Abstract
A system for the tetracycline-inducible regulation of gene expression in mycobacteria has been developed. We have sub-cloned the tetRO region from the Corynebacterium glutamicum TetZ locus into a mycobacterial shuttle plasmid, making expression of genes cloned downstream of tetRO responsive to tetracycline. Using the luxAB-encoded luciferase from Vibrio harveyi as a reporter (pMind-Lx), we observed a 40-fold increase in light output from Mycobacterium smegmatis cultures 2 h after adding 20 ng ml−1 of tetracycline. Similarly, exposure to the drug resulted in up to 20-fold increase in relative light units from M.bovis BCG carrying the reporter construct, and a 10-fold increase for M.tuberculosis. Tetracycline induction was demonstrated in log and stationary phase cultures. To evaluate whether this system is amenable to use in vivo, J774 macrophages were infected with M.bovis BCG[pMind-Lx], treated with amikacin to kill extracellular bacteria, and then incubated with tetracycline. A 10-fold increase in light output was measured after 24 h, indicating that intracellular bacteria are accessible and responsive to exogenously added tetracycline. To test the use of the tetracycline-inducible system for conditional gene silencing, mycobacteria were transformed with a pMind construct with tetRO driving expression of antisense RNA for the ftsZ gene. Bacterial cells containing the antisense construct formed filaments after 24 h exposure to tetracycline. These results demonstrate the potential of this tetracycline-regulated system for the manipulation of mycobacterial gene expression inside and outside cells.
INTRODUCTION
An estimated 2 billion people carry a latent infection with Mycobacterium tuberculosis, the causative agent of tuberculosis (1). There are 8 million new cases of disease every year and 2 million deaths, many of those in patients already infected with HIV. There is a need for new drugs and vaccines to treat active disease, and also to prevent development of disease in individuals with latent infection. Progress is restricted by the lack of information about the physiological status of tuberculosis bacteria within infected tissues; particularly during latent infection. The current consensus is that the bacteria persist predominantly in a non-dividing form which may be partially mimicked by in vitro models involving exposure to low oxygen tension or nutrient depletion (2,3). Recent experiments with non-human primates suggests that these may provide a useful model of latent tuberculosis, with ∼50% of Macaca fascicularis given a low bronchial dose of M.tuberculosis developing a stable asymptomatic infection that can be reactivated to cause clinical disease by immuno-suppressing the animals (4,5). A major challenge is to identify genes that are essential for maintaining bacterial viability during latent infection in such models, and to test their suitability as drug targets. To identify such genes, we need to be able to switch them on or off at selected times during infection.
Research into the molecular microbiology of mycobacteria has been hampered by a lack of the tools that most bacterial geneticists take for granted. Members of the M.tuberculosis complex are neither naturally transformable nor do they commonly exchange DNA by conjugation, therefore many of the systems used for enteric bacteria have had to be extensively re-engineered for use in mycobacteria. Inducible systems have remained problematic, with reports in the literature limited to the use of the heat-shock promoter (6), the Ptra promoter and temperature-sensitive TraR repressor from Streptomyces (7), and the inducible acetamidase gene from Mycobacterium smegmatis (8,9). The acetamidase system has been used to control gene expression (10–14) and to make a conditional antisense system (15). However the inducer, acetamide requires special growth media for optimum activity, is not suitable for use in vivo, and depends on the presence of a large 1.4 kb operator region, which lies 1.5 kb upstream of the promoter (9,16). The hsp60 promoter has also been used for protein over-expression (6,12,17,18), but it is not tightly repressed, with a high basal level of activity in multi-copy and only limited induction after heat shock.
The aim of the present study was to develop an inducible system for mycobacteria that meets the criteria of tight repression in the absence of inducer, low likelihood of gratuitous induction, and with an inducer that shows good bioavailability in vitro and in vivo. These requirements have been met in a range of eukaryotes and prokaryotes by regulation of gene expression by tetracyclines, based on Tet efflux systems such as that encoded by Tn10, (19–21). Ehrt et al. have recently described the potential use of the Tn10 system in mycobacteria (22). In the present study, we have explored the use of an alternative Tet efflux system identified on a plasmid isolated from Corynebacterium glutamicum (23). We reasoned that the close taxonomic relation between corynebacteria and mycobacteria might allow us to avoid problems often encountered in adapting E.coli expression systems for use in high GC organisms. Sequence analysis of the R-plasmid pAG1 from C.glutamicum identified a novel tetracycline-resistance determinant, TetZ, comprising a predicted tetA-like efflux pump divergently transcribed from a tetR-like transcriptional repressor (Figure 1A). This is the first repressor-regulated Tet efflux system to be found in gram-positive bacteria (23), and it is of interest to determine whether similarities identified at the level of sequence comparison are borne out by functional analysis.
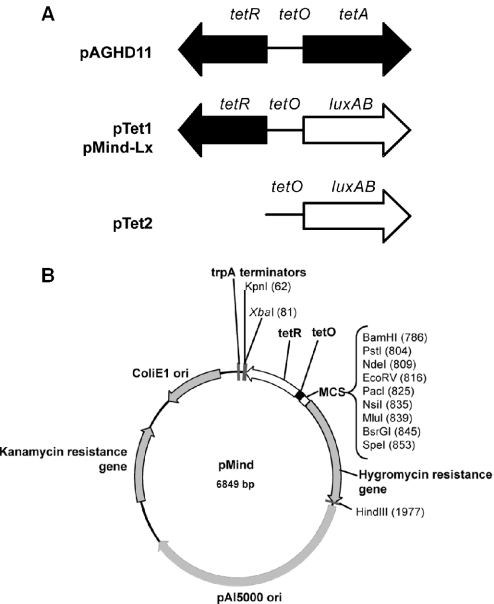
Figure 1.
(A) The organization of the TetZ region from pAGHD1. pTet1 and pMind-Lx contained the tetR and tetO regions cloned in front of the luxAB reporter; tetR was omitted from pTet2. (B) A map of the pMind vector, illustrating origins of replication and selectable markers for both E.coli and mycobacteria, and the tetRO operator–repressor region upstream of a multiple cloning site.
In this study, we report the first application of the corynebacterial Tet-efflux system and describe the development of a tetracycline-inducible system for use in mycobacteria. We demonstrate its application to the regulation of gene expression in extracellular and intracellular mycobacteria.
MATERIALS AND METHODS
Bacterial strain and growth conditions
Mycobacterium smegmatis mc2155 was grown at 37°C, in Hartmans–de-Bont minimal medium (supplemented with 0.08% glycerol and 0.05% Tween-80) as described previously (3), or on Luria-agar plates. Mycobacterium bovis BCG (Pasteur) and M.tuberculosis H37Rv were grown in Middlebrook 7H9 liquid broth or on Middlebrook 7H11 solid media, prepared according to the manufacturer's instructions, and supplemented with OADC (Difco). All E.coli strains were grown on LB-agar plates or in LB broth. Hygromycin was added as required at a concentration of 250 μg ml−1 for E.coli and 50 μg ml−1 for mycobacteria. Growth curves were performed using triplicate cultures in Hartmans–de-Bont minimal medium (3) (supplemented with 0.08% glycerol and 0.05% Tween-80) at 37°C in 125 ml conical flasks shaken at 100 or 200 r.p.m. in an orbital shaker for BCG and M.smegmatis, respectively. Tetracycline (Tc) was added as appropriate. Samples were processed for luminescence as described below.
Plasmid construction
Plasmid pAGHD1 containing the 11 kb HindIII fragment of pAG1 encompassing the TetZ determinant was a kind gift from Dr Andreas Tauch, Department of Genetics, University of Bielefeld, Germany (23). The tetRO region from TetZ was amplified by PCR using primers TetRFor (5′-CGGGATCCTCACGATTCGCTCGAGGTC-3′) and TetORev1 (5′-CGCATATGTGTCAGGATTCCACGATGAG-3′); the tetO determinant was amplified using TetOFor (5′-CGGGATCCAGTTGCACTTTATCATCGATAAC-3′) and TetORev1. The forward primers contain BamHI restriction sites and the reverse primer contains an NdeI restriction site (underlined). Both products were cloned upstream of the luxAB genes from Vibrio harveyi in the vector pSMT1 (24), to make pTet1 and pTet2 (Figure 1A). For further work, the tetRO region was amplified by PCR using TetRO-F (5′-GCTCTAGATCACGATTCGCTCGAGGTC-3′) and TetRO-R (5′-CGGGATCCTGTCAGGATTCCACGATGAG-3′) containing XbaI and BamHI sites (underlined) respectively, and cloned into the XbaI–BamHI sites upstream of the luxAB genes in pSHKLx (25) to generate pMindLx (Figure 1A). Plasmid pSHKLx is an E.coli–mycobacteria shuttle plasmid containing kanamycin and hygromycin selectable markers and the luxAB genes from Vibrio harveyi as a reporter. In pMind the luxAB genes have been replaced by a multiple cloning site (Figure 1B) to facilitate cloning of other genes of interest. Vectors were introduced into mycobacteria by electroporation (26).
Luciferase assay
Bioluminescence of M.smegmatis and M.bovis BCG was measured in a Berthold AutoLumat LB953 tube luminometer; for M.tuberculosis a Turner-Designs 20/20 tube luminometer was used. Previous work has shown that the Turner-Designs reader gives relative light unit (RLU) values approximately 1000-fold lower than the Berthold instrument, but the study confirmed linearity between machines (24). To measure RLUs, 0.1 ml of the substrate, 1% n-decyl aldehyde (Sigma) in ethanol, was injected automatically into tubes containing cells in a final volume of 1 ml in phosphate-buffered saline (PBS). Raw data were collected in duplicate or triplicate over a 20 s period and mean values calculated. The luminescent output was normalized to the cell density and expressed as RLU. Samples were diluted 10-fold in PBS to minimize quenching of luminescence.
Construction of an ftsZ antisense knock-down in M.smegmatis
The M.tuberculosis ftsZ gene (Rv2150c) was amplified by PCR using primers ftsZantisense1 (5′-GGACTAGTATGACCCCCCCGCACAACTA-3′) and ftsZantisense2 (5′-CGGGATCCTCAGCGGCGCATGAAGGGCG-3′), containing SpeI and BamHI restriction sites (underlined), respectively. This product was cloned into the BamHI–SpeI sites of pMind, to generate pMind-FtsZ-antisense, which was transformed into M.smegmatis. Tetracycline (20 ng ml−1) was added to mid-log phase bacteria (OD600nm ∼0.5) and culture continued for 24 h (OD600nm ∼ 1.6). Samples were acid fast stained and slides were viewed at 100× magnification with a Nikon Eclipse E600 microscope. Images were captured using a Nikon DXM1200 camera with ACT-1 software.
Infection of macrophages in vitro and induction of luxAB reporter
The J774A.1 murine macrophage-like cell line was seeded overnight at 5 × 105 cells per well in NUNC 12-well plates at 37°C and 5% CO2 in HI-glucose DMEM (Sigma), supplemented with 10% heat-inactivated foetal calf serum and 2 mM L-glutamine. Log-phase cultures of mycobacteria were centrifuged at 3000 g for 10 min, and resuspended in DMEM to obtain the required multiplicity of infection (MOI). Infections were performed at an MOI of 10–20 bacteria per cell for 6 h at 37°C, and then the monolayer was washed three times with Dulbecco's PBS (Invitrogen). Cells were treated for 2 h with 200 μg ml−1 amikacin in DMEM to eliminate extracellular mycobacteria (27). The cells were washed again three times with Dulbecco's PBS and incubated in fresh DMEM. After 24 h of further incubation, 100 ng ml−1 Tc was added to the medium. Samples were taken from triplicate wells at 24, 48 and 72 h after the addition of tetracycline. RLUs and colony forming units (CFUs) were determined at each time point; eukaryotic cells were lysed by the addition of 1800 μl of sterile Dulbecco's PBS containing 0.1% Triton X-100 per well.
RESULTS
Functional activity of the repressor-regulated C.glutamicum TetZ
During sequence analysis of pAG1 plasmid from Corynebacterium glutamicum, Tauch et al. identified a set of genes with homology to tetracycline-regulated efflux systems previously characterized in gram-negative bacteria (Figure 1A) (23). This comprises the gene predicted to encode a transcriptional repressor (TetR) together with a divergently transcribed efflux pump (TetA). The intervening operator region (tetO) is predicted to include the promoter sequence for each gene, as well as repressor sites for binding of TetR. By analogy with the Tn10 system, it is anticipated that binding of TetR to tetO will block expression from the tetA and tetR promoters in the absence of tetracycline, and that this inhibition will be relieved when drug is added to the system.
To test this model, we amplified the gene encoding the transcriptional regulator (tetR) along with the operator region (tetO). This region was then cloned upstream of the luxAB genes encoding the luciferase enzyme from Vibrio harveyi, in the mycobacteria–E.coli shuttle vector pSMT1 (24) to construct pTet1 (Figure 1A), which was introduced into Mycobacterium smegmatis by electroporation. A second construct, pTet2, was generated in the same way, but without the tetR gene (Figure 1A). We anticipated that luciferase activity conferred by pTet2 would provide a measure of gene expression from the tetA promoter, and that pTet1 expression would provide a measure of the efficiency of repression by TetR. In fact, the luminescence output for both constructs was ∼105 RLU per ml, a similar level to that obtained with luxAB in the absence of any added promoter. This background expression is thought to be due to readthrough from sequences within the plasmid backbone (24). The absence of enhanced luminescence in pTet2 suggested either that the tetA promoter was inactive in mycobacteria, or that promoter activity required sequence elements present within the tetR gene. Results obtained by addition of tetracycline are consistent with the second explanation. Cultures containing the two constructs were grown in the presence or absence of 10 ng ml−1 Tc for 2.5 h and luminescence was compared. Addition of Tc to the pTet1 cultures resulted in a 68-fold increase in luminescence output (∼107 RLU ml−1). Tc had no effect on luminescence of pTet2 cultures. These results show that TetR is expressed as a functional repressor in mycobacteria, and that it regulates activity from a promoter region that includes tetO and part of the tetR gene itself.
Dose-dependent Tc regulation of gene expression in mycobacteria
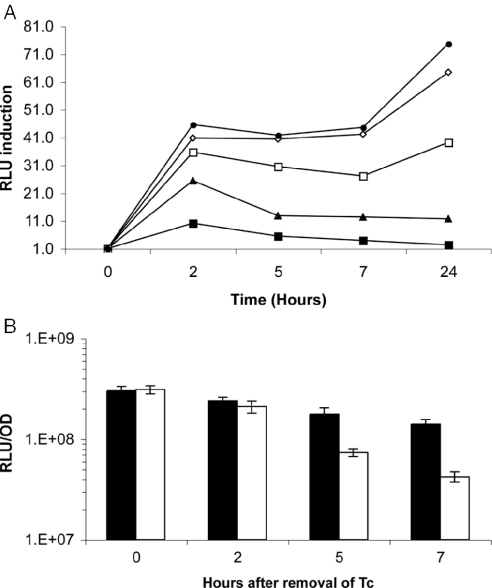
For further work, the tetRO region was recloned upstream of luxAB in pSHKLx (25) to construct pMind-Lx, or without the luciferase genes to make pMind (Figure 1B). A dose–response curve was established by adding doubling dilutions of Tc from 20 to 1.25 ng ml−1 to triplicate samples from a log phase culture of M.smegmatis harbouring pMind-Lx (OD600nm = 0.5, ∼109 CFU ml−1). Figure 2A illustrates the kinetics of induction over a 24 h time period. At the 24 h time point, luminescence in the absence of Tc was 6.3 × 105 RLU per OD unit; this rose to 4.6 × 107 RLU per OD unit after 24 h growth in the presence of 20 ng ml−1 of Tc. Background luminescence from a promoterless luxAB in this vector is 2 × 105 RLU per OD unit, indicating repression is incomplete. A concentration of 20 ng ml−1 gave the maximum levels of induction and was chosen for future experiments. This is well below the minimal inhibitory concentration of Tc for M.smegmatis (80 ng ml−1; data not shown), and no effect on mycobacterial growth was observed in the dose–response experiment.
Figure 2.
(A) The kinetics of luciferase induction following addition of varying concentrations of Tc to M.smegmatis [pMind-Lx]. Tc was added to bacterial cultures and luminescence was measured after different time intervals. Tc was added at 1.25 ng ml−1 (filled square), 2.5 ng ml−1 (filled triangle), 5 ng ml−1 (open square), 10 ng ml−1 (open diamond) and 20 ng ml−1 (filled circle). Y-axis values are the ratio of RLU measured in the presence and absence of Tc. (B) The decrease in luminescence over time after Tc has been removed. Open bars show the loss of luminescence from induced M.smegmatis [pMind-Lx] that were washed and resuspended in fresh medium. The corresponding induced culture was washed then resuspended in its original Tc-containing medium (dark bars). Y-axis values are RLU per OD unit.
As observed previously (24), luminescence output was lower in stationary phase bacteria. To test the effect of Tc-induction in non-dividing M.smegmatis[pMind-Lx], 20 ng ml−1 Tc was added to a culture that had been allowed to grow into stationary phase (OD600nm = 1.6, ∼8 × 109 CFU ml−1). A 10-fold increase in light output was observed after 2 h.
Restoration of repression of the system once Tc is removed was tested by inducing bacteria with 20 ng ml−1 of Tc for 18 h, washing the cells, then continuing growth in fresh broth. There was a progressive reduction in luminescence, with a 70% decrease in light output after 7 h (Figure 2B).
Activity of tetracycline derivatives
Other members of the tetracycline family are able to induce the Tn10-based Tet system (19). We tested six tetracyclines for their ability to induce light production by M.smegmatis [pMind-Lx] at a concentration of 20 ng ml−1. After 18–24 h exposure, Tc showed the highest level of induction (Table 1), with relatively poor levels of induction by all the other derivatives, including doxycycline and anhydrotetracycline, both of which have proved successful in Tn10-based systems (19). Doxycycline and chlortetracycline were found to be toxic in this system, and bacterial death, as indicated by a decrease in optical density, was observed at both time points.
Table 1.
Luciferase induction in response to 20 ng ml−1 different tetracyline-related compounds compared to non-induced control
| Time | Control | Tetracycline | Minocycline | Doxycycline | Chlortetracycline | Oxytetracycline | Anhydrotetracycline |
|---|---|---|---|---|---|---|---|
| 18 h | 1 | 38 | 1 | 3 | 8 | 1 | 1 |
| 24 h | 1 | 108 | 1 | 6 | 41 | 1 | 2 |
Tetracycline-dependent luminescence in slow-growing mycobacteria
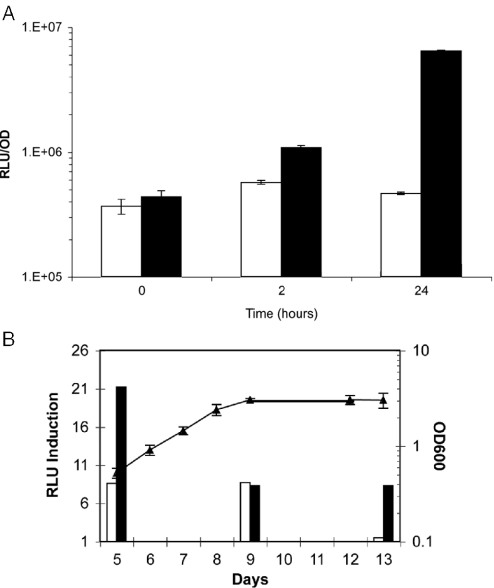
M.smegmatis is a rapid-growing saprophytic mycobacterium with a doubling time of ∼3 h. To test the function of this system in slow-growing mycobacteria (which have a doubling time of ∼24 h), 20 ng ml−1 Tc was added to a log phase culture (OD600nm 0.4–0.5) of M.tuberculosis harbouring the pMind-Lx plasmid, and light output was measured after 2 and 24 h. Luminescence of the treated culture was 2-fold higher than that in the control culture at the 2 h time point, rising to 13-fold after 24 h (Figure 3A). Similarly, Tc induction of an M.bovis BCG[pMind-Lx] culture during log phase (day 5), resulted in 8-fold and 21-fold increase in luminescence after 2 and 24 h (from a level of 1.8 × 105 RLU per OD unit in the uninduced culture). Luminescence output and Tc induction decreased as cultures entered stationary phase with a maximum 8-fold induction observed after 24 h in day 9 and day 13 cultures (from a level of 4.3 × 104 and 6.1 × 103 RLU per OD unit, respectively, in the uninduced cultures) (Figure 3B). These results show that the Tc regulated promoter is active in logarithmic and stationary phase cultures of slow-growing as well as fast-growing mycobacteria.
Figure 3.
(A) The light output from M.tuberculosis [pMind-Lx] after 2 and 24 h induction with Tc. Samples were incubated with 20 ng ml−1 Tc (filled bars) and controls without Tc (open bars). Y-axis values are RLU per OD unit. (B) Luciferase induction from M.bovis BCG[pMind-Lx] at various times during growth. Replicate cultures were induced with 20 ng ml−1 Tc on day 5 (log phase growth), day 9 (early stationary phase) and day 13 (late stationary phase) and the light output measure after 2 h (open bars) and 24 h (filled bars). The ratio of RLU in the presence and absence of Tc induction is plotted on the Y-axis. The mean OD600nm for all the replicate cultures is also shown (filled triangles).
Tetracycline-dependent luminescence of intracellular M.bovis BCG
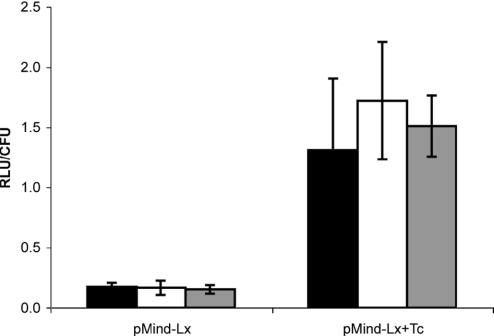
Pathogenic mycobacteria are able to persist during infection within a phagosomal compartment of host macrophages, and an important property of a tool for mycobacterial gene regulation is the ability to control gene expression within this in vivo environment. To test the bioavailability of tetracycline to mycobacteria inside phagosomes, we infected J774 macrophages with M.bovis BCG[pMind-Lx]. After 6 h, cells were treated for 2 h with 200 μg ml−1 of amikacin. This procedure has been shown to be effective in killing the majority of extracellular mycobacteria with minimal effect on bacteria that have already been taken up by the macrophage (27). Medium containing amikacin and dead bacteria was then removed and replaced with fresh tissue culture medium and incubation continued. After 18 h, the medium was removed and replaced with fresh medium containing 100 ng ml−1 Tc. This procedure allowed us to focus predominantly on the luminescence signal from intracellular bacteria. Infected cultures were then lysed, and the number of viable bacteria (CFU) and light output (RLU) was measured from triplicate wells after 24, 48 and 72 h induction (Figure 4). The number of bacteria was similar in all cultures (∼104 CFU ml−1). There was a 10-fold increase in luminescence 48 h after exposure to Tc. As a control, we also measured light output from BCG carrying a construct in which luciferase is constitutively expressed under control of the hsp60 promoter (24). Luminescence output from this construct (1 × 102 RLU per CFU after 24 h) was unaffected by addition of Tc (data not shown).
Figure 4.
Luminescence produced by intra-phagosomal M.bovis BCG[pMind-Lx]. J774 macrophages infected with M.bovis BCG[pMind-Lx] were exposed to 100 ng ml−1 Tc and light output was measured in samples taken after 24, 48 or 72 h. Addition of Tc resulted in an ∼10-fold increase in light output from BCG[pMind] but had no effect on output from the constitutively expressed luxAB in BCG[pSHK-Lx] (data not shown). There was no significant difference in the number of viable bacteria present in drug-treated or control cultures. Y-axis values are mean RLU per CFU from triplicate wells; error bars represent SD values. Filled bars, 24 h; open bars, 48 h; grey bars, 72 h induction.
Tetracycline-regulated expression of antisense mRNA
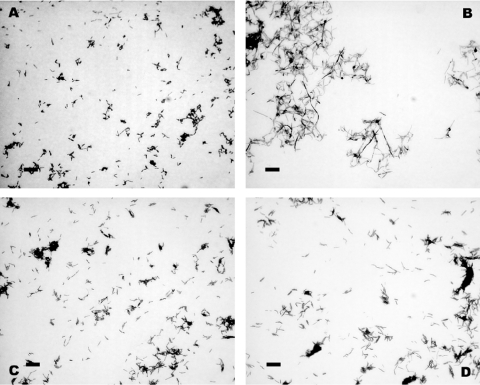
A further important property of a tool for gene regulation is its application in experiments involving down regulation of selected genes. One way to achieve this is to induce expression of antisense mRNA. To test the function of the pMind construct in this system, we focused on regulation of the ftsZ gene in mycobacteria. The FtsZ protein forms a ring at the site of bacterial cell division that acts as a key focus for many of the proteins involved in septum formation and cell division. Decreased production of FtsZ means that the septum fails to form and results in a filamentous phenotype that is easily assessed by visual examination (14). We constructed a strain of M.smegmatis carrying pMind-FtsZ-antisense, which contains the whole of the ftsZ gene in the antisense orientation under the control of the tetRO region. Logarithmic phase cultures were grown for 24 h in the presence or absence of Tc, and then examined by microscopy. Panel A of Figure 5 illustrates the normal morphology of M.smegmatis in the absence of antisense induction, and panel B shows the characteristic filamentation associated with reduced expression of ftsZ after antisense induction. Panels C and D illustrate results with control cultures of M.smegmatis without the inducible construct, demonstrating that filamentation is not simply a consequence of exposure to Tc. This experiment shows that the TetZ system has application in the construction of conditional phenotypes for otherwise essential genes.
Figure 5.
Micrographs showing the filamented morphology of M.smegmatis after induction of ftsZ antisense. (A) M.smegmatis [pMind-FtsZ-antisense] after 24 h without Tc; (B) M.smegmatis [pMind-FtsZ-antisense] after 24 h induction with 20 ng ml−1 Tc; (C) M.smegmatis without Tc; (D) M.smegmatis with 20 ng ml−1 Tc. Images taken under a 100× objective, the scale bar is 2 μm.
DISCUSSION
Repressor-regulated efflux systems for tetracycline resistance have been extensively characterized from gram-negative bacteria and have been widely used in development of tools for conditional gene regulation. In the present study, we have demonstrated the functional activity of an analogous regulatory system from a gram-positive bacterium, Corynebacterium glutamicum, and demonstrated its potential as a tool for gene regulation in mycobacteria.
In contrast to Tn10-based Tet-systems we found that tetracycline, rather than other members of the family such as doxycycline, was the most efficient inducer of the C.glutamicum TetZ-based system. The TetR(Z) protein may have a different Tc-binding specificity to the corresponding Tn10 repressor, and some support for this comes from an examination of the protein sequence. TetR(Z) shares 43% identical amino acid residues with the two other members of the Tet-efflux family TetR(A) and TetR(G), but TetR(Z) is missing alpha-helix 9, which contains 2 out of 15 amino acids residues that are in contact with the magnesium-Tc inducer (28). It may be that these residues are important for efficient binding of other tetracyclines.
An important feature of an inducible system is the tightness of the control of expression in the absence of inducer. Incomplete repression of tetA expression by TetR is indicated by the fact that luminescence output from the pMind-Lx construct in the absence of Tc was ∼10-fold higher than that observed for luxAB in the absence of tetRO insert. A potential explanation for incomplete repression is suggested by comparison of the TetR(Z) and TetR(B) sequences. TetR(Z) contains a mutation in the DNA binding α2 helix–turn–helix motif; Thr40 of TetR(B), which contacts the GC base pair at position 6 of the operator, is changed to Gly in TetR(Z), a substitution previously shown to lead to 40-fold less efficient repression in a tetR-lacZ system in E.coli (29). It is possible that re-engineering the tetR(Z) gene to reverse this change will enhance repressor binding.
A second important feature is the magnitude of the induced response, which is in turn a function of the selected promoter region. In the present study, we have relied on the tetA promoter present in the original C.glutamicum plasmid. Luminescence output from unrepressed tetA in BCG was significantly lower than that obtained with the strong mycobacterial hsp60 promoter, and it is likely that substitution of tetA by alternative mycobacterial promoter sequences could be used to enhance the induced response.
While there is clearly scope to improve the TetZ regulatory system, in its present form it represents a significant advance on previous acetamide-based inducible expression systems available for mycobacterial genetics. It does not require specific growth media and functions in slow-growing as well as rapid-growing mycobacteria. The magnitude of induction (∼10-fold) was lower in M.tuberculosis and M.bovis BCG than in M.smegmatis (∼50-fold), and the kinetics of induction were slower. This difference may be due to a combination of factors such as the metabolic status of the cell and its permeability to Tc, and may reflect differences both in gene expression and in the enzymic activity of the luciferase reporter resulting from differences in cofactor availability. A reduction in magnitude and speed of the induction response was also observed as bacteria entered the stationary phase of growth.
We have demonstrated two features of the TetZ system that will be of particular importance in future applications to studying mycobacterial pathogenesis. First, it can be used to regulate the expression of genes in mycobacteria that are present inside eukaryotic cells during infection. The initial studies presented here demonstrate Tc-induced expression of luciferase by BCG in macrophage cultures, and comparison with other systems suggests that an extension of this approach can be used to regulate M.tuberculosis gene expression during infection in whole animal models (30–33). Second, we have shown that TetZ can be used to regulate expression of antisense mRNA, allowing knockdown of selected gene products. Using the essential ftsZ gene as proof of principle, our results show that the level of background expression in the absence of Tc is sufficiently low that it does not have a phenotypic effect, and also that the level of induction achieved by the TetZ system is sufficiently high that it results in a very marked biological phenotype. The combination of these two features of the TetZ system—allowing us to switch off target genes during infection in animal models—will have broad application in studying the biology of latent tuberculosis.
Acknowledgments
The authors thank Sabine Ehrt and Dirk Schnappinger for helpful discussions and a critical review of the manuscript. This work was funded by the Biotechnology and Biological Sciences Research Council and the Wellcome Trust. Funding to pay the Open Access publication charges for this article was provided by the Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Dye C., Scheele S., Dolin P., Pathania V., Raviglione R.C. Global burden of tuberculosis—estimated incidence, prevalence, and mortality by country. J. Am. Med. Assoc. 1999;282:677–686. doi: 10.1001/jama.282.7.677. [DOI] [PubMed] [Google Scholar]
- 2.Wayne L.G., Sohaskey C.D. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- 3.Smeulders M.J., Keer J., Speight R.A., Williams H.D. Adaptation of Mycobacterium smegmatis to stationary phase. J. Bacteriol. 1999;181:270–283. doi: 10.1128/jb.181.1.270-283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Capuano S.V., III, Croix D.A., Pawar S., Zinovik A., Myers A., Lin P.L., Bissel S., Fuhrman C., Klein E., Flynn J.L. Experimental Mycobacterium tuberculosis infection of cynomolgus macaques closely resembles the various manifestations of human M.tuberculosis infection. Infect. Immun. 2003;71:5831–5844. doi: 10.1128/IAI.71.10.5831-5844.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flynn J.L., Capuano S.V., Croix D., Pawar S., Myers A., Zinovik A., Klein E. Non-human primates: a model for tuberculosis research. Tuberculosis (Edinb.) 2003;83:116–118. doi: 10.1016/s1472-9792(02)00059-8. [DOI] [PubMed] [Google Scholar]
- 6.O'Donnell M.A., Aldovini A., Duda R.B., Yang H., Szilvasi A., Young R.A., DeWolf W.C. Recombinant Mycobacterium bovis BCG secreting functional interleukin-2 enhances gamma interferon production by splenocytes. Infect. Immun. 1994;62:2508–2514. doi: 10.1128/iai.62.6.2508-2514.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim A., Boon C., Dick T. Inducibility of the Streptomyces traRts107-Ptra expression cassette in Mycobacterium smegmatis. Biol. Chem. 2000;381:517–519. doi: 10.1515/BC.2000.066. [DOI] [PubMed] [Google Scholar]
- 8.Mahenthiralingam E., Draper P., Davis E.O., Colston M.J. Cloning and sequencing of the gene which encodes the highly inducible acetamidase of Mycobacterium smegmatis. J. Gen. Microbiol. 1993;139:575–583. doi: 10.1099/00221287-139-3-575. [DOI] [PubMed] [Google Scholar]
- 9.Parish T., Mahenthiralingam E., Draper P., Davis E.O., Colston M.J. Regulation of the inducible acetamidase gene of Mycobacterium smegmatis. Microbiology. 1997;143:2267–2276. doi: 10.1099/00221287-143-7-2267. [DOI] [PubMed] [Google Scholar]
- 10.Gordon S., Parish T., Roberts I.S., Andrew P.W. The application of luciferase as a reporter of environmental regulation of gene expression in mycobacteria. Lett. Appl. Microbiol. 1994;19:336–340. doi: 10.1111/j.1472-765x.1994.tb00469.x. [DOI] [PubMed] [Google Scholar]
- 11.Triccas J.A., Parish T., Britton W.J., Gicquel B. An inducible expression system permitting the efficient purification of a recombinant antigen from Mycobacterium smegmatis. FEMS Microbiol. Lett. 1998;167:151–156. doi: 10.1111/j.1574-6968.1998.tb13221.x. [DOI] [PubMed] [Google Scholar]
- 12.Dziadek J., Madiraju M.V., Rutherford S.A., Atkinson M.A., Rajagopalan M. Physiological consequences associated with overproduction of Mycobacterium tuberculosis FtsZ in mycobacterial hosts. Microbiology. 2002;148:961–971. doi: 10.1099/00221287-148-4-961. [DOI] [PubMed] [Google Scholar]
- 13.Greendyke R., Rajagopalan M., Parish T., Madiraju M.V. Conditional expression of Mycobacterium smegmatis dnaA, an essential DNA replication gene. Microbiology. 2002;148:3887–3900. doi: 10.1099/00221287-148-12-3887. [DOI] [PubMed] [Google Scholar]
- 14.Dziadek J., Rutherford S.A., Madiraju M.V., Atkinson M.A., Rajagopalan M. Conditional expression of Mycobacterium smegmatis ftsZ, an essential cell division gene. Microbiology. 2003;149:1593–1603. doi: 10.1099/mic.0.26023-0. [DOI] [PubMed] [Google Scholar]
- 15.Parish T., Stoker N.G. Development and use of a conditional antisense mutagenesis system in mycobacteria. FEMS Microbiol. Lett. 1997;154:151–157. doi: 10.1111/j.1574-6968.1997.tb12637.x. [DOI] [PubMed] [Google Scholar]
- 16.Narayanan S., Selvakumar S., Aarati R., Vasan S.K., Narayanan P.R. Transcriptional analysis of inducible acetamidase gene of Mycobacterium smegmatis. FEMS Microbiol. Lett. 2000;192:263–268. doi: 10.1111/j.1574-6968.2000.tb09392.x. [DOI] [PubMed] [Google Scholar]
- 17.Curcic R., Dhandayuthapani S., Deretic V. Gene expression in mycobacteria: transcriptional fusions based on xylE and analysis of the promoter region of the response regulator mtrA from Mycobacterium tuberculosis. Mol. Microbiol. 1994;13:1057–1064. doi: 10.1111/j.1365-2958.1994.tb00496.x. [DOI] [PubMed] [Google Scholar]
- 18.DeMaio J., Zhang Y., Ko C., Bishai W.R. Mycobacterium tuberculosis sigF is part of a gene cluster with similarities to the Bacillus subtilis sigF and sigB operons. Tuber. Lung Dis. 1997;78:3–12. doi: 10.1016/s0962-8479(97)90010-1. [DOI] [PubMed] [Google Scholar]
- 19.Berens C., Hillen W. Gene regulation by tetracyclines. Constraints of resistance regulation in bacteria shape TetR for application in eukaryotes. Eur. J. Biochem. 2003;270:3109–3121. doi: 10.1046/j.1432-1033.2003.03694.x. [DOI] [PubMed] [Google Scholar]
- 20.Corbel S.Y., Rossi F.M. Latest developments and in vivo use of the Tet system: ex vivo and in vivo delivery of tetracycline-regulated genes. Curr. Opin. Biotechnol. 2002;13:448–452. doi: 10.1016/s0958-1669(02)00361-0. [DOI] [PubMed] [Google Scholar]
- 21.Yin D., Ji Y. Genomic analysis using conditional phenotypes generated by antisense RNA. Curr. Opin. Microbiol. 2002;5:330–333. doi: 10.1016/s1369-5274(02)00315-6. [DOI] [PubMed] [Google Scholar]
- 22.Ehrt S., Guo Z.V., Hickey C.M., Ryou M., Monteleone M., Riley L.W., Schnappinger D. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 2005;33:e21. doi: 10.1093/nar/gni013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tauch A., Puhler A., Kalinowski J., Thierbach G. TetZ, a new tetracycline resistance determinant discovered in gram-positive bacteria, shows high homology to gram-negative regulated efflux systems. Plasmid. 2000;44:285–291. doi: 10.1006/plas.2000.1489. [DOI] [PubMed] [Google Scholar]
- 24.Snewin V.A., Gares M.P., Gaora P.O., Hasan Z., Brown I.N., Young D.B. Assessment of immunity to mycobacterial infection with luciferase reporter constructs. Infect. Immun. 1999;67:4586–4593. doi: 10.1128/iai.67.9.4586-4593.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiles S., Ferguson K., Stefanidou M., Young D.B., Robertson B.D. An alternative luciferase for monitoring bacterial cells under adverse conditions. Appl. Env. Microbiol. 2005 doi: 10.1128/AEM.71.7.3427-3432.2005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wards B.J., Collins D.M. Electroporation at elevated temperatures substantially improves transformation efficiency of slow-growing mycobacteria. FEMS Microbiol. Lett. 1996;145:101–105. doi: 10.1111/j.1574-6968.1996.tb08563.x. [DOI] [PubMed] [Google Scholar]
- 27.Mehta P.K., King C.H., White E.H., Murtagh J.J., Jr., Quinn F.D. Comparison of in vitro models for the study of Mycobacterium tuberculosis invasion and intracellular replication. Infect. Immun. 1996;64:2673–2679. doi: 10.1128/iai.64.7.2673-2679.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saenger W., Orth P., Kisker C., Hillen W., Hinrichs W. The tetracycline repressor-A paradigm for a biological switch. Angew. Chem. Int. Ed. Engl. 2000;39:2042–2052. doi: 10.1002/1521-3773(20000616)39:12<2042::aid-anie2042>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 29.Baumeister R., Helbl V., Hillen W. Contacts between Tet repressor and tet operator revealed by new recognition specificities of single amino acid replacement mutants. J. Mol. Biol. 1992;226:1257–1270. doi: 10.1016/0022-2836(92)91065-w. [DOI] [PubMed] [Google Scholar]
- 30.Saville S.P., Lazzell A.L., Monteagudo C., Lopez-Ribot J.L. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell. 2003;2:1053–1060. doi: 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji Y., Zhang B., Van Horn S.F., Warren P., Woodnutt G., Burnham M.K., Rosenberg M. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science. 2001;293:2266–2269. doi: 10.1126/science.1063566. [DOI] [PubMed] [Google Scholar]
- 32.Ji Y., Marra A., Rosenberg M., Woodnutt G. Regulated antisense RNA eliminates alpha-toxin virulence in Staphylococcus aureus infection. J. Bacteriol. 1999;181:6585–6590. doi: 10.1128/jb.181.21.6585-6590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bateman B.T., Donegan N.P., Jarry T.M., Palma M., Cheung A.L. Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect. Immun. 2001;69:7851–7857. doi: 10.1128/IAI.69.12.7851-7857.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]