Abstract
MicroRNAs (miRNAs) play important regulatory roles in animals and plants by targeting mRNAs for cleavage or translational repression. They have diverse expression patterns and might regulate various developmental and physiological processes. Profiling miRNA expression is very helpful for studying biological functions of miRNAs. We report a novel miRNA profiling microarray, in which miRNAs were directly labeled at the 3′ terminus with biotin and hybridized with complementary oligo-DNA probes immobilized on glass slides, and subsequently detected by measuring fluorescence of quantum dots labeled with streptavidin bound to miRNAs through streptavidin–biotin interaction. The detection limit of this microarray for miRNA was ∼0.4 fmol, and the detection dynamic range spanned about 2 orders of magnitude. We made a model microarray to profile 11 miRNAs from leaf and root of rice (Oryza sativa L. ssp. indica) seedlings. The analysis results of the miRNAs had a good reproducibility and were consistent with the northern blot result. To avoid using high-cost detection equipment, colorimetric detection, a method based on nanogold probe coupled with silver enhancement, was also successfully introduced into miRNA profiling microarray detection.
INTRODUCTION
MicroRNAs (miRNAs) are a highly evolutionarily conserved class of small noncoding RNAs that can play important regulatory roles in animals and plants by targeting mRNAs for cleavage or translational repression (1,2). They are ∼22 nt transcripts cleaved from ∼70 nt hairpin precursors in animals or ∼100 nt in plants with signature 3′-hydroxyl and 5′-monophosphate termini of an RNase III cleavage event that leaves a 2 nt overhang. They have diverse expression patterns and might regulate various developmental and physiological processes. Misregulation of miRNA function might contribute to human disease (2). So, profiling miRNA expression is very important in studying the biological functions of miRNAs.
Large-scale studies on miRNA expression profiles were carried out in many model organisms by using northern blot analysis and miRNA cloning (3–8). Both of these methods are time consuming. DNA microarray technology has resulted in profiling expression of thousands of genes simultaneously. Applying similar technology would make miRNA profiling more efficient. To profile expression patterns of neuronal miRNAs, Krichevsky et al. designed an oligonucleotide array on charged nylon membranes and detected the miRNAs by labeling filtered low molecular weight RNA with radioactive isotopes (9). To avoid the need of large amounts of total RNA and using radioactive isotopes as in the above methods, Liu et al. developed an oligonucleotide microchip on glass slides to profile miRNA by detecting the 5′ biotin-labeled cDNAs of miRNAs introduced through a random primer (10,11). However, this labeling method may be ineffective because of the inability for 8 nt random primer to bind on such short templates as ∼22 nt miRNAs. Recently, Miska et al. developed a microarray for miRNA expression analysis, in which miRNAs were ligated to 3′ and 5′ adaptor oligonucleotides followed by reverse transcription and amplified by PCR with Cy3-labeled primer to label the sense strand of PCR products (12). Yet, miRNA population may be distorted by hybridization with cDNAs or sense strands of PCR product of miRNAs because of enzymatic reverse transcription and/or amplification (13). To direct-label RNA, Babak et al. used a platinum-based chemical labeling reagent for nucleic acids (Ulysis Alexa Fluor from Molecular Probes) (14). However, this reagent cannot label miRNAs lacking G residuals, and labeling of residuals in miRNA may interfere the following hybridization. The best target for miRNA profiling microarray hybridization should be fluorescently labeled miRNAs directly at either 3′ adjacent hydroxyl or 5′-monophosphate terminus. One method to direct-label RNA at 3′ terminus is by using T4 RNA ligase to couple the 3′ end of RNA to a fluorescent-modified ribodinucleotide (13,15). This method has been successfully used to investigate the role of small noncoding RNAs by directly labeling total RNA and hybridizing the target to whole-genome querying microarrays (16). Recently, Thomson et al. used this method in direct-labeling of miRNAs on a microarray platform for analysis of miRNA gene expression (17). However, this method needs labeled donor ribodinucleotide and RNA ligase that have a poor reputation for reliability and differential ligation efficiency toward the acceptor nucleotide on the miRNA.
We have developed a method for small RNA sequencing by direct fluorescence labeling in which 3′ adjacent hydroxyl of RNA were periodate-oxidized into a dialdehyde followed by conjugation with fluorescein-5-thiosemicarbazide through a condensation reaction, resulting in that trace amounts (<10 fmol) of RNA, which can be detected on sequencing PAGE by measuring fluorescence (18). Quantum dots (QDs) are a new type of fluorescence probe with important advantages over classical organic dyes (19,20). In particular, QDs have high extinction coefficient and high quantum yield, which should dramatically increase the sensitivity for microarray detection in theory. QDs have been successfully conjugated to DNA and used in many applications (21,22). Therefore, it was thought that the direct-labeling of miRNA with QDs could be well used in miRNA detection and applied in microarray. Here, we report a novel miRNA profiling microarray in which miRNA targets were biotinylated at 3′ terminus, hybridized with corresponding complementary oligo-DNA probes immobilized on glass slides, then detected by measuring fluorescence of QDs labeled on miRNA. Analysis of a model microarray indicated that the detection limit for miRNA was ∼0.4 fmol and detection dynamic range spanned about 2 orders of magnitude, from 156 to 20 000 pM. In addition, miRNAs from leaf and root of rice (Oryza sativa L. ssp. indica) seedlings were profiled using a model microarray for 11 miRNAs in rice. On the other hand, a low-cost colorimetric detection method based on nanogold probe labeling coupled with silver enhancement demonstrated equal detection ability as fluorescent dyes in DNA and protein microarray (23–26). In this method, a flat scanner for DNA microarray or a commercial digital camera for protein chip were used as detector, resulting in great decrease in cost. Therefore, this method was also introduced into miRNA profiling microarray detection.
MATERIALS AND METHODS
Principle of the miRNA profiling microarray
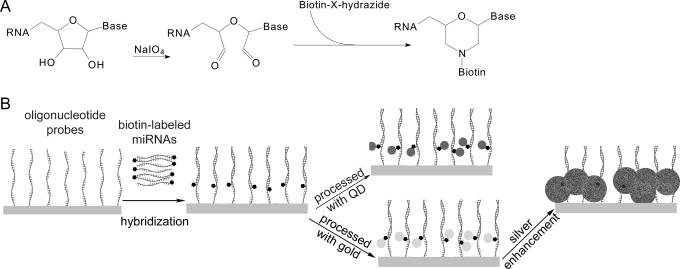
The principle of miRNA profiling microarray includes two parts (Figure 1). First, miRNAs were oxidized with sodium periodate to convert 3′ terminal adjacent hydroxyl groups (2′ and 3′ position of ribose) into dialdehyde, which was then reacted with biotin-X-hydrazide through a condensation reaction resulting in biotinylated miRNA. Second, 5′ amine-modified oligonucleotide probes antisense to miRNAs were immobilized on amine-reactive glass slides. Then the biotinylated miRNAs were captured on the microarray by oligonucleotide probes in hybridization. Quantum dots were labeled on the captured miRNAs through the strong specific interaction of streptavidin and biotin. QDs have a high extinction coefficient and a high quantum yield, so trace amounts of miRNAs are easily detected with a laser confocal scanner. As an alternative, the colorimetric gold–silver detection method was used: captured miRNAs were labeled with streptavidin-conjugated gold followed by silver enhancement. During silver enhancement, the gold nanoparticles bound to miRNAs catalyzed the reduction of silver ions to metallic silver, which further autocatalyzed the reduction of silver ions to form metallic silver precipitation on gold, resulting in a signal enhancement (27). This process allowed straightforward detection of the microarray with an ordinary charge-coupled device (CCD) camera mounted on a microscope.
Figure 1.
Schematic principles of the miRNA profiling microarray. (A) Principle of labeling miRNA at the 3′ terminus with biotin. (B) Principle of the miRNA profiling microarray detected with QD or colorimetric method.
Microarray fabrication
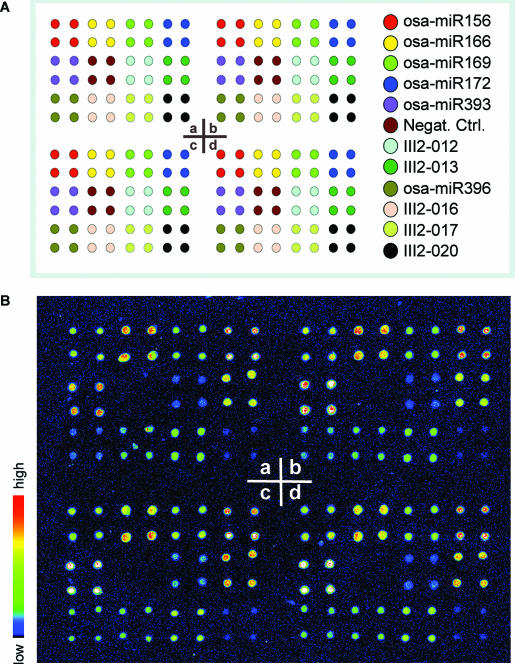
Standard 1″ × 3″ microscope glass slides from Sigma were activated with glycidyloxipropyltrimethoxysilane (GOPTS) as previously described (26). The activated glass slides immobilize amine-containing molecules such as amino-modified oligonucleotide DNA. Eleven rice miRNAs were selected for the model miRNA profiling microarrays. Five of the 11 miRNAs were recently cloned in our lab (Li, Y., Li, W., Zhao, B.T. et al., in preparation); the remaining six were from the miRNA Registry (28). The specific oligonucleotide DNA probes for these miRNAs were designed as shown in Table 1, along with a negative control RNA, all purchased from Beijing SBS Genetech Co. (China). Each DNA probe was complementary to a corresponding full length of mature miRNA and contained 10 deoxyadenosines in the 5′ terminus to minimize the spatial obstacle in hybridization and detection. The probes were dissolved in 50 mM phosphate buffer (pH 8.0, with 10 mM EDTA) at 50 μM concentration, then printed onto the GOPTS-activated slides with PixSys 5500 spotting robot (Cartesian Technology), in which ArrayIt SMP3 spotting pin from TeleChem was used; humidity was maintained at 80%. The diameter of spots was about 80 μm, and the distance between spots was set as 0.2 mm. The pattern of the model microarrays are shown in Figure 3A. Each microarray contained four subarray replicates (a, b, c and d), and each subarray contained 48 spots of the 12 probes in quadruplicate. The fabricated microarrays were processed with 1% BSA in phosphate-buffered saline (PBS) containing 10 mM EDTA to block intact amine-reactive groups on slides before hybridization. To examine the detection limit and dynamic range of the miRNA profiling microarrays, a synthesized 21 nt single-strand siRNA (5′-GATAATGGACCCCAATCAAAC-3′) was used as a model miRNA. Similarly, an amino-modified oligonucleotide DNA probe complementary to this siRNA with 10 deoxyadenosines in the 5′ terminus was synthesized and printed pentaplicately onto activated glass slides at 50 μM as mentioned above.
Table 1.
Oligonucleotide probes in the model miRNA profiling microarray
| miRNAs | Oligonucleotide probes |
|---|---|
| osa-miR156 | amino-5′-(A)10GTGCTCACTCTCTTCTGTCA-3′ |
| osa-miR166 | amino-5′-(A)10GGGGAATGAAGCCTGGTCCGA-3′ |
| osa-miR169 | amino-5′-(A)10TCGGCAAGTCATCCTTGGCTG-3′ |
| osa-miR172 | amino-5′-(A)10TGCAGCATCATCAAGATTCT-3′ |
| osa-miR393 | amino-5′-(A)10GATCAATGCGATCCCTTTGGA-3′ |
| osa-miR396 | amino-5′-(A)10CAGTTCAAGAAAGCCTGTGGA-3′ |
| III2-012a | amino-5′-(A)10TCCCCTCTCCCACAACCCCG-3′ |
| III2-013a | amino-5′-(A)10GCCAGGGAAGAGGCAGTGCAG-3′ |
| III2-016a | amino-5′-(A)10GTTGCATCTGCCTCTGCAC-3′ |
| III2-017a | amino-5′-(A)10GTAGGTGCAGGTGCAAATGCA-3′ |
| III2-020a | amino-5′-(A)10CTACCATCTGAGCTACATCCCC-3′ |
| Negaive Ctrl. | amino-5′-(A)10GTGTGTGTGTGTGTGTGTGTGT-3′ |
amiRNAs recently cloned from rice by Li, Y., Li, W., Zhao, B.T., Yao, C.G., Qin, W.M, and Jin, Y.X. (in preparation) without official names assigned yet.
Figure 3.
Model miRNA profiling microarray for 11 rice miRNAs. (A) Pattern of the model miRNA profiling microarray for 11 rice miRNAs. The microarray contains four replicate subarrays (a, b, c and d). Each subarray contains spots of probes for 11 miRNAs and 1 negative control in quadriplicate. (B) Image of the model miRNAs profiling microarray detected with QD. The microarray was hybridized with biotinylated miRNAs from rice seedling leaves.
miRNA labeling
Total RNA was extracted from leaf and root of liquid nitrogen-frozen rice seedlings by using TRIZOL reagent (Invitrogen) according to the manufacturer's instructions. miRNAs were enriched from total RNA (termed enriched miRNA) according to a protocol from Drs Natalie Doetsch and Richard Jorgensen (29).
The enriched miRNAs were labeled with biotin-X-hydrazide (Sigma) according to a protocol described previously and modified slightly (18). Briefly, 18 μl of 0.5 μg/μl enriched miRNAs from 90 μg total RNA was diluted with 9 μl labeling buffer (0.25 M sodium acetate, pH 5.6) and 4 μl doubly distilled water (DEPC treated), then 5 μl of 5 mM sodium periodate was added. The oxidization of the 3′ terminus of RNA was carried out at 25°C in the dark for 90 min. Then, 2-fold excess of sodium sulphite over sodium periodate was added to the reaction mixture followed by 15 min incubation at 25°C. Finally, 37.5 nmol of biotin-X-hydrazide was added and incubated at 37°C for 3 h. The biotinylated miRNAs were precipitated with ethanol and stored in −80°C. The labeling of 21 nt siRNA was performed in the same way.
Microarray hybridization and detection
Each miRNA profiling microarray was hybridized with 1.5 μg biotinylated miRNAs in 10 μl formamide prehybridization/hybridization solution at 37°C overnight. The hybridized microarray was washed with 1× SSC/0.5% SDS at 37°C for 10 min. For detection with QD, the microarray was incubated with 10 μl of 2 nM Qdot 655 streptavidin conjugate (QD-streptavidin, from Quantum Dot Corp.) for 1 h at room temperature. After a thorough washing, the microarray was scanned on a PerkinElmer ScanArray 5000 Scanner with the laser 1 (633 nm) and filter 8, power at 100%, photomultiplier at 80%, and a scan resolution of 5 μm. For simplification, this QD-based detection method is termed the detection with QD in this report. For detection with the colorimetric gold–silver method, the microarray was incubated with 10 μl of 0.5 OD520 streptavidin-conjugated gold (gold-streptavidin, from Sigma). After a thorough washing, the microarray analysis was then performed with silver enhancer kit (Sigma) for 20 min, and detected with a commercial CCD camera (Olympus C-4000Z digital camera) mounted on a microscope. This method is termed the colorimetric method.
Northern blot
Total RNA from rice seedling leaves and roots were loaded on a 12.5% denaturing polyacrylamide gel. The resolved RNA was transferred to a Zeta-Probe GT blotting membrane (Bio-Rad) overnight. Oligodeoxynucleotides labeled at the 5′ end with [γ-32P]ATP were used as probes. Prehybridization and hybridization were carried out using ExpressHyb Hybridization Solution (Clontech) according to the manufacturer's instruction. The sequences of probes were same as shown in Table 1, unless the 5′ amino and 10 deoxyadenosines were removed.
Data analysis
Images of miRNA microarray obtained with QD detection were quantified by QuantArray software (PerkinElmer). Meanwhile, grey-scale images of miRNA microarray obtained in the colorimetric method were processed with Photoshop 7.0 (Adobe System) to map the lightest and darkest pixels into black and white before quantifying with QuantArray software. Signal intensities of each spot in images obtained by both detection methods were calculated by subtracting local background from total intensities of the spot.
Radioactivity contained in northern blot bands was quantified with the NIH ImageJ software, and was expressed as a ratio of background subtracted signal in the band of hybridization to background subtracted signal in the band of stained 5S rRNA as loading control.
RESULTS
Detection limit and dynamic range of the miRNA microarrays in QD detection
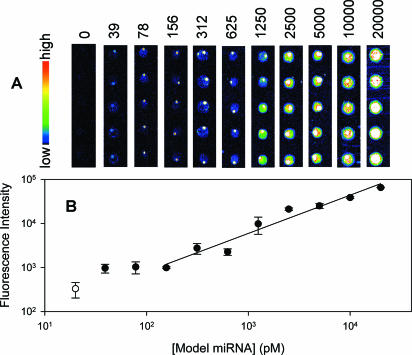
Figure 2 shows a set of images for various concentrations of miRNA (21 nt siRNA) detected by QD. The signals become gradually weaker with the decrease in miRNA concentration. When the miRNA concentration was as low as 39 pM, the fluorescence signal could be detected, indicating that the lower detection limit of miRNA microarrays is at least 0.4 fmol. As shown in Figure 2B, the fluorescence intensity of the spots is linear to the model miRNA in logarithm from 156 to 20 000 pM, and the dynamic range is about 2 orders of magnitude, which implies that this method can be used to quantify miRNAs with broad concentration range. In Miska's method in which signal amplification with 10 rounds of PCR was employed, the lower detection limit of input miRNA was 0.1 fmol and the dynamic range was also 2 orders of magnitude (from 0.1 to 10 fmol) (12). While in quantitative northern blot, the lower detection limit of miRNA was 1 fmol and dynamic range was about 3 orders of magnitude, due to the high capacity of membrane in which the miRNA was immobilized (7).
Figure 2.
Detection limit and dynamic range of the model miRNA detection microarray. (A) Image sets of microarrays hybridize with various concentrations of miRNAs from 20 nM to 39 pM and the backgound. The 50 μM concentration of oligonucleotide probes printed on slides pentaplicately. The volume of model miRNA needed to hybridize with microarray was 10 μl. (B) Correlation between fluorescence intensity of spots and concentrations of model miRNA. The values were calculated from image in (A). Open circle represents the background.
Model miRNA profiling microarray for 11 rice miRNAs
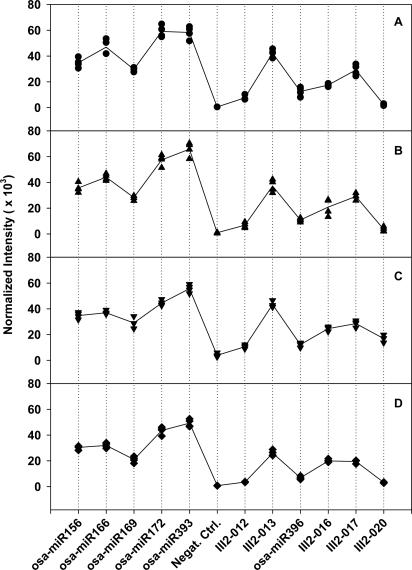
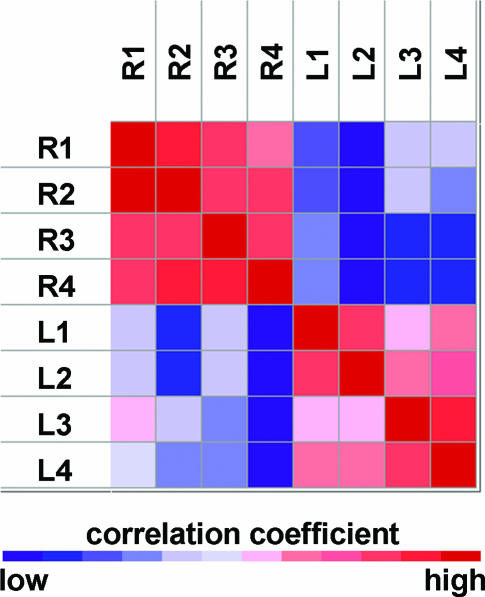
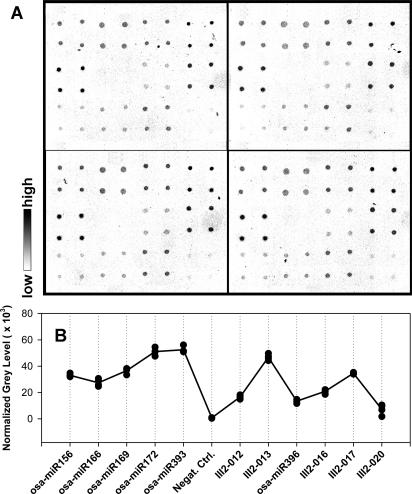
When the microarray was applied to profile the 11 miRNAs in rice seedling leaves with QD detection, the image clearly showed different fluorescence intensities corresponding to these miRNAs, while the negative control was almost zero (see Figure 3B). This indicated that rice seedling leaves do contain these miRNAs. Figure 4 shows the reproducibility of the modeling microarrays for the 11 miRNA profiling. The Panels A and B were for the reproducibility among four subarrays in a microarray and four micrarrays (slides), respectively. The correlation coefficients obtained in Panel A were 0.98, 0.96 and 0.96, showing a high reproducibility between these subarray replicates. The correlation coefficients in Panel B were very similar, indicating high reproducibility between microarrays. Furthermore, when a two-cluster self-organization map was used, GeneCluster software (30) automatically clustered the eight samples shown in Figure 4B and C into two classes based on the expression profile of 11 miRNAs, one cluster contained the exact same four samples from leaf and the other four from root (Figure 5). These results indicate that our miRNA profiling microarray is reliable and sensitive. Although 1.5 μg of enriched miRNAs were mostly used for hybridization in this work giving a good result (see Figures 3B and 4A–C), 0.2 μg of enriched miRNAs was also tested and the normalized fluorescence intensities of the 11 miRNA in leaves are shown in Figure 4D. A correlation coefficient of 0.99 between the average intensity values of corresponding miRNAs in Figure 4B (1.5 μg miRNAs) and Figure 4D (0.2 μg miRNAs) was obtained, indicating that as low as 0.2 μg enriched miRNAs can be used. As known, at least 20 μg of total RNA was ordinarily used for each northern blot and the corresponding enriched miRNAs were about 2 μg. Consequently, about 10% of total RNA used in northern blots would be enough for one miRNA profile microarray experiment.
Figure 4.
Reproducibility of miRNA profiling microarray detected with QD. (A) Mean normalized intensities of four subarray replicates in the microarray shown in Figure 3B. Straight-line plot represents the mean normalized intensities of four subarray replicates for each miRNA. The signal intensities of each four-spot replicates were normalized by using a per-subarray normalize to total method, which allowed comparison among subarrays in a chip. (B) Mean normalized intensities of four microarrays for 11 miRNA from seedling leaves. Straight-line plot represents the mean normalized intensities of the four microarrays for each miRNA. As in (A), a per-chip normalize to total method was used in comparison among chips. (C) As in (B), except that the miRNA was from seedling roots. (D) As in (A), except that 0.2 μg instead of 1.5 μg of enriched miRNA was used for hybridization.
Figure 5.
Cluster of samples from leaves and roots of rice seedling based on the expression profile of 11 miRNAs. Correlation coefficients between every two samples are presented. L1 to L4 and R1 to R4 represent four independent assays to profiling miRNAs from leaves and roots of rice seedlings respectively.
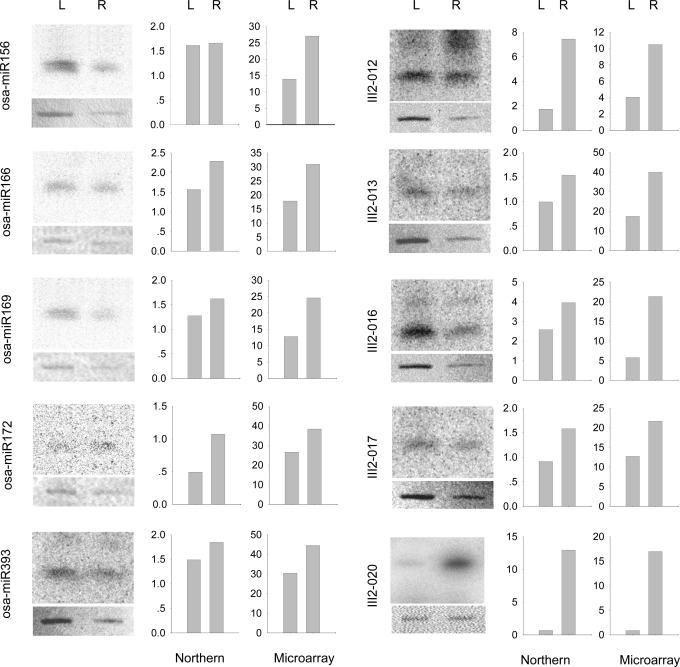
As the hybridization efficiency between different miRNA with its DNA probe is different, it is hard to estimate the molecular abundance of each miRNA according to the corresponding fluorescence intensities in microarray. However, for one miRNA, its fluorescence intensity do quantitatively relate to its amount as shown in Figure 2, therefore the miRNA microarray can be used to profile the same miRNA in different samples, for instance, to profile the 11 miRNAs in roots and leaves of the rice. The changes in fluorescence intensities of each miRNA should reflect the relative changes of each miRNA expression in these two samples. Figure 6 shows the fluorescence intensities of the 10 miRNAs in roots and leaves of rice seedling, indicating that the expression of the 10 miRNAs in root were higher than that in leaves. Among them, the III2-020 miRNA was almost undetectable in seedling leaves, but was strongly expressed in seedling roots, suggesting us to further explore it in future. These results were validated with northern blots. From Figure 6, it can be seen that most results from microarray and northern blots showed a similar pattern, indicating that the miRNA microarray described above could offer a high-throughput method that generally captures changes in miRNA expression. Only for osa-miR156, the relative levels of expression of osa-miR156 in root and leaf obtained from the microarray differed from that of the northern blot. As is the case for mRNAs, small differences may be seen between these methods, and northern blot analysis is superior to microarrays for quantitative analysis (31). This result reminds us to take care in using the results from microarrays.
Figure 6.
Comparison of microarray data with northern blots of miRNAs from leaves and roots of rice seedlings. Left panel for the five miRNAs selected from miRNA Registry and right panel for the five miRNAs cloned in our laboratory. The y-axis for the microarray data refers to the averaged mean fluorescence intensities, and y-axis for northern blots data to radioactivity. 5S rRNA immobilized on membrane was stained as control for comparison.
Detection with colorimetric gold–silver method
Microarray hybridized with biotin-labeled miRNAs from rice seedling leaves was also detected by gold-streptavidin with silver enhancement. Figure 7A shows the image taken from the colorimetric-detected microarray. Different miRNAs show different grey levels, while the negative control is indistinguishable from the background. Figure 7B shows the quantitative analysis of the colorimetric detection, in which the grey level profile is very similar to that seen in Figure 4A. This means that the colorimetric detection has a similar detection sensitivity as that of the QDs method. The correlation coefficient between the fluorescence detection and colorimetric detection was 0.93, a reasonable value for two different detection methods. Reproducibility between subarrays was also evaluated. The correlation coefficients between the subarray replicates were 0.97, 0.99 and 0.98, showing the high reproducibility between subarrays in the microarray. Alexander et al. (24) proved that when using the colorimetric method to detect multibiotinylated target DNA with DNA microarray, the lower detection limit was 0.1 fmol and dynamic range was from 0.1 to 10 fmol. These results indicate that colorimetric gold–silver detection can be used in miRNA profiling microarray detection in a low-cost and efficient manner.
Figure 7.
Detection of microarray for 11 rice miRNAs from seedling leaves with colorimetric gold–silver method. (A) Image of the model miRNAs profiling microarray detected with colorimetric method. (B) Quantitative analysis of the microarray image in panel (A). Filled circles represent the mean normalized intensities of the four spots for each miRNA in the four subarrays shown in (A). Straight-line plot represents the mean normalized intensities of the four subarray replicates for each miRNA.
DISCUSSION
Although northern blot can effectively profile the expression of a miRNA in many different conditions simultaneously, it is inefficient in profiling the expression of hundreds of miRNAs. The reason is that in northern blots, the miRNA mixture is immobilized on the membrane and hybridized with one certain probe. Such a characteristic also makes it inconvenient to evaluate molecular amounts of various miRNAs. To the contrary, in miRNA profiling microarray, a number of known probes are immobilized in addressable spots and hybridized with corresponding miRNAs in the sample, thereby providing a parallel and high throughput method of detecting thousands of miRNAs simultaneously. The linear dependence of fluorescence intensities on miRNA concentration in 2 orders of magnitude described above indicates that the microarrays can be used effectively to describe the miRNA expression profile. The results that the expression of the 11 miRNA in root were mostly higher than that in leaves, which is in agreement with the results obtained from northern blot experiment provided a solid evidence to prove that this microarray can be used to profile miRNA. Of course, this microarray cannot be used to compare the relative expression of different miRNAs at the moment as mentioned above, but there is no doubt that the microarray can be used to compare the expression of the same miRNAs in different sample or in different physiological condition for a same sample. And we believe that the microarrays could reveal the molecular abundance of different miRNAs after correcting the binding efficiency of each miRNA with its DNA probe. Until recently, a total of 1345 miRNAs from 12 species have been deposited in the miRNA Registry (Release 5.0); 207 miRNAs have been identified in human and 125 in rice. A bioinformatic study has suggested that there exist 200–255 miRNAs in human (32). Our miRNA profiling microarray can be easily expanded to profile thousands of features of miRNAs. Also it should not be difficult to make a universal microarray for several species. This universal microarray could be used in trans-species miRNA expression profiling for each known miRNA under various conditions.
Sensitivity to detect target miRNA is a very important parameter for miRNA profiling microarrays. We found that only 0.4 fmol of miRNA was needed for QD detection with microarrays when a 633 nm laser was used as excitation light. In fact, we speculate that the detection sensitivity could easily be raised, using a 488 nm laser. The extinction coefficient of QD 655 at 488 nm is 2.9 × 106 cm−1 M−1, about 4-fold higher than that at 633 nm (0.85 × 106 cm−1 M−1). So, for the same concentration of miRNA, the fluorescent signal excited at 488 nm would be about 4-fold higher than that excited at 633 nm. Even with the 633 nm excitation, we would expect that the detection limit would be lower than 0.4 fmol because the signal obtained at 633 nm excitation (Figure 2) would be 2-fold higher than the minimum readable level of the laser confocal scanner. Therefore, one can expect that the detection limit can reach as low as 0.05–0.1 fmol of miRNA when a 488 nm laser is used. Such a high sensitivity is essential to detect trace amounts of miRNAs. Molecular amounts of miRNAs have been estimated to be 1000–50 000 molecules per cell (7). For the lowest amount of miRNAs (i.e. 1000 miRNA molecules per cell), only 6 × 104 cells would be required to detect these miRNAs with a detection limit of 0.1 fmol, and only 2.5 × 105 cells with a detection limit of 0.4 fmol. This translates to only 5 mg of tissue or one well of a 24-well plate, which makes high throughput assay feasible.
Based on quantum dot or colorimetric method, miRNA is measured by detecting fluorescence or grey-level of labeled miRNA, in which miRNAs were captured by corresponding antisense oligonucleotide probes. This method has at least three advantages. First, fluorescence coming from directly labeled miRNA can accurately reveal relative miRNA amounts in a sample, whereas the miRNA population might be distorted with labeling cDNA of miRNA through reverse transcription and/or enzymatic amplification (13). Second, preparation of the sample and the procedure of microarray hybridization and detection are relatively simple. Third, the miRNA profiling microarrays can be used to evaluate amounts of both the miRNAs and their targets. For the latter, the miRNA targets should be reverse-transcribed into cDNA. These advantages of miRNA profiling microarray will make miRNome deciphering more efficient and will contribute much to the studies of miRNA target identification, miRNA expression regulation and even pathological studies of diseases.
Acknowledgments
We thank Dr David Armbruster for his critical reading. This work was funded by National Key Basic Research and Development Program (No. 2002CB713802), National 863 Program (030431002), National Natural Science Foundation of China (No. 30430210), and a key basic research grant (No. 04DZ14006) from the Shanghai Council of Science and Technology. The Open Access publication charges for this article were waived by Oxford University Press.
REFERENCES
- 1.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nature Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 3.Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 4.Sempere L.F., Sokol N.S., Dubrovsky E.B., Berger E.M., Ambros V. Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev. Biol. 2003;259:9–18. doi: 10.1016/s0012-1606(03)00208-2. [DOI] [PubMed] [Google Scholar]
- 5.Houbaviy H.B., Murray M.F., Sharp P.A. Embryonic stem cell-specific MicroRNAs. Dev. Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 6.Aravin A.A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- 7.Lim L.P., Lau N.C., Weinstein E.G., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sempere L.F., Freemantle S., Pitha-Rowe I., Moss E., Dmitrovsky E., Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krichevsky A.M., King K.S., Donahue C.P., Khrapko K., Kosik K.S. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA. 2003;9:1274–1281. doi: 10.1261/rna.5980303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C.G., Calin G.A., Meloon B., Gamliel N., Sevignani C., Ferracin M., Dumitru C.D., Shimizu M., Zupo S., Dono M., et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl Acad. Sci. USA. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calin G.A., Liu C.G., Sevignani C., Ferracin M., Felli N., Dumitru C.D., Shimizu M., Cimmino A., Zupo S., Dono M., et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl Acad. Sci. USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miska E.A., Alvarez-Saavedra E., Townsend M., Yoshii A., Sestan N., Rakic P., Constantine-Paton M., Horvitz H.R. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole K., Truong V., Barone D., McGall G. Direct labeling of RNA with multiple biotins allows sensitive expression profiling of acute leukemia class predictor genes. Nucleic Acids Res. 2004;32:e86. doi: 10.1093/nar/gnh085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Babak T., Zhang W., Morris Q., Blencowe B.J., Hughes T.R. Probing microRNAs with microarrays: tissue specificity and functional inference. RNA. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Igloi G.L. Nonradioactive labeling of RNA. Anal. Biochem. 1996;233:124–129. doi: 10.1006/abio.1996.0016. [DOI] [PubMed] [Google Scholar]
- 16.Kampa D., Cheng J., Kapranov P., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., et al. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomson J.M., Parker J., Perou C.M., Hammond S.M. A custom microarray platform for analysis of microRNA gene expression. Nature Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 18.Wu T.P., Ruan K.C., Liu W.Y. A fluorescence-labeling method for sequencing small RNA on polyacrylamide gel. Nucleic Acids Res. 1996;24:3472–3473. doi: 10.1093/nar/24.17.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan W.C., Maxwell D.J., Gao X., Bailey R.E., Han M., Nie S. Luminescent quantum dots for multiplexed biological detection and imaging. Curr. Opin. Biotechnol. 2002;13:40–46. doi: 10.1016/s0958-1669(02)00282-3. [DOI] [PubMed] [Google Scholar]
- 20.Tan C.Y., Liang R.Q., Ruan K.C. Application of quantum dot to life science. Acta Biochim. Biophys. Sin. 2002;34:1–5. [PubMed] [Google Scholar]
- 21.Kim J.H., Morikis D., Ozkan M. Adaptation of inorganic quantum dots for stable molecular beacons. Sens. Actuators B: Chem. 2004;102:315–319. [Google Scholar]
- 22.Patolsky F., Gill R., Weizmann Y., Mokari T., Banin U., Willner I. Lighting-up the dynamics of telomerization and DNA replication by CdSe–ZnS quantum dots. J. Am. Chem. Soc. 2003;125:13918–13919. doi: 10.1021/ja035848c. [DOI] [PubMed] [Google Scholar]
- 23.Taton T.A., Mirkin C.A., Letsinger R.L. Scanometric DNA array detection with nanoparticle probes. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 24.Alexandre I., Hamels S., Dufour S., Collet J., Zammatteo N., Longueville F.D., Gala J.-L., Remacle J. Colorimetric silver detection of DNA microarrays. Anal. Biochem. 2001;295:1–8. doi: 10.1006/abio.2001.5176. [DOI] [PubMed] [Google Scholar]
- 25.Huber M., Wei T.F., Muller U.R., Lefebvre P.A., Marla S.S., Bao Y.P. Gold nanoparticle probe-based gene expression analysis with unamplified total human RNA. Nucleic Acids Res. 2004;32:e137. doi: 10.1093/nar/gnh133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang R.Q., Tan C.Y., Ruan K.C. Colorimetric detection of protein microarrays based on nanogold probe coupled with silver enhancement. J. Immunol. Methods. 2004;285:157–163. doi: 10.1016/j.jim.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 27.Holgate C.S., Jackson P., Cowen P.N., Bird C.C. Immunogold–silver staining: new method of immunostaining with enhanced sensitivity. J. Histochem. Cytochem. 1983;31:938–944. doi: 10.1177/31.7.6189883. [DOI] [PubMed] [Google Scholar]
- 28.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park W., Li J., Song R., Messing J., Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taniguchi M., Miura K., Iwao H., Yamanaka S. Quantitative assessment of DNA microarrays—comparison with Northern blot analyses. Genomics. 2001;71:34–39. doi: 10.1006/geno.2000.6427. [DOI] [PubMed] [Google Scholar]
- 31.Reich M., Ohm K., Angelo M., Tamayo P., Mesirov J.P. GeneCluster 2.0: an advanced toolset for bioarray analysis. Bioinformatics. 2004;20:1797–1798. doi: 10.1093/bioinformatics/bth138. [DOI] [PubMed] [Google Scholar]
- 32.Lim L.P., Glasner M.E., Yekta S., Burge C.B., Bartel D.P. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]