 50 mM HEPES, pH 8.3, 0.2 mM EDTA). A sample of 1800 ng of the driver DNA was also purified, precipitated and dissolved in 5 μl of hybridization buffer.
The tracer A/driver and tracer B/driver samples were denatured at 96°C for 5 min and hybridized separately at 68°C for 3 h to remove the most abundant Alu flanks from each of the two tracer DNA portions. Then 5 μl of driver DNA was denatured at 96°C for 5 min, incubated at 68°C for 5 min, and mixed first with the tracer A/driver and then with the tracer B/driver samples. The resulting mixture was incubated at 68°C overnight and diluted with 100 μl dilution buffer (50 mM NaCl, 5 mM HEPES, pH 8.3, 0.2 mM EDTA). An aliquot of 1 μl of this diluted mixture was pre-incubated at 72°C for 3 min (to fill in the ends of DNA duplexes by AdvanTaq DNA Polymerase) and then PCR amplified with 0.4 μM each of A3 and A5 primers (Table 1) as follows: 94°C for 20 s, 65°C for 20 s, 72°C for 80 s, 22 cycles. The PCR product was 500-fold diluted and re-amplified with A4 and A6 primers in the second nested PCR round (18 cycles at 94°C for 20 s, 68°C for 20 s, 72°C for 80 s).
Library construction and analysis
The subtracted PCR product obtained was cloned in E.coli DH5α using a TA-cloning system (Promega), and 288 individual clones were arrayed on 96-well microtiter plates. Clones were sequenced by the dye termination method using an ABI Prism 3100-Avant Genetic Analyzer automatic DNA sequencer. Homology searches against GenBank and mapping of the clones were done using the BLAST Web-server at NCBI (http://www.ncbi.nlm.nih.gov/BLAST) and the UCSC Human Genome Browser (http://genome.ucsc.edu/goldenPath/hgTracks.html).
The primers to verify Alu insertion polymorphism were designed against human genomic sequences surrounding each of Alu Ya5 or Ya8 integration sites. They were used for genomic PCR with the driver and 10 tracer DNA samples. Alu-containing PCR products for the polymorphic insertions represented in GenBank by only Alu-lacking homologs (see below) were cloned and sequenced. For each individual insertion, the PCR product selected for cloning was obtained by amplification of DNA from individuals homozygous for the Alu-containing allele. Otherwise, DNA of heterozygous individuals was amplified, and the longer PCR product was cloned. The clones were then tested for the length of inserts by PCR with standard M13 For/Rev primers, and the Alu-containing inserts were selected for sequencing.
RESULTS
Outline of the method
The technique is based on subtractive hybridization of whole-genome fractions of sequences flanking retroelement integration sites in the genomes to be compared. Recently, we successfully applied a similar approach to the identification of HERV-K LTR and L1 insertions distinguishing the human and chimpanzee genomes (7,8,21). In contrast to other known techniques, the present research is aimed at the identification of RE insertion polymorphisms in the human genome. The technique was adapted to reveal differences between individual human genomes and used in this study to identify Alu Ya5/Ya8 insertion polymorphisms.
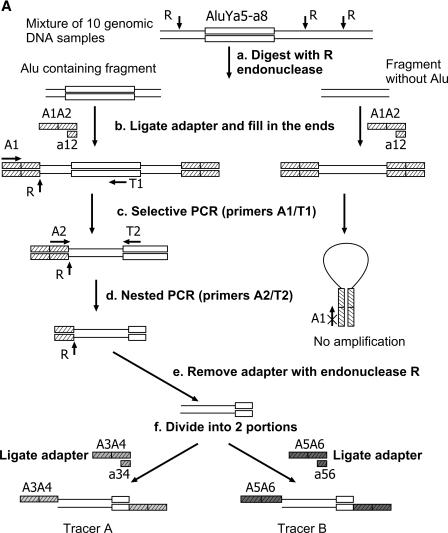
A principal distinction of our approach from that developed for interspecies comparison is that an individual human genome was compared to a mixture of ten other individual genomes. This makes it possible to detect even rare polymorphisms. The approach used includes two principal stages (outlined in Figure 1):
A whole-genome selective amplification of the flanks adjacent to Alu elements belonging to the young Ya5 and Ya8 branches in all the 11 genomic DNAs under comparison (Figure 1A). Ten of the genomes were mixed before the amplification and then used as tracer in the subsequent subtractive hybridization steps. The eleventh genome was amplified separately and then used as driver. For selective amplification, the genomes were first digested with a restriction endonuclease, and the restriction fragments were ligated to a terminal adapter. After this, the fragments with Alu inserts were selectively amplified using a universal primer against a conservative Alu sequence and a primer against the adapter attached.
A subtractive hybridization of the amplicons obtained (Figure 1B). This stage takes into account an unequal abundance of various Alu flanking sequences in the mixture of the amplified DNA fragments. This inequality is due to different frequencies of various Alu containing alleles in the human genome. As a result, the number of copies of particular flanks varies in the range from 0 for Alu-lacking loci to 2n× (haploid genome equivalents in the initial mixture of the ten genomes), where n is the number of PCR cycles used for the amplification of flanks of those Alu inserts present in the given locus of all the genomes used. This variation is supposed to result in the loss of rare and over-representation of frequent polymorphisms in the final complex amplicon. To avoid this effect, we used an equalizing DNA subtraction approach developed by us for cDNA subtraction (22) and utilized in a subtractive hybridization scheme (Figure 1B). The approach equalizes the abundance of various flanks in the mixture in the course of subtraction.
Figure 1.
(A) Tracer DNA preparation. Double lines denote genomic DNA. R, RsaI restriction sites; open box represents individual Alu Ya5 or Ya8 element. A1A2 (hatched boxes), the first oligonucleotide adapter ligated to the restricted DNA; A1/T1 and A2/T2, primers used in the first and second PCR rounds, respectively. A3A4 (gray boxes) and A5A6 (dark boxes), ligated adapters forming Tracer A and Tracer B, respectively. Note: Driver DNA was prepared from a single sample exactly in the same way as Tracer but omitting stage f. (B) Scheme of subtractive hybridization. Wavy lines depict flanking sequences of Alu present in one or more of 10 Tracer genomes and absent from Driver. Straight lines mark flanks of Alu common for Driver and Tracer. Open boxes represent the remainder of Alu Ya5 or Ya8 elements; A3/A5 and A4/A6, primers used in the first and second PCR rounds, respectively. Other designations are as in (A).
A more detailed description of the experiment is presented below.
Driver and tracer amplicons
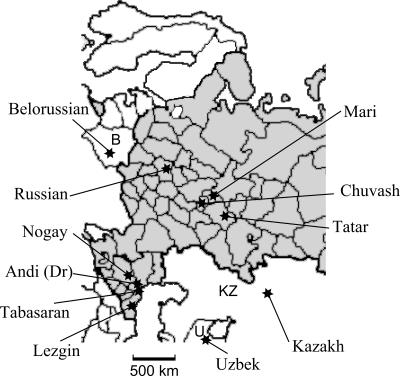
The tracer amplicon was derived from a mixture of genomic DNAs isolated from 10 human individuals different in their geographic (Figure 2), ethnic and language origin, as described in Materials and Methods. Mixing several genomes increases the probability of finding polymorphic insertions. The driver amplicon was prepared by selective amplification of the genomic DNA from an Andi male. The DNA of male was chosen to avoid enrichment in non-polymorphic Alu repeats located on Y chromosome.
Figure 2.
Geographic origin of populations used for individual DNAs sampling. Territory of Russian Federation is in gray and neighboring countries are white (B, Belarus; U, Uzbekistan; KZ, Kazakhstan). Scale is indicated at the bottom. Dr indicates that Andi genomic DNA sample was used as driver.
In order to improve the specificity of selective amplification of Alu containing fragments, we used the ‘PCR-suppression’ effect (PS-effect) (23). Briefly, it includes digestion of genomic DNAs (Figure 1A, stage a) with a frequent cutter restriction enzyme (RsaI in this case) and ligation of the DNA fragments to the A1A2 ‘pan-handle’ forming adapter (Figure 1A, stage b) followed by two steps of selective PCR. This procedure allows to obtain a specific set of genomic sequences flanking REs of interest (Alu in this case). At this stage, a pair of A1 primer specific to the adapter and T1 target primer corresponding to fragments of the Alu Ya5 and Ya8 consensus sequences was used (Figure 1A, stage c). These fragments included three diagnostic nucleotides of the Alu Y subfamily and two nucleotides characteristic of the Alu Ya5–Ya8 branch. Presumably, this branch includes about 2500 members in the human genome (4,12). The fragments obtained in this way contained rather long residual Alu sequences able to cross-hybridize during subtractive hybridization thus affecting the subsequent analysis. To avoid this undesirable effect, the residual sequences were shortened by means of repeated PCR with T2 primer targeted at the utmost 5′ end of Alu repeats and A2 primer against an internal part of A1A2 adapter (Figure 1A, stage d), the RsaI restriction site being preserved. Both the driver and tracer amplicons were treated with RsaI endonuclease to remove the remainder of A2 adapter (Figure 1A, stage e).
For the subtraction (Figure 1B), the tracer amplicon was divided into two portions (Figure 1A, stage f) each of which was ligated to one of two different ‘pan-handle’ forming adapters (A3A4 or A5A6, respectively). Then, each of the two samples in separate tubes was mixed with an approximately 50-fold excess of the driver DNA, denatured and incubated under hybridization conditions for 3 h (Figure 1B, stage a). This stage is an equalizing step during which highly abundant fractions of single-stranded (ss) flanks reassociate at a higher rate than low abundant fractions. As a result, with time the abundances of both the fractions in the ss form become approximately equal. Since the ss fragments also readily reassociate with their driver counterparts, the ss fraction becomes enriched with differential sequences distinguishing driver and tracer DNAs.
After this step, the contents of the two tubes were mixed together and allowed to reassociate (Figure 1B, stage b) after addition of a new driver portion. Here, only ss and not double-stranded (ds) fragments were capable of reassociation, and only ss fragments from different tubes reassociated with each other to form ds fragments with different termini. All the ds fragments of other origin possessed either two identical termini or one end lacking the adapter. Whereas the fragments with different termini could be further selectively PCR amplified (Figure 1B, stage d), PCR of the fragments with identical ends was suppressed due to the formation of pan-handle structures between terminal complementary sequences as illustrated in Figure 1A. Therefore, the resulting amplicon is supposed to be enriched with tracer-specific DNA fragments. We cloned and arrayed the selectively amplified fragments in three 96-well microtiter plates representing a library enriched with polymorphic Alu flanks (Figure 1B, stage e).
Analysis of the library
Some 125 randomly selected clones of the library were sequenced. All of them contained A4 and A6 oligo sequences at their ends. Five clones lacked a small (20 nt) 5′ part of Alu sequences, while the other 120 clones had this sequence adjacent to either A4 or A6 oligo: 12 of these 120 clone inserts were presented by 2 clones each, 5 by 3 clones, 2 by 4 clones and one sequence by 5 clones. Thus, 120 clones represented 88 individual sequences (Table 2).
Table 2.
The numbers of Alu inserts studied and proportions of polymorphic AluYa5 inserts
| Total | Alu inserts | ||
|---|---|---|---|
| Polymorphic | Fixed | ||
| Clones arrayed | 288 | ||
| Clones sequenced | 120 | 78 | 21 |
| The number of different Alu flanking sequences | 88 | ||
| Flanking sequences unambiguously mapped on the human genome | 77 | ||
| Alu insertions characterized | 46 | 21 | |
| Previously described | 8 | 5 | 3 |
| Tested by PCR in this work | 64a | 41 | 18 |
| The number of polymorphic Alu insertions present in databases | 23 | ||
| The number of polymorphic Alu insertions absent from databases | 18 | ||
aFive of 64 PCR assays resulted in uncertain results (see ‘Alu polymorphisms identification’ and notes to Table 3).
All these 88 sequences were mapped on the human genome using the NCBI human genome databases and the UCSC Genome Browser. Eleven sequences could not be unambiguously mapped due to their small size, presence of low-diverged repetitive elements or chimeras sequences whose parts are homologous to different genomic loci, and/or extended chromosomal duplications. Other 11 sequences corresponded to the loci of young Alu Yb8 (2 sequences), Alu Yc1 (1 sequence) and Alu Ya5 (8 sequences) described earlier (4,14,24,25). Five of 8 Alu Ya5 insertions (B65, HS2.25, Ya5NBC203, Ya5NBC327 and Ya5DP71) were previously reported to be polymorphic, and three insertions (HS4.21, Ya5NBC46 and Ya5NBC76) were found to be fixed in human populations (Table 2).
The remaining 66 of 88 sequences containing 5′ portion of Alu corresponded to human genomic loci known to contain Alu insertions or described as not containing any Alu Ya5 or Ya8 insertions. In the contradictory cases, we confirmed the presence of Alu repeats within the identified sites by direct sequencing (see below).
Alu polymorphisms identification
Sixty-four sequences were analyzed to confirm the Alu insertion polymorphism, and two Alu elements appeared to be integrated into clusters of frequent low-divergent repeats that made impossible reliable selective amplification of the corresponding genomic regions. Sixty-four pairs of PCR primers specific to both 5′ and 3′ flanking sequences of each Alu were designed and used for PCR assay of the 11 human genomic DNAs used as tracer and driver.
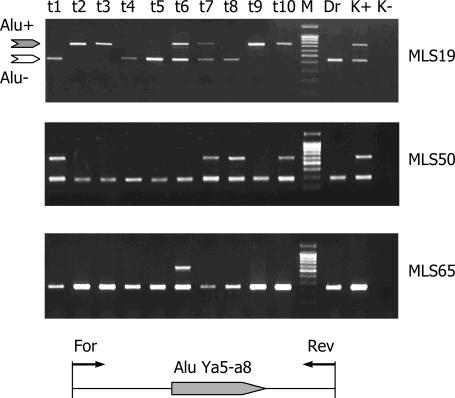
Examples of individual locus-specific PCRs are shown in Figure 3. A total of 41 Alu repeats were identified as polymorphic, while other 18 were found in all the genomes analyzed. It was technically impossible to unambiguously identify the rest five insertions as polymorphic (additional data are available in Supplementary Material).
Figure 3.
Examples of PCR amplification with three individual polymorphic Alu-containing loci (MLS 19, MLS 50 and MLS 65) in ten tracer (lines t1–t10) and one driver (line Dr) DNA samples. Lines K+ and K− represent positive and negative controls, respectively. M, DNA fragments of a 100 bp ladder length marker (SybEnzime). Gray and white arrows to the left of the electrophoregrams indicate the predicted locations of the Alu containing and Alu-free PCR products, respectively. Scheme of genomic primers location is represented at the bottom.
Frequencies of Alu positive alleles in various loci of the genomes used as tracer components varied from 1/20 to 18/20 (see Table 3 and Supplementary Material for details). As mentioned above (Table 2), 18 of 41 polymorphic Alu repeats were not found at the expected positions in the draft human genome sequence. To verify whether the PCR length polymorphism is actually due to Alu inserts, the corresponding PCR-products were cloned and sequenced (AY736285–AY736302). For 5 of these 18 polymorphic insertions Alu positive alleles were present in only one genome of the complex tracer, and all of them were heterozygous (see Table 3). For 2 of these 5 insertions (Ya5-MLS14 and Ya5-MLS20) Alu positive alleles were identified also in other non-tracer human DNA samples, used for optimization of PCR conditions (see Materials and Methods). The remaining three Alu repeats (Ya5-MLS51, Ya5-MLS58 and Ya5-MLS65) can be either polymorphic or represent rare de novo insertions.
Table 3.
Alu Ya5 genomic location, neighboring genes and polymorphism frequency
| Name | Accession | Chromosome | Neighboring genesb | Polymorphism frequencyc | |
|---|---|---|---|---|---|
| Alu+a | Alu− | ||||
| Ya5-MLS09 | AK056306 | AL162431 | 1q25.3 | 2k down XPR1 | 7/20 |
| Ya5-MLS33 | AL513546 | 1q21.3 | No genes | N/A | |
| Ya5-MLS51 | AY736296a | AL592148 | 1q41 | In AK096526 | 1/20 |
| Ya5-MLS58 | AY736298a | AL390117 | 1p12 | No genes | 1/20 |
| Ya5-MLS59 | AL356501 | AL365175 | 1p31.1 | No genes | 7/20 |
| Ya5-MLS60 | AL928921 | 1p36.22 | No genes | N/A | |
| Ya5-MLS48 | AC073577 | AC073046 | 2p13.1 | In ACTG2 | 14/20 |
| Ya5-MLS26 | AY736289a | AC099331 | 3p22.1 | In MYRIP | 9/20 |
| Ya5-MLS29 | AC011325 | 3q29 | No genes | 13/20 | |
| Ya5-MLS47 | AC024248 | 4q26 | No genes | 3/20 | |
| Ya5-MLS57 | AY736297a | AC010683 | 4q31.22 | No genes | 2/20 |
| Ya5-MLS05 | AC105756 | AC087107 | 4q34.3 | No genes | 5/20 |
| Ya5-MLS50 | AY736295a | AC115540 | 4q35.2 | No genes | 4/20 |
| Ya5-MLS04 | AC114316 | AC116332 | 5q14.3 | No genes | 10/20 |
| Ya5-MLS06 | AC020921 | AC007554 | 5q14.3 | No genes | 8/20 |
| Ya5-MLS31 | L43392 | 5q23.3 | In RAD50 | N/A | |
| Ya5-MLS44 | AY736294a | AL365508 | 6q22.31 | In c6orf170 | 2/18 |
| Ya5-MLS19 | AC010942 | 6q22.33 | In LAMA2 | 10/20 | |
| Ya5-MLS10 | AY736285a | AL121969 | 6p12.2 | No genes | 5/20 |
| Ya5-MLS14 | AY736286a | AC019066 | 7p12.3 | In HUS1, PKD1L1 | 1/20 |
| Ya5-MLS39 | AC005377 | 7q34 | No genes | 6/20 | |
| Ya5-MLS23 | AP003357 | AC012482 | 8q22.1 | 4k up LAPTM4B | 5/20 |
| Ya5-MLS41 | AP005354 | AC103853 | 8q23.1 | In ZFPM2 | 14/20 |
| Ya5-MLS37 | AY736292a | AC022389 | 10q23.1 | 4k down PCDH21 | 6/20 |
| Ya5-MLS28 | AY736290a | AL358033 | 10p13 | No genes | 2/20 |
| Ya5-MLS56 | AL139818 | 10p15.3 | No genes | N/A | |
| Ya5-MLS07 | AC025427 | 10q21.1 | No genes | 3/20 | |
| Ya5-MLS35 | AL731574 | 10q25.1 | No genes | 18/20 | |
| Ya5-MLS36 | AC090832 | 11p11.2 | In BHC80 | 11/16 | |
| Ya5-MLS18 | AC080183 | AC023869 | 11p14.3 | In AK127695 | 12/20 |
| Ya5-MLS45 | AC079363 | 12q21.31 | In PPFIA2 | N/A | |
| Ya5-MLS34 | AY736291a | AC009721 | 12q24 | In RPC2 | 6/18 |
| Ya5-MLS70 | AY736302a | AL159153 | 13q34 | In COL4A2 | 14/20 |
| Ya5-MLS12 | AL138681 | 13q12.3 | No genes | 14/20 | |
| Ya5-MLS46 | AL390722 | 13q14.3 | No genes | 14/20 | |
| Ya5-MLS69 | AY736301a | AL390964 | 13q21.1 | No genes | 4/20 |
| Ya5-MLS22 | AL161897 | AE014308 | 13q33.2 | No genes | 4/20 |
| Ya5-MLS68 | AY736300a | AL132990 | 14q32.13 | 3k up SERPINA 4 | 5/20 |
| Ya5-MLS65 | AY736299a | AL445883 | 14q13.1 | No genes | 1/20 |
| Ya5-MLS11 | AC010674 | 15q21.2 | In MYO5C | 5/20 | |
| Ya5-MLS21 | AC021958 | AC026583 | 15q23 | No genes | 9/20 |
| Ya5-MLS63 | AC130456 | AC003108 | 16p12.3 | In TMC5 | 3/18 |
| Ya8-MLS32 | AC139236 | AC140504 | 16p13.1 | No genes | 3/20 |
| Ya5-MLS16 | AY736287a | AC108483 | 16q23.2 | No genes | 3/16 |
| Ya5-MLS30 | AC016586 | 19p13.3 | 8k up EEF2 | 16/18 | |
| Ya5-MLS20 | AY736288a | AL135935 | 20p12.2 | In PAK7 | 1/20 |
| Ya5-MLS66 | AC023275 | 1q31.1 | In FIBL-6 | FP | |
| Ya5-MLS13 | AL590082 | 1p21.1 | No genes | FP | |
| Ya5-MLS62 | AC079300 | 2p16.3 | No genes | FP | |
| Ya5-MLS49 | AC108068 | 2q32.3 | No genes | FP | |
| Ya5-MLS52 | AC016903 | 2q33.3 | No genes | FP | |
| Ya5-MLS24 | AC019130 | 2q37.1 | In MGC42174 | FP | |
| Ya5-MLS61 | AC099339 | 4q31.3 | In ARFIP1 | FP | |
| Ya5-MLS55 | AC094098 | 5q33.2 | No genes | FP | |
| Ya5-MLS25 | AL139093 | 6p22.3 | No genes | FP | |
| Ya5-MLS38 | AC006006 | 7q34 | In BRAF | FP | |
| Ya5-MLS15 | AL355592 | 9q33.1 | No genes | FP | |
| Ya5-MLS53 | AC069540 | 10q23.1 | No genes | FP | |
| Ya5-MLS42 | AC022878 | 11p15.3 | No genes | FP | |
| Ya5-MLS43 | AP001782 | 11q24.1 | No genes | FP | |
| Ya5-MLS01 | AC023795 | 12q12 | In CPNE8 | FP | |
| Ya5-MLS27 | AC078860 | 12q21.1 | 2k down GPR49 | FP | |
| Ya5-MLS67 | AC007432 | 17q24.3 | No genes | FP | |
| Ya5-MLS54 | AL031657 | 20q11.23 | 4k up PPP1R16B | FP | |
aFor Alu repeats that were sequenced in this work for the first time accession numbers obtained in this work are given.
bNeighboring genes were detected by Human Genome Browser under RefSeq Genes. Up, Alu is N kb upstream gene; down, Alu is N kb downstream gene; in, Alu is into intron of gene; no genes, no neighboring genes were detected within 10 kb of Alu.
cFor polymorphic Alu repeats fraction of Alu-positive haplotypes to the total haplotypes tested for tracer DNA. Driver DNA for polymorphic insertions is always Alu negative. FP (fixed present), all the DNA samples tested were Alu-positive; N/A, PCR products were not detected for driver DNA sample, or the samples were Alu negative, or no specific PCR products detected for driver and tracer DNAs.
DISCUSSION
As mentioned above, RE polymorphisms are very useful markers for human population studies and medical genetics. However, most known RE polymorphisms were predicted by in silico analysis of human genomic sequences. Although this approach has opened a great opportunity for identification of a great number of polymorphic REs, the published human genome sequences probably lack a significant portion of polymorphic RE insertions [also suggested in (15)] because they represent only a few human genomes and do not cover all genetic variations in humans.
We have developed a technique that enables direct experimental isolation of sequences flanking polymorphic RE insertions. The technique allows one to specifically analyze selected subfamilies of certain REs. A high specificity of the technique was confirmed in this study. Here, the technique was used to detect polymorphic insertions of the youngest Alu Ya5 and Ya8 subfamilies' members. Although Ya5 is an abundant Alu subfamily that includes about 2500 members as compared to several tens of Ya8, there are other young subfamilies practically of similar abundance. For example, Yb8 and Yc1 have quite comparable copy numbers (about 2000 and 400–500, respectively). Nevertheless, only 3 (2.5%) of 120 sequenced Alu-containing clones were found to be flanks of Alu repeats other than the members of the Alu Ya branches. The achieved selectivity of amplification thus seems to be very high and close to 97.5%.
The efficiency of the method with regard to detection of polymorphic insertions seems to be also rather high. Only 21 of 120 sequenced clones (17.5%) were flanks of Alu repeats fixed in the populations, which means about 82.5% ‘efficiency’ of the technique. According to results of the PCR assay, 78 sequenced clones contain genomic fragments adjacent to Alu repeats that are polymorphic. Some of these 78 clones were redundant and corresponded to 46 independent individual sequences. Five of these sequences were flanks of Alu repeats earlier characterized as insertionally polymorphic (for the numbers see Table 2). All the remaining 41 identified polymorphic Alu elements belonged to the Alu Ya5 subfamily except one Alu Ya8 element. It is important that 18 (45%) of them originated from Alu containing alleles located in loci known only before our study as Alu-lacking.
The technique permits one to analyze at least 10 different individual DNAs at once (more than the number of completely sequenced genomes available in databases) and identify RE insertions absent from one selected genome and present in at least one of other genomes. The sensitivity of the technique is sufficient to isolate insertions present in only one genome and in a heterozygous state. Since 60–80% of library was represented by polymorphic RE flanks, the use of subtractive hybridization highly increased the efficiency of the subsequent PCR screening. One of major advantages of the technique is the possibility to identify polymorphic Alu repeats not available in human genome databases. The technique allows detecting polymorphisms in wide range of their frequencies in population. Frequencies observed in this study varied in the range of 5–90% (Table 3).
We have also added some increments to the most recently published (12) quantitative data on the distribution of polymorphic Alu Ya insertions among human chromosomes (Table 4). For example, for chromosome 15 we added two new polymorphisms to four reported earlier. Similar increase was found for chromosome 16.
Table 4.
Chromosomal distribution of polymorphic Alu Ya5 and Ya8 repeats found in this work compared to the distribution of other polymorphic REs detected by in silico approach
| Chromosome | Polymorphic (other works)a | Polymorphic (this work)b | Total polymorphic |
|---|---|---|---|
| 1 | 24 | 4 | 28 |
| 2 | 29 | 1(1) | 29 |
| 3 | 18 | 2 | 20 |
| 4 | 20 | 4(1) | 23 |
| 5 | 18 | 2(1) | 19 |
| 6 | 27 | 3(1) | 29 |
| 7 | 37 | 2 | 39 |
| 8 | 10 | 2 | 12 |
| 9 | 6 | 0 | 6 |
| 10 | 13 | 4(1) | 16 |
| 11 | 12 | 2 | 14 |
| 12 | 12 | 1 | 13 |
| 13 | 18 | 5(2) | 21 |
| 14 | 17 | 2 | 19 |
| 15 | 4 | 2 | 6 |
| 16 | 7 | 3(1) | 9 |
| 17 | 10 | 0 | 10 |
| 18 | 7 | 0 | 7 |
| 19 | 5 | 1 | 6 |
| 20 | 12 | 1 | 13 |
| 21 | 4 | 0 | 4 |
| 22 | 3 | 0 | 3 |
| Total | 313 | 41 | 346 |
It is difficult to precisely estimate the number of polymorphic insertions absent from one certain genome and present in at least one of other 10 genomes. The total number of Alu Ya5 family members was evaluated to be about 2500, and ∼25% of them are polymorphic in human populations (4,12). Thus, the total number of polymorphic Alu repeats that can be found in the human genome databases is about 625. Here, we showed that 45% (18 of 41) of the identified polymorphic Alu insertions were lacking from the human genome databases, so the total number of Alu polymorphic insertions can be estimated as high as over 900 in populations of different origin.
The developed technique can be successfully applied to comprehensive searches for polymorphic Alu insertions of other subfamilies, as well as polymorphisms resulted from retroposition of other retroelements. Other possible applications of the technique are identification of specific RE insertion polymorphisms that distinguish human subpopulations or groups and identification of RE polymorphisms presumably associated with hereditary multigene diseases. Moreover, the technique can be easily adapted for using microarrays and to other advanced high throughput formats. Finally, it should be emphasized that the technique can be also successfully used for searching polymorphic RE insertions in genomes with unknown or partially known sequences, e.g. those of various primate species.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We would like to express our gratitude to Dr B. Glotov for helpful and constructive discussion and help in manuscript preparation. We also thank Dr E. Khusnutdinova for providing DNA samples and Drs V. Potapov and N. Skaptsova for oligonucleotides synthesis. This work was supported by INTAS-2001-0759, the Russian Foundation for Basic Research 02-04-48614 and 2006.20034 grants, and the Molecular and Cellular Biology Program of the Presidium of the Russian Academy of Sciences. The Open Access publication charges for this article were waived by Oxford University Press.
REFERENCES
- 1.van de Lagemaat L.N., Landry J.R., Mager D.L., Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003;19:530–536. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Kazazian H.H., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 3.Hughes J.F., Coffin J.M. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc. Natl Acad. Sci. USA. 2004;101:1668–1672. doi: 10.1073/pnas.0307885100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll M.L., Roy-Engel A.M., Nguyen S.V., Salem A.H., Vogel E., Vincent B., Myers J., Ahmad Z., Nguyen L., Sammarco M., et al. Large-scale analysis of the Alu Ya5 and Yb8 subfamilies and their contribution to human genomic diversity. J. Mol. Biol. 2001;311:17–40. doi: 10.1006/jmbi.2001.4847. [DOI] [PubMed] [Google Scholar]
- 5.Turner G., Barbulescu M., Su M., Jensen-Seaman M.I., Kidd K.K., Lenz J. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 2001;11:1531–1535. doi: 10.1016/s0960-9822(01)00455-9. [DOI] [PubMed] [Google Scholar]
- 6.Roy-Engel A.M., Carroll M.L., Vogel E., Garber R.K., Nguyen S.V., Salem A.H., Batzer M.A., Deininger P.L. Alu insertion polymorphisms for the study of human genomic diversity. Genetics. 2001;159:279–290. doi: 10.1093/genetics/159.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buzdin A., Khodosevich K., Mamedov I., Vinogradova T., Lebedev Y., Hunsmann G., Sverdlov E. A technique for genome-wide identification of differences in the interspersed repeats integrations between closely related genomes and its application to detection of human-specific integrations of HERV-K LTRs. Genomics. 2002;79:413–422. doi: 10.1006/geno.2002.6705. [DOI] [PubMed] [Google Scholar]
- 8.Mamedov I., Batrak A., Buzdin A., Arzumanyan E., Lebedev Y., Sverdlov E.D. Genome-wide comparison of differences in the integration sites of interspersed repeats between closely related genomes. Nucleic Acids Res. 2002;30:e71. doi: 10.1093/nar/gnf071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boissinot S., Chevret P., Furano A.V. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol. Biol. Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- 10.Myers J.S., Vincent B.J., Udall H., Watkins W.S., Morrish T.A., Kilroy G.E., Swergold G.D., Henke J., Henke L., Moran J.V., et al. A comprehensive analysis of recently integrated human Ta L1 elements. Am. J. Hum. Genet. 2002;71:312–326. doi: 10.1086/341718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mamedov I., Lebedev Y., Hunsmann G., Khusnutdinova E., Sverdlov E. A rare event of insertion polymorphism of a HERV-K LTR in the human genome. Genomics. 2004;84:596–599. doi: 10.1016/j.ygeno.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Otieno A.C., Carter A.B., Hedges D.J., Walker J.A., Ray D.A., Garber R.K., Anders B.A., Stoilova N., Laborde M.E., Fowlkes J.D., et al. Analysis of the Human Alu Ya-lineage. J. Mol. Biol. 2004;342:109–118. doi: 10.1016/j.jmb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 13.Carter A.B., Salem A.-H., Hedges D.J., Nguyen Keegan C., Kimball B., Walker J.A., Watkins W.S., Jorde L.B., Batzer M.A. Genome wide analysis of the human Alu Yb lineage. Human Genomics. 2004;1:167–178. doi: 10.1186/1479-7364-1-3-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Callinan P.A., Hedges D.J., Salem A.H., Xing J., Walker J.A., Garber R.K., Watkins W.S., Bamshad M.J., Jorde L.B., Batzer M.A. Comprehensive analysis of Alu-associated diversity on the human sex chromosomes. Gene. 2003;317:103–110. doi: 10.1016/s0378-1119(03)00662-0. [DOI] [PubMed] [Google Scholar]
- 15.Boissinot S., Entezam A., Young L., Munson P.J., Furano A.V. The insertional history of an active family of L1 retrotransposons in humans. Genome Res. 2004;14:1221–1231. doi: 10.1101/gr.2326704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Badge R.M., Alisch R.S., Moran J.V. ATLAS: a system to selectively identify human-specific L1 insertions. Am. J. Hum. Genet. 2003;72:823–838. doi: 10.1086/373939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheen F.M., Sherry S.T., Risch G.M., Robichaux M., Nasidze I., Stoneking M., Batzer M.A., Swergold G.D. Reading between the LINEs: human genomic variation induced by LINE-1 retrotransposition. Genome Res. 2000;10:1496–1508. doi: 10.1101/gr.149400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lavrentieva I., Broude N.E., Lebedev Y., Gottesman I.I., Lukyanov S.A., Smith C.L., Sverdlov E.D. High polymorphism level of genomic sequences flanking insertion sites of human endogenous retroviral long terminal repeats. FEBS Lett. 1999;443:341–347. doi: 10.1016/s0014-5793(99)00004-6. [DOI] [PubMed] [Google Scholar]
- 19.Tambets K., Rootsi S., Kivisild T., Help H., Serk P., Loogvali E.L., Tolk H.V., Reidla M., Metspalu E., Pliss L., et al. The western and eastern roots of the Saami—the story of genetic ‘outliers’ told by mitochondrial DNA and Y chromosomes. Am. J. Hum. Genet. 2004;74:661–682. doi: 10.1086/383203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lebedev Y.B., Belonovitch O.S., Zybrova N.V., Khil P.P., Kurdyukov S.G., Vinogradova T.V., Hunsmann G., Sverdlov E.D. Differences in HERV-K LTR insertions in orthologous loci of humans and great apes. Gene. 2000;247:265–277. doi: 10.1016/s0378-1119(00)00062-7. [DOI] [PubMed] [Google Scholar]
- 21.Buzdin A., Ustyugova S., Gogvadze E., Lebedev Y., Hunsmann G., Sverdlov E. Genome-wide targeted search for human specific and polymorphic L1 integrations. Hum. Genet. 2003;25:25. doi: 10.1007/s00439-002-0904-2. [DOI] [PubMed] [Google Scholar]
- 22.Gurskaya N.G., Diatchenko L., Chenchik A., Siebert P.D., Khaspekov G.L., Lukyanov K.A., Vagner L.L., Ermolaeva O.D., Lukyanov S.A., Sverdlov E.D. Equalizing cDNA subtraction based on selective suppression of polymerase chain reaction: cloning of Jurkat cell transcripts induced by phytohemaglutinin and phorbol 12-myristate 13-acetate. Anal. Biochem. 1996;240:90–97. doi: 10.1006/abio.1996.0334. [DOI] [PubMed] [Google Scholar]
- 23.Diatchenko L., Lau Y.F., Campbell A.P., Chenchik A., Moqadam F., Huang B., Lukyanov S., Lukyanov K., Gurskaya N., Sverdlov E.D., et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl Acad. Sci. USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arcot S.S., Adamson A.W., Lamerdin J.E., Kanagy B., Deininger P.L., Carrano A.V., Batzer M.A. Alu fossil relics—distribution and insertion polymorphism. Genome Res. 1996;6:1084–1092. doi: 10.1101/gr.6.11.1084. [DOI] [PubMed] [Google Scholar]
- 25.Arcot S.S., DeAngelis M.M., Sherry S.T., Adamson A.W., Lamerdin J.E., Deininger P.L., Carrano A.V., Batzer M.A. Identification and characterization of two polymorphic Ya5 Alu repeats. Mutat. Res. 1997;382:5–11. doi: 10.1016/s1383-5726(97)00002-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.