Abstract
We describe a new method for the assay of sequence-specific DNA-binding proteins in this paper. In this method, the sensitive fluorescence resonance energy transfer (FRET) technology is combined with the common DNA footprinting assay in order to develop a simple, rapid and high-throughput approach for quantitatively detecting the sequence-specific DNA-binding proteins. We named this method as exonuclease III (ExoIII) protection assay with FRET probe. The FRET probe used in this assay was a duplex DNA which was designed to contain one FRET pair in the center and two flanking protein-binding sites. During protein detection, if a target protein exists, it will bind to the two protein-binding sites of the FRET probe and thus protect the FRET pair from ExoIII digestion, resulting in high FRET. However, if the target protein does not exist, the FRET pair on the naked FRET probe will be degraded by ExoIII, resulting in low FRET. Three kinds of recombinant transcription factors including NF-κB, SP1 and p50, and the target protein of NF-κB in HeLa cell nuclear extracts, were successfully detected by the assay. This assay can be extensively used in biomedical research targeted at DNA-binding proteins.
INTRODUCTION
The sequence-specific DNA-binding proteins play critical roles in many cellular processes, including gene transcription regulation (1), DNA replication (2), recombination (3), repair and restriction (4). Among the various sequence-specific DNA-binding proteins, those that are directly involved in the regulation of gene transcription, e.g. DNA-binding transcription factors, attract increasing interest because they play pivotal roles in the pathways and networks of gene expression regulation, and become potential targets in medical diagnosis and drug development (5). For example, NF-κB (6), a transcription factor that is involved in the regulation of a large number of genes and closely related to multiple diseases, has already become an important and popular target for drug development (7). The research and development of sequence-specific DNA-binding proteins attracts fast-growing attention in genomics, proteomics and biomedicine. Therefore, there is a need for robust methods to detect the presence of these proteins and monitor their DNA-binding activities. However, the most common methods for detecting the sequence-specific DNA-binding proteins including gel-shift assays (8) and DNA footprinting assays (9,10) are laborious, radioactive and time-consuming, which hinder them from more extensive applications. In addition, both assays depend on gel electrophoresis, making them unadaptable to high-throughput technology.
Fluorescence-based methods have been exploited for the detection of sequence-specific DNA-binding proteins (11–14). Fluorescence resonance energy transfer (FRET) was found to be a powerful technique for the detection of biological interactions (13,14). The FRET technique describes the transfer of excitation energy from a donor fluorophore to an acceptor chromophore when the two dye molecules are separated by <100 Å and an overlap occurs between the donor emission and acceptor absorption spectra (15). Two strategies using the FRET technology have already been successfully applied to the detection of DNA–protein interactions (13,14). In the first strategy, the DNA is labeled with one fluorochrome and the protein with another fluorochrome. FRET was produced due to the proximity between the DNA and the protein in the protein–DNA complex (13). In the other strategy, known as the molecular beacon assay, the DNA is labeled with two fluorochromes, each appearing in one duplex DNA of the two half sites of a DNA-binding protein. FRET was produced due to the protein-driven annealing of the two half sites (14). However, the complicated procedures in labeling the protein with fluorescence or in designing the proper half-sited DNA molecular beacon may suppress their applications. Therefore, there is still a need for the development of new methods based on the superiority of FRET for detecting the sequence-specific DNA-binding proteins and monitoring their DNA-binding activities.
We have been working on developing double-stranded DNA microarray-based methods for this purpose (16). However, we found that it was difficult to analyze DNA-binding proteins in sensitive and high-throughput format using the double-stranded DNA microarray, although it was helpful in studying DNA-binding activities of a protein to multiple DNA targets (17). Based on our previous studies using molecular beacons for detecting DNA mutations (18), we introduced the sensitive FRET technology to the common exonuclease III (ExoIII) footprinting assay to develop a new, general, inexpensive FRET-based approach for assaying sequence-specific DNA-binding proteins. This method allows simple, rapid and high-throughput detection of the sequence-specific DNA-binding proteins, and can be extensively used in biomedical research targeted at DNA-binding proteins.
MATERIALS AND METHODS
FRET probes
Oligonucleotides were synthesized using the standard phosphoramidate chemistry and were purified by HPLC (BIOASIA, Shanghai). The following oligonucleotides were synthesized for the NF-κB FRET probe (NF-κB probe): 5′-AGTTGAGGGGACTTTCCCAACTAGGAAFTCTACCTGGGGACTTTCCCAGGC-3′ and 3′-TCAACTC CCCTGAAAGGGTTGATCCTDAAGATGGACCCCTGAAAGGGTCCG-5′ (F, dT-FAM; D, dT-Dabcyl). The oligonucleotides used for the SP1 FRET probe (SP1 probe) were: 5′-ATTCGATCGGGGCGGGGCGAAAGCGA AFTCGATTCTCGGGGCGGGGCGTCG-3′ and 3′-TAAGCTAGCCCCGCCCCGCTTTCGCTDAAGCTA AGAGCCCCGCCCCGCAGC-5′ (F, dT-FAM; D, dT-Dabcyl). The oligonucleotides used for the p53 FRET probe (p53 probe) were: 5′-GTCACAGACATGCCTAGACATGCCTTAACTAGGAAF/JTCTACCTCAGACATGCCTAGA CATGCCTTGCAG-3′ and 3′-CAGTGTCTGTACGGATCTGTACGGAATTGATCCTDAAGATGGAGTCTGTACGGATCTGTACGGAACGTC-5′ (F, dT-FAM; J, dT-JOE; D, dT-Dabcyl). The italic sequence represents the protein-binding sites. FAM is 6-carboxyfluorescein with a maximum excitation wavelength of 494 nm and a maximum emission wavelength of 520 nm; JOE is 6-carboxy-4′,5′-dichloro-2′,7′-dimethoxyfluorescein with a maximum excitation wavelength of 520 nm and a maximum emission wavelength of 548 nm; Dabcyl is 4-(4′-dimethylaminophenylazo) benzoic acid with an absorbance wavelength of 453 nm and no fluorescence emission. To obtain FRET probes of duplex DNA, FAM or JOE-labeled donor oligonucleotides and Dabcyl-labeled acceptor oligonucleotides were mixed in the same molar ratios at the final concentration of 50 μM in 100 μl of 10 mM Tris–HCl (pH 8.0), 100 mM NaCl and 1 mM EDTA. The mixture was heated for 5 min at 95°C and cooled slowly to 25°C. The following oligonucleotides were prepared to be used as cold NF-κB specific competition probe: 5′-AGTTGAGGGGACTTTCCCAGGCTTTTT-3′ and 3′-TTTTTTCAACTC CCCTGAAAGGGTCCG-5′. The oligonucleotides used as cold NF-κB nonspecific competition probe were: 5′-AGTTGAGATTACTTTCACCAGGCTTTTT-3′ and 3′-TTTTTTCAACTCTAATGAAAGTGTCCG-5′. The competitor DNAs were synthesized with five protruding bases at each 3′ end for protection from ExoIII digestion. The duplex competitor DNAs were prepared in a manner similar to the FRET probes.
DNA-binding proteins
Purified NF-κB p50 (rhNF-κB p50) and SP1 (rhSP1) were purchased from Promega (Madison, WI). Human recombinant NF-κB p50 was expressed in bacteria from a human cDNA derived from the full-length p105. p50, a processed product of its p105 precursor, is described as the first DNA-binding subunit of the NF-κB transcription factor. The SP1 recombinant protein was expressed in a human baculovirus cDNA clone in Sf9 cells. Purified p53 was purchased from ProteinOne (College Park, MD). The wild-type p53 (393 amino acids) was expressed in a baculovirus system and purified by an affinity column in combination with fast performance liquid chromatography. The TNF-α-induced HeLa cell nuclear extract was prepared using a Nuclear Extract Kit (ActiveMotif, Carlsbad, CA). The HeLa cell was cultivated and incubated with or without TNF-α (R&D System, Minneapolis, MN; 20 ng/ml) for 30 min. The nuclear extract was collected according to the manufacturer's instructions and the protein content was determined using a Bradford-based assay. The activity of NF-κB p50 in the TNF-α-induced HeLa cell nuclear extract was confirmed by electrophoretic mobility shift assay (EMSA) and super shift assay with a DIG-labeled oligonucleotide (5′-AGTTGAGGGGACTTTCCCAGGC-3′) (upper strand) and NF-κB (p50) antibody (Abcam, Cambridge, UK) using a DIG Gel Shift Kit (Roche Molecular Biochemicals, Mannheim, Germany). EMSA was performed by incubating 10 μg of the HeLa cell nuclear extract in a 9 μl binding reaction mixture containing 10 mM Tris–HCl (pH 7.5), 50 mM NaCl, 0.5 mM EDTA, 0.5 mM DTT, 3 mM MgCl2, 0.05 mg/ml poly (dI-dC) and 10% (v/v) glycerol at 37°C for 10 min. The binding reaction mixture for the super shift assay containing 1 μl of the non-diluted antibody of NF-κB (p50) (Abcam, Cambridge, UK) was added to 1 μl of DIG-labeled double-stranded oligonucleotide and was incubated at 37°C for 20 min, followed by the addition of 1 μl of the gel loading 10× buffer (250 mM Tris–HCl, pH 7.5, 40% glycerol) at room temperature. This mixture was then loaded on a pre-run 4% polyacrylamide gel and run at 10°C in 0.5× TBE buffer at 350 V until the bromophenol blue dye was three-fourth of the way down the gel. After electrophoresis, the DIG-labeled DNA was electroblotted on the positively-charged nylon membrane (Roche Molecular Biochemicals, Mannheim, Germany). The blotted nylon membrane was fixed by baking at 120°C for 15 min and the chemiluminescent detection was performed according to the manufacturer's instructions.
Fluorescence spectroscopy
All fluorescence measurements were carried out at 25°C in 200 μl quartz cuvettes using an LS 55 spectrofluorometer (PerkinElmer, Boston, MA). Spectra for single-color and two-color detection experiments with purified recombinant proteins were obtained using the DNA-binding buffer containing 10 mM Tris–HCl (pH 7.5), 50 mM NaCl, 0.5 mM EDTA, 0.5 mM DTT, 3 mM MgCl2, 0.05 mg/ml poly (dI-dC) (Roche, Basel, Switzerland), 10% v/v glycerol, 0.5 mg/ml BSA and 0.05% NP-40. All proteins used in this experiment were pre-incubated with DNA-binding buffer at 37°C for 10 min. The FRET probe was then added and incubated at the same temperature for further 20 min. At the end of the DNA–protein-binding reaction, a sufficient amount of ExoIII was added and kept for 5min at 37°C. About 200 U of ExoIII (MBI Fermentas, Hanover, MD) was used for all the FRET probes of 25 nM concentration. EDTA was added to a final concentration of 20 mM to terminate the ExoIII reaction, and the reaction mixture was immediately subjected to fluorescence measurement.
For single-color detection of the target transcription factor NF-κB in the HeLa cell nuclear extract, the nuclear extract stimulated with or without TNF-α was pre-incubated in the binding buffer containing 10 mM Tris–HCl (pH 7.5), 50 mM NaCl, 0.5 mM EDTA, 0.5 mM DTT, 3 mM MgCl2, 0.05 mg/ml poly (dI-dC), 10% v/v glycerol, 2 mM sodium phosphate, pH 7.0, 20 ng/μl HaeIII-cut E.coli DNA and 25 ng/μl of yeast tRNA at 37°C for 10 min. The NF-κB probe was then added and incubated at the same temperature for a further 20 min. After the binding step, ExoIII was added and the probe was allowed to digest at 37°C for 5 min. The reaction was terminated by the addition of EDTA to a final concentration of 20 mM. The assays of nuclear extract included an additional ExoIII-negative control with nuclear extract, which was used to detect any destruction of the FRET probe by endogenous nuclease. The reaction mixture was immediately subjected to fluorescence measurement.
FRET analysis
In the FRET analysis, the following formula was used: Relative FRET = −(Fe − Fs)/(Fe − Fn), where Fe was the fluorescence signal intensity of the free FRET probe digested by ExoIII, Fs was the fluorescence signal intensity of the FRET probe bound by protein and digested by ExoIII, and Fn was the fluorescence signal intensity of the FRET probe not digested by ExoIII. Fe was subtracted from Fs as background. As a result, the relative FRET signal of the ExoIII-negative control (Fn), the ExoIII-positive control (Fe) and the detected samples (Fs) was 1, 0, and 0–1, respectively.
RESULTS
Mechanism
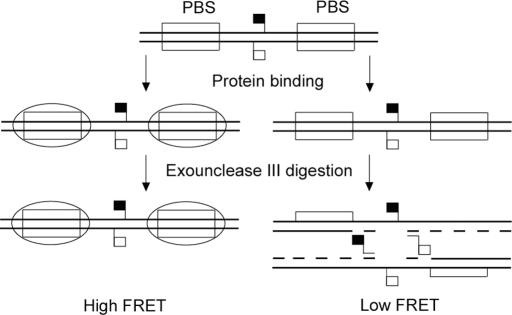
Figure 1 illustrates the general design of the FRET probe and the mechanism of detection of the sequence-specific DNA-binding proteins by our assay. The linear duplex DNA FRET probe consists of two reverse complementary oligonucleotides, one was labeled with a fluorochrome as donor and the other was labeled with a fluorochrome as acceptor. The donor and acceptor fluorochromes were brought into close proximity by the annealing of the two oligonucleotides, resulting in a high FRET probe signal. For protein detection, the FRET probe harbors one complete protein-binding site at both sides of the FRET pair. In essence, the detection of DNA-binding proteins with the FRET probe is a special ExoIII protection assay. In this assay, if a target protein is present, the protein will bind to two protein-binding sites of the FRET probe and produce a physical hindrance to ExoIII, which protects the FRET pair from digestion by ExoIII and results in a high FRET signal. On the contrary, in the absence of the target protein, the naked FRET probe will be degraded by ExoIII and the close proximity of the FRET pair is thus destroyed, resulting in a low FRET signal.
Figure 1.
Schemes of ExoIII protection assay with the FRET probe. PBS, protein-binding sites of the FRET probe. The solid and open flags on the FRET probe represent the fluorescence donor and acceptor. The ellipse represents DNA-binding protein.
It should be noted that the following aspects are important in the preparation of the FRET probe. First, there should be enough flanking sequences on both sides of the two protein-binding sites, as the DNA sequence covered by protein in the DNA–protein complex is often larger than the protein-binding site. Second, the FRET pair should be sufficiently far away from both the protein-binding sites, lest it results in steric hindrance to protein binding. Third, ExoIII preferentially binds to a free 3′ hydroxyl on the double-stranded DNA and then cleaves inward in a semiprocessive fashion. The FRET probe should therefore have a recessed or blunt 3′ hydroxyl group at both ends. The protruded 3′ end larger than four bases is not permitted in the FRET probe.
Feasibility
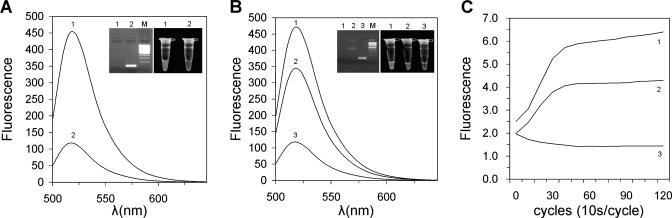
To verify our assay (Figure 1) experimentally, we first prepared a FRET probe for detecting NF-κB, a transcription factor that binds to the consensus DNA of GGGACTTTCC in a sequence-specific manner (6). The NF-κB probe consisted of two reverse complementary oligonucleotides. One oligonucleotide was labeled with FAM as donor and the other was labeled with Dabcyl as acceptor. The FAM and Dabcyl were introduced into oligonucleotides using dT-fluorescein and dT-Dabcyl. Two complete 10 bp NF-κB-binding sites of GGGACTTTCC were contained in the NF-κB probe, which flanked the FRET pair of FAM/Dabcyl. A 10 and 8 bp sequence was, respectively, stuffed between the two NF-κB-binding sites and a FRET pair. The FAM and Dabcyl were brought into close proximity by the annealing of two oligonucleotides to obtain a high FRET signal (Figure 2A, curve 2). Once ExoIII was added into the NF-κB probe solution, dT-FAM and dT-Dabcyl were released as mononucleotides from the NF-κB probe by ExoIII digestion. The close proximity of the FRET pair of FAM/Dabcyl was thus destroyed, resulting in a low FRET signal of the NF-κB probe (Figure 2A, curve 1). The ExoIII digestion of the NF-κB probe was also confirmed by gel electrophoresis and UV transilluminator imaging (inset of Figure 2A).
Figure 2.
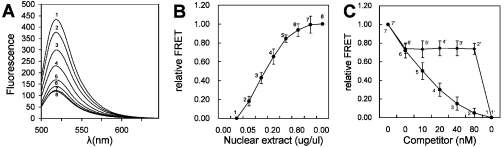
ExoIII digestion of the FRET probe. (A) Emission spectra of free NF-κB probe digested by ExoIII. Curve 1, the 25 nM NF-κB probe digested by ExoIII (ExoIII-positive control); Curve 2, the 25 nM NF-κB probe not digested by ExoIII (ExoIII-negative control). (B) Emission spectra of p50-bound NF-κB probe digested by ExoIII. Insets are gel electrophoresis and UV transilluminator images. (C) Dynamics of ExoIII digestion of NF-κB probes. In (B) and (C), curve 1, the ExoIII-positive control of 25 nM NF-κB probe; curve 2, the 25 nM NF-κB probe bound by 15 nM p50 and digested by ExoIII; curve 3, the ExoIII-negative control of the 25 nM NF-κB probe.
Although the half-life of the protein–DNA complex is longer than the time required for the ExoIII reaction, ExoIII can wait until the protein falls off its site and passes through it in footprinting (19). To avoid producing a false negative result, we assumed complete ExoIII digestion of the NF-κB probe in short time to eliminate the ExoIII activity. We investigated the efficiency of ExoIII digestion of the NF-κB probe by performing a real-time PCR. In the experiment, the NF-κB probe solution was taken in the PCR tube, followed by addition of the ExoIII solution in the lids of PCR tubes. Under a simple PCR program of incubation at 37°C and detection at 470 nm on RotorGene 2000 (Corbett Research, Australia), the ExoIII solutions were centrifuged into the NF-κB probe solutions, and the dynamics of ExoIII digestion of the NF-κB probes were therefore recorded (Figure 2C). It was demonstrated that the ExoIII digestion was completed in 5 min at the proper ratio of DNA and FRET probe. Before performing other experiments, the ratio of DNA to ExoIII was optimized by carrying out a real-time PCR and gel electrophoresis.
Based on the principle of DNA footprinting with ExoIII, we designed our assay for the detection of DNA-binding proteins. We assumed that the binding of target protein to the FRET probe could protect the FRET pair from being degraded by ExoIII. Figure 2B and its insets show that, as predicted, in the presence of NF-κB p50, a portion of the NF-κB probe bound by NF-κB p50 was protected from ExoIII digestion, resulting in the increase of the FRET signal when compared with the negative control containing no p50 protein. The binding of p50 to the NF-κB probe and ExoIII digestion were also confirmed by gel electrophoresis and UV transilluminator imaging (insets of Figure 2B). The dynamics of ExoIII digestion of the p50-bound probe was monitored by real-time PCR (Figure 2C), which revealed that the ExoIII digestion of the solutions with and without p50 could reach equilibrium at the same time. After 5 min of digestion, ExoIII activity was completely terminated by adding EDTA. The EDTA-terminated reaction was immediately analyzed using a spectrofluorometer. All these data verified the design of our assay.
Specificity and applicability
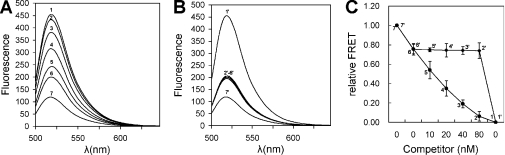
In addition, we investigated the specificity of our assay. For performing competitive assays, two non-labeled competitor duplex DNAs were prepared, one containing a consensus NF-κB binding site of GGGACTTTCC as a specific competitor, and the other not containing any NF-κB binding site as the non-specific competitor. The results of competition assays are shown in Figure 3. It was demonstrated that the specific competitor DNA reversed the FRET signal produced by p50 (Figure 3A and C), whereas the nonspecific competitor DNA had no effect (Figure 3B and C). These results confirmed the specificity of our assay. It also suggests that this method can be used for the rapid assessment of the relative binding affinity of the protein to various DNAs.
Figure 3.
Specificity of protein detection. (A) Emission spectra of the 25 nM NF-κB probes in the presence of 20 nM NF-κB p50 and increasing amounts of cold specific probe. (B) Emission spectra of the 25 nM NF-κB FRET probes in the presence of 20 nM NF-κB p50 and increasing amounts of cold nonspecific probe. (C) Relative FRET of the competition assay. The final concentrations of the competitor probe were 0 nM (6 and 6′), 10 nM (5 and 5′), 20 nM (4 and 4′), 40 nM (3 and 3′) and 80 nM (2 and 2′). ExoIII-positive and ExoIII-negative controls of the 25 nM NF-κB probe were (1 and 1′) and (7 and 7′), respectively.
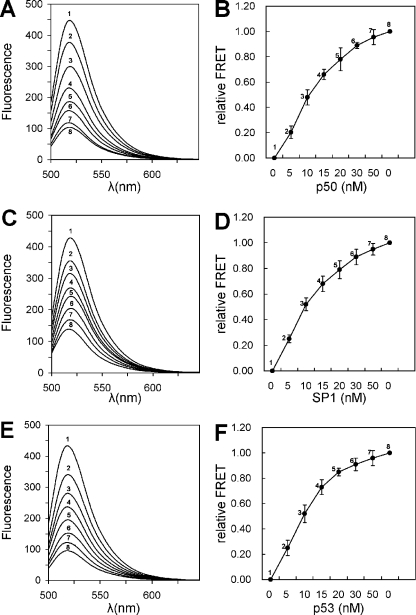
We next investigated the possibility of the assay for the quantitative detection of the DNA-binding proteins. By adding increasing amounts of NF-κB p50 to a series of same molar NF-κB probes and then performing simultaneous ExoIII digestion, we found that the FRET signal of the NF-κB probe increased with the increase in NF-κB p50 (Figure 4A and B). This result implies that under optimized experimental conditions, it is possible to employ the assay to quantify DNA-binding proteins. We performed similar quantification assays with two other sequence-specific DNA-binding proteins, the human transcription factor SP1 (20,21) and p53 (22). For both proteins, we observed an increase in the FRET signal with the increase in protein concentration (Figure 4C–F). These data provide additional evidence for the applicability of the assay. The assay can be generally applied to the detection of the fast-growing numbers of the known DNA-binding proteins.
Figure 4.
Protein concentration dependence of the FRET signal. (A) Emission spectra of the 25 nM NF-κB probes in the presence of increasing amounts of NF-κB p50. (B) NF-κB p50 concentration–dependent relative FRET. (C) Emission spectra of the 25 nM SP1 probes in the presence of increasing amounts of SP1. (D) SP1 concentration–dependent relative FRET. (E) Emission spectra of the 25 nM p53 probes in the presence of increasing amounts of p53. (F) p53 concentration–dependent relative FRET. The final concentration of proteins were 0 nM (1), 5 nM (2), 10 nM (3), 15 nM (4), 20 nM (5), 30 nM (6) and 50 nM (7). ExoIII-negative control of the 25 nM FRET probe was (8).
Nuclear extracts detection
ExoIII was demonstrated to be capable of detecting specific DNA–protein interactions in crude extracts in the DNA protection assay (23–28). It prompted us to investigate the applicability of our assay to cell nuclear extracts. However, interference may arise in the assay due to the endogenous nuclease activity upon ExoIII exposure (24). To avoid this problem, sodium phosphate and carrier nucleic acids including poly(dI-dC), HaeIII-cut E.coli DNA and yeast tRNA were included in our assay for suppressing endogenous nuclease activities in the crude extracts (24,26). For the detection of the target transcription factor NF-κB in the HeLa cell nuclear extract, the HeLa cell was induced with TNF-α to increase the amount of NF-κB in the HeLa cell nuclear extract (29). The NF-κB p50 activity of the harvested nuclear extracts was confirmed by EMSA and super shift assay to provide a nuclear extract sample for NF-κB detection by our assay. The NF-κB protein in the HeLa cell nuclear extract was detected by adding increasing amounts of nuclear extracts to a series of NF-κB probes of the same molar concentration. As shown in Figure 5, the relative FRET increased with an increase in the amount of extract (Figure 5A and B). The competition assays with cold specific and nonspecific probes demonstrated that the extract-dependent variation of the FRET signal was NF-κB-specific (Figure 5C). The ExoIII-negative control with the nuclear extract revealed that the addition of sodium phosphate and carrier nucleic acids suppressed the possible endogenous nuclease activity which may destroy the FRET probe (sample 7 in Figure 5A–C). These data indicate that ExoIII protection assay with the FRET probe can be used to detect proteins in crude extracts.
Figure 5.
Nuclear extract detection by the assay. (A) Emission spectra of 25nM NF-κB probes in the presence of increasing amounts of the TNF-α-induced HeLa cell nuclear extract. The final concentrations of extract were 0 μg/μl (1), 0.05 μg/μl (2), 0.1 μg/μl (3), 0.2 μg/μl (4), 0.4 μg/μl (5) and 0.8 μg/μl (6). The ExoIII-negative control of the 25 nM NF-κB probe was (8). An additional ExoIII-negative control which contained 0.8 μg/μl extract was (7). (B) Nuclear extract concentration dependence of the FRET signal. (C) Relative FRET signal of the competition assay. The assay solutions contained 25 nM NF-κB probes, the 0.4 μg/μl TNF-α-induced HeLa cell nuclear extract and different concentrations of the cold specific (2–6) and nonspecific (2′–6′) competitor probes. The final concentration of competitor probes were 0 nM (6 and 6′), 10 nM (5 and 5′), 20 nM (4 and 4′), 40 nM (3 and 3′) and 80 nM (2 and 2′). ExoIII-positive and -negative controls of the 25 nM NF-κB probe were (1 and 1′) and (7 and 7′), respectively.
Simultaneous detection of two proteins
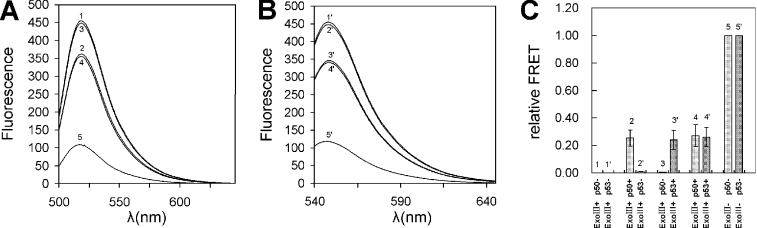
To demonstrate that our assay can be used for multicolor detection, we prepared a JOE-labeled p53 probe for carrying out the simultaneous, independent two-color detection of p50 and p53 proteins in a single assay. The probe solution used in this experiment consisted of equal amounts of the FAM-labeled p50 probe and the JOE-labeled p53 probe. Both probes contained Dabcyl as acceptor. The maximum excitation wavelengths of FAM and JOE are 494 nm and 520 nm, respectively. The maximum emission wavelengths of FAM and JOE are 520 nm and 548 nm, respectively. Thus, excitation at 494 nm provides the fluorescence signal only for the NF-κB probe at the maximum wavelength of 520 nm, and excitation at 520 nm provides the fluorescence signal only for the p53 probe at the maximum wavelength of 548 nm. We mixed equal amounts of the two probes and aliquots were distributed into five tubes. In two tubes (A and B), no protein was added, in two other tubes p50 (C) and p53 (D), respectively, were added and in the last tube (E), both p50 and p53 were added. After the binding reaction, same units of ExoIII were added into tubes B, C, D and E. Addition of p50 to the double-probe solution resulted in an increase in the FRET signal at the wavelength of 520 nm (Figure 6A), whereas addition of p53 resulted in an increase in the FRET signal at a wavelength of 548 nm (Figure 6B), and addition of both the proteins resulted in increases in the FRET signal at both wavelengths of 520 nm and 548 nm (Figure 6A–C). These data provide evidence for the feasibility of two-color detection of two different proteins simultaneously by our assay. If multiple fluorochromes are employed in the assay, it should be possible to detect multiple proteins in high-throughput format.
Figure 6.
Simultaneous detection of two proteins. (A) Emission spectra with excitation at 480 nm (absorption wavelength of FAM). (B) Emission spectra with excitation at 520 nm (absorption wavelength of JOE). (C) Relative FRET signal of the assay. 1: no p50, no p53; 2: 15 nM p50, no p53; 3: no p50, 15 nM p53; 4: 15 nM p50, 15 nM p53. The concentrations of the NF-κB and p53 FRET probes used are 25 nM.
DISCUSSION
FRET (15) provides a powerful technique for the detection of biological interactions (13,30–33). It has already been successfully used in the detection of DNA–protein interactions (13,14,34). In the detection of DNA–protein interactions using FRET technology, the method by which the DNA or protein is labeled is very important. In general, two labeling strategies were adopted in previous studies. In the first strategy, the DNA is labeled with one fluorochrome and the protein with another fluorochrome; FRET is produced by the proximity between DNA and protein in the protein–DNA complex (13,35). In the other strategy, known as the molecular beacon assay, the DNA is labeled with two fluorochromes, each appearing in one duplex DNA of the two half sites of a DNA-binding protein; FRET is produced by the protein-driven annealing of two half sites (14,35). In the first FRET strategy, detection of DNA-binding proteins depends on FRET between the two fluorochromes in a DNA–protein complex (34). The first limitation of this approach is that both the DNA and the protein have to be labeled with fluorochromes, but protein labeling is not always easy as it may affect the native features of the protein. The second limitation is that large proteins may increase the distance between the donor and the acceptor in the protein–DNA complex; therefore, the FRET signal may not be measurable. Finally, the optimum formation of a protein–DNA complex often requires an excess of one labeled component over the other, which does not ensure a 1:1 stoichiometry of fluorescence donor and acceptor groups, and thus produce difficulties in the detection of the fluorescence signal. In the second FRET strategy using molecular beacons, detection of the DNA-binding protein relies on the protein-driven FRET between the two fluorochromes in the complex of the protein and the two DNA half sites (14). The assay does not require labeling of the protein and circumvents the deficiencies of the first FRET strategy. It develops a simple, rapid, homogeneous and high-throughput fluorescence assay for the detection and quantification of DNA-binding proteins. However, the molecular beacon assay may be limited by some difficulties. First, it is difficult to design molecular beacons for those proteins with small binding sites. Second, it may be not easy to prepare the optimal DNA–protein binding buffer which keeps DNA half sites apart without the protein but provides the optimal condition for DNA–protein interaction. Finally, fluorochromes present in the protein-binding site may interfere with the protein–DNA interaction.
In comparison with the two existing FRET-based methods, our assay provides a more easy and simple approach for the detection of DNA-binding proteins. In our assay, the FRET probe was employed as a substitute of a radiolabeled probe in a common ExoIII protection assay. This assay has several advantages. First, the design, preparation and application of the FRET probe are easy and flexible. This assay does not require labeling of the protein, which helps to preserve the natural features of the target protein and allows the detection of crude cell extracts. The labeling of the FRET probe with a fluorescence donor and acceptor is very simple. The donor and acceptor can be easily designed to be in close proximity in the FRET probe, and they are still sufficiently separated from each other after ExoIII digestion, resulting in a large dynamic range of the signal change before and after ExoIII digestion. The dynamic range and sensitivity can be further optimized by adding more than one FRET pairs in the FRET probe. The FRET pair is designed to be sufficiently separated from the protein-binding sites to prevent possible steric hindrance from the FRET pair from exerting its influence on the formation of the protein–DNA complex. The FRET probe consisting of complete protein-binding sites is helpful for natural and optimal interaction between DNA and protein. For optimal detection of proteins with small binding sites, more than one protein-binding site on both sides of FRET pairs can be placed in the FRET probe. The same protein-binding sites on both sides of the FRET pair can be changed into two different binding sites for two different proteins. This type of FRET probe can be used to investigate whether the two proteins are expressed coordinately in cells and also to study their relationship. If multiple FRET probes are labeled with different FRET pairs, the assay allows detection of multiple proteins simultaneously. Besides using Dabcyl as a black acceptor, the acceptor can be another fluorescein molecule with an absorption wavelength that overlaps the emission wavelength of the donor fluorescein. In this case, protein detection can be accomplished by measuring either the decrease in the donor signal or the increase in the acceptor signal, or by assessing the ratio between the two signals (14). Other proximity-based luminescence signals can be flexibly used in place of FRET as the detection signal (11,36–42). Second, it is easy to establish the optimal conditions for DNA–protein binding and ExoIII digestion. In our assay, since the state of the DNA probes is not as important as in the molecular beacon assay, and the ExoIII activity is rather stable over a wide range of ionic strengths, the DNA-binding buffer is easy to prepare for obtaining the optimum yield of specific protein–DNA complexes. We found that the optimized DNA-binding buffer was also suitable for the ExoIII reaction. Thus, there is no need to add any other component besides ExoIII for performing the protection assay. The ExoIII activity is high at 37°C, the most common reaction temperature for DNA–protein interaction. Finally, the procedure of our assay is very simple and time-saving. The protocol includes three steps, including DNA–protein binding, ExoIII digestion and stopping. The whole assay can be completed in 30 min. As a result, our assay provides an easy, simple, sensitive, flexible and high-throughput technique for the detection of DNA-binding proteins.
ExoIII is a 28 kDa monomeric enzyme with 3′ to 5′ exonuclease activity (43). It is used in the construction of nested unidirectional deletions of DNA fragments (44), generation of a single-stranded template for dideoxy sequencing of DNA (45), site-directed mutagenesis (46), cloning of PCR products (47) and DNA footprinting assays (10,24,48–50). In DNA footprinting assays, after a protein specifically binds to a DNA fragment containing its recognition site, ExoIII is used to remove mononucleotides from both DNA strands in a processive fashion, beginning from the 3′ ends. The specifically bound protein blocks the action of ExoIII and leaves double-stranded DNA only in the region bound by the protein, but any free-DNA is fully digested. The successful use of ExoIII in DNA footprinting assays depends on several factors. First, the half-life of the protein–DNA complex is longer than the time required for the ExoIII reaction and, in most cases, the half-life of the complex is not changed by the ExoIII digestion of the DNA (24,51,52). Second, ExoIII activity is rather stable over a wide range of ionic strengths between 0 and 100 mM NaCl (or KCl) (19). However, other common nucleases with exonuclease activity of Bal31 (53) require very high ionic strengths of 600 mM NaCl; it is therefore not applicable in our assay. Finally, ExoIII possesses high activity, releasing mononucleotides as many as 420 bases per minute at 37°C (Data sheet of exonuclease III, MBI Fermentas, Hanover, MD). These features of ExoIII make it easily adaptable to our assay. Another important advantage of ExoIII is that it can be used to detect specific DNA–protein interactions in crude extracts (23–27), which makes ExoIII more suitable for the clinical sample assay, such as tissue extracts. In footprinting, ExoIII footprints are slightly smaller than DNase I footprints. For example, ExoIII generates a 21 bp footprint for GAL4, whereas the footprint of DNase I is 27 bp (50). The shorter footprint of ExoIII is preferable for the FRET probe synthesis at lower cost in our assay. The major disadvantage of ExoIII is that it sometimes nibbles into a protein-bound DNA segment for proteins with lower binding affinity, and even displaces some DNA-binding proteins from DNA as the GAGA factor (19). To avoid this problem, more than one protein-binding site can be harbored on both the sides of FRET pairs, or a selective natural or artificial binding site with high binding affinity can be used in our assay. In total, ExoIII is a very suitable nuclease for our assay.
CONCLUSIONS
We have described a novel method to detect sequence-specific DNA-binding proteins. We named this method as ExoIII protection assay with FRET probe, which combines FRET technology with an ExoIII protection assay, to develop a simple, sensitive and high-throughput method for detecting sequence-specific DNA-binding proteins, such as transcription factors. We employed this method to successfully detect three different transcription factors, NF-κB, SP1 and p53, in quantitative and specific formats. The results also revealed that the assay could be used to detect DNA-binding proteins in crude cell nuclear extracts. Therefore, the assay has wide potential application in research and biomedicine. In research, the assay provides a convenient tool for detecting the presence and monitoring the DNA-binding activity of the fast-growing numbers of DNA-binding proteins. In biomedicine, the assay can be applied to identify the presence or the activity of a DNA-binding protein in disease samples (tissue extracts) in medical diagnosis, and to screen potential drugs targeted at a DNA-binding protein in medicine development.
Acknowledgments
We appreciate the critical readings of the two anonymous reviewers of our manuscript. We also acknowledge the detailed editing of the anonymous editor of our paper. This work was partially supported by the grants 2003AA2Z2012 from the National High Tech Program (863) and Projects 60471019 and 90408027 of National Natural Science Foundation of China. Funding to pay the Open Access publication charges for this article was provided by National Natural Science Foundation of China, and the Ministry of Science and Technology, People's Republic of China.
REFERENCES
- 1.Pabo C.O., Sauer R.T. Transcription factors: structural families and principles of DNA recognition. Annu. Rev. Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 2.Pingoud A., Jeltsch A. Recognition and cleavage of DNA by type-II restriction endonucleases. Eur. J. Biochem. 1997;246:1–22. doi: 10.1111/j.1432-1033.1997.t01-6-00001.x. [DOI] [PubMed] [Google Scholar]
- 3.Craig N.L. The mechanism of conservative site-specific recombination. Annu. Rev. Genet. 1988;22:77–105. doi: 10.1146/annurev.ge.22.120188.000453. [DOI] [PubMed] [Google Scholar]
- 4.Margulies C., Kaguni J.M. Ordered and sequential binding of DNA protein to oriC, the chromosomal origin of Escherichia coli. J. Biol. Chem. 1996;271:17035–17040. doi: 10.1074/jbc.271.29.17035. [DOI] [PubMed] [Google Scholar]
- 5.Pandolfi P.P. Transcription therapy for cancer. Oncogene. 2001;20:3116–3127. doi: 10.1038/sj.onc.1204299. [DOI] [PubMed] [Google Scholar]
- 6.Sen R., Baltimore D. Inducibility of kappa immunoglogulin enhancer-binding protein NF-κB by a posttranslational mechanism. Cell. 1986;47:921–928. doi: 10.1016/0092-8674(86)90807-x. [DOI] [PubMed] [Google Scholar]
- 7.Baldwin A.S. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-kB. J. Clin. Invest. 2001;107:241–246. doi: 10.1172/JCI11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fried M.G., Crothers D.M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981;9:6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galas D.J., Schmitz A. DNase footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shalloway D., Kleinberger T., Livingston D.M. Mapping of SV 40 DNA replication origin region binding sites for the SV 40 DNA replication antigen by protection against Exonuclease III digestion. Cell. 1980;20:411–422. doi: 10.1016/0092-8674(80)90627-3. [DOI] [PubMed] [Google Scholar]
- 11.Selvin P.R., Rana T.M., Hearst J.E. Luminescence energy transfer. J. Am. Chem. Soc. 1994;116:6029–6030. [Google Scholar]
- 12.Hill J.J., Royer C.A. Fluorescence approaches to study of protein-nucleic acid complexation. Methods Enzymol. 1997;278:390–416. doi: 10.1016/s0076-6879(97)78021-2. [DOI] [PubMed] [Google Scholar]
- 13.Furey W.S., Joyce C.M., Osborne M.A., Klenerman D., Peliska J.A., Balasubramanian S. Use of fluorescence resonance energy transfer to investigate the conformation of DNA substrates bound to the Klenow fragment. Biochemistry. 1998;37:2979–2990. doi: 10.1021/bi9719758. [DOI] [PubMed] [Google Scholar]
- 14.Heyduk T., Heyduk E. Molecular beacons for detecting DNA binding proteins. Nat. Biotechnol. 2002;20:171–176. doi: 10.1038/nbt0202-171. [DOI] [PubMed] [Google Scholar]
- 15.Forster T. Zwischenmoleculare Energiewanderung und Fluorescenz [Title translation: Intermolecular energy transfer and fluorescence. J. Ann. Phys. 1948;2:57–75. [Google Scholar]
- 16.Wang J.K., Li T.X., Bai Y.F., Lu Z.H. Evaluating the binding affinities of NF-κB p50 homodimer to the wild-type and single-nucleotide mutant Ig-κB sites by the unimolecular dsDNA microarray. Anal. Biochem. 2003;316:192–201. doi: 10.1016/s0003-2697(03)00049-6. [DOI] [PubMed] [Google Scholar]
- 17.Wang J.K., Bai Y.F., Li T.X., Lu Z.H. DNA microarrays with unimolecular hairpin double-stranded DNA probes: fabrication and exploration of sequence-specific DNA/protein interactions. J. Biochem. Biophy. Methods. 2003;55:215–232. doi: 10.1016/s0165-022x(03)00048-4. [DOI] [PubMed] [Google Scholar]
- 18.Wang H., Li J., Liu H.P., Liu Q.J., Mei Q., Wang Y.J., Zhu J.J., He N.Y., Lu Z.H. Label-free hybridization detection of a single nucleotide mismatch by immobilization of molecular beacons on an agarose film. Nucleic Acids Res. 2003;30:e61. doi: 10.1093/nar/gnf061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metzger W., Heumann H. Footprinting with exonuclease III. In: Moss T., editor. DNA-Protein Interactions: Principles and Protocols, 2nd edn. Vol. 148. Totowa, NJ: Humana Press Inc.; 2001. pp. 39–47. Methods in Molecular Biology. [Google Scholar]
- 20.Dynan W.S., Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the Sv40 early promoter. Cell. 1983;35:79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- 21.Briggs M.R., Kadonga J.T., Bell S.P., Tjian R. Purification and biochemical characterization of the promoter specific transcription factor, Sp1. Science. 1985;234:47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- 22.Ko L.L., Prives C. p53—puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 23.Wu C. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980;286:854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- 24.Wu C. Two protein-binding sites in chromatin implicated in the activation of heat-shock genes. Nature. 1984;309:229–234. doi: 10.1038/309229a0. [DOI] [PubMed] [Google Scholar]
- 25.Wu C. An exonuclease protection assay reveals heat-shock element and TATA box binding proteins in crude nuclear extracts. Nature. 1985;317:84–87. doi: 10.1038/317084a0. [DOI] [PubMed] [Google Scholar]
- 26.Ng R., Carbon J. Mutational and in vitro protein-binding studies on centromere DNA from Saccharomyces cerevisiae. Mol. Cell. Biol. 1987;7:4522–4534. doi: 10.1128/mcb.7.12.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilmour D.S., Dietz T.J., Elgin S.C.R. TATA box-dependent protein–DNA interactions are detected on heat shock and histone gene promoters in nuclear extracts derived from Drosophila melanogaster embryos. Mol. Cell. Biol. 1988;8:3204–3214. doi: 10.1128/mcb.8.8.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Vries E., van Driel W., Bergsma W.G., Arnberg A.C., van der Vliet P.C. HeLa nuclear protein recognizing DNA termini and translocating on DNA forming a regular DNA-multimeric protein complex. J. Mol. Biol. 1989;208:65–78. doi: 10.1016/0022-2836(89)90088-0. [DOI] [PubMed] [Google Scholar]
- 29.DiDonato J.A., Hayakawa M., Rothwarf D.M., Zandi E., Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 30.Heyduk E., Heyduk T. Architecture of a complex between the σ70 subunit of Escherichia coli RNA polymerase and the nontemplate strand oligonucleotide. Luminescence resonance energy transfer study. J. Biol. Chem. 1999;274:3315–3322. doi: 10.1074/jbc.274.6.3315. [DOI] [PubMed] [Google Scholar]
- 31.Callaci S., Heyduk E., Heyduk T. Core RNA polymerase from E.coli induces a major change in the domain arrangement of the σ70 subunit. Mol. Cell. 1999;3:229–238. doi: 10.1016/s1097-2765(00)80313-5. [DOI] [PubMed] [Google Scholar]
- 32.Ghosh S.S., Eis P.S., Blumeyer K., Fearon K., Millar D.P. Real time kinetics of restriction endonuclease cleavage monitored by fluorescence resonance energy transfer. Nucleic Acids Res. 1994;22:3155–3159. doi: 10.1093/nar/22.15.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyagi S., Bratu D.P., Kramer F.R. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 34.Heyduk T., Lee J.C. Application of fluorescence energy transfer and polarization to monitor E.coli cAMP receptor protein and lac promoter interaction. Proc. Natl Acad. Sci. USA. 1990;87:1744–1748. doi: 10.1073/pnas.87.5.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heyduk E., Knoll E., Heyduk T. Molecular beacons for detecting DNA binding proteins: mechanism of action. Anal. Biochem. 2003;316:1–10. doi: 10.1016/s0003-2697(03)00004-6. [DOI] [PubMed] [Google Scholar]
- 36.Hart H.E., Greenwald E.B. Scintillation proximity assay (SPA)—a new method of immunoassay. Direct and inhibition mode detection with human albumin and rabbit antihuman albumin. Mol. Immunol. 1979;16:265–267. doi: 10.1016/0161-5890(79)90065-8. [DOI] [PubMed] [Google Scholar]
- 37.Campbell A.K., Patel A. A homogeneous immunoassay for cyclic nucleotides based on chemiluminescence energy transfer. Biochem. J. 1983;216:185–194. doi: 10.1042/bj2160185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tyagi S., Kramer F.R. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 39.Packard B.Z., Toptygin D.D., Komoriya A., Brand L. Characterization of fluorescence quenching in bifluorophoric protease. Biophys. Chem. 1997;67:167–176. doi: 10.1016/s0301-4622(97)00036-7. [DOI] [PubMed] [Google Scholar]
- 40.Nolan J.P., Sklar L.A. The emergence of flow cytometry for sensitive, real-time measurements of molecular interactions. Nat. Biotechnol. 1998;16:633–638. doi: 10.1038/nbt0798-633. [DOI] [PubMed] [Google Scholar]
- 41.Xu Y., Piston D.W., Johnson C.H. A bioluminescence resonance energy transfer (BRET) system: application to interacting circadian clock proteins. Proc. Natl Acad. Sci. USA. 1999;96:151–156. doi: 10.1073/pnas.96.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rippe K. Simultaneous binding of two DNA duplexes to the NtrC-enhancer complex studied by two-color fluorescence cross-correlation spectroscopy. Biochemistry. 2000;39:2131–2139. doi: 10.1021/bi9922190. [DOI] [PubMed] [Google Scholar]
- 43.Rogers S.G., Weiss B. Exonuclease III of Escherichia coli K-12, an AP endonuclease. Methods Enzymol. 1980;65:201–211. doi: 10.1016/s0076-6879(80)65028-9. [DOI] [PubMed] [Google Scholar]
- 44.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 45.Guo L.H., Wu R. New rapid methods for DNA sequencing based on exonuclease III digestion followed by repair synthesis. Nucleic Acids Res. 1982;10:2065–2084. doi: 10.1093/nar/10.6.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vandeyar M., Weiner M.P., Hutton C., Batt C. A simple and rapid method for the selection of oligodeoxynucleotide-directed mutants. Gene. 1988;65:129–133. doi: 10.1016/0378-1119(88)90425-8. [DOI] [PubMed] [Google Scholar]
- 47.Li C., Evans R.M. Ligation independent cloning irrespective of restriction site compatibility. Nucleic Acids Res. 1997;25:4165–4166. doi: 10.1093/nar/25.20.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siebenlist U., Simpson R.B., Gilbert W. E.coli RNA polymerase interacts homologously with two different promoters. Cell. 1980;20:269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- 49.Siebenlist U., Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc. Natl Acad. Sci. USA. 1980;77:122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carey M., Kakidani H., Leatherwood J., Mostashari F., Ptashne M. An amino-terminal fragment of GAL4 binds DNA as a dimer. J. Mol. Biol. 1989;209:423–432. doi: 10.1016/0022-2836(89)90007-7. [DOI] [PubMed] [Google Scholar]
- 51.Loh T.P., Sievert L.L., Scott R.W. Evidence for a stem cell-specific repressor of Moloney murine leukemia virus expression in embryonic carcinoma cells. Mol. Cell. Biol. 1990;10:4045–4057. doi: 10.1128/mcb.10.8.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carnevali F., La Porta C., Ilardi V., Beccari E. Nuclear factors specifically bind to upstream sequences of a Xenopus laevis ribosomal protein gene promoter. Nucleic Acids Res. 1989;17:8171–8184. doi: 10.1093/nar/17.20.8171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual, 2nd edn. NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]