Abstract
The mechanisms of response to radiation exposure are conserved in plants and animals. The DNA damage response (DDR) pathways are the predominant molecular pathways activated upon exposure to radiation, both in plants and animals. The conserved features of DDR in plants and animals might facilitate interdisciplinary studies that cross traditional boundaries between animal and plant biology in order to expand the collection of biomarkers currently used for radiation exposure monitoring (REM) in environmental and biomedical settings. Genes implicated in trans-kingdom conserved DDR networks often triggered by ionizing radiation (IR) and UV light are deposited into biological databases. In this study, we have applied an innovative approach utilizing data pertinent to plant and human genes from publicly available databases towards the design of a ‘plant radiation biodosimeter’, that is, a plant and DDR gene-based platform that could serve as a REM reliable biomarker for assessing environmental radiation exposure and associated risk. From our analysis, in addition to REM biomarkers, a significant number of genes, both in human and Arabidopsis thaliana, not yet characterized as DDR, are suggested as possible DNA repair players. Last but not least, we provide an example on the applicability of an Arabidopsis thaliana—based plant system monitoring the role of cancer-related DNA repair genes BRCA1, BARD1 and PARP1 in processing DNA lesions.
Keywords: DNA damage repair, in silico analysis, bioinformatics, ionizing radiation, ultraviolet radiation, plant radiation biodosimeter
1. Introduction
Both prokaryotic and eukaryotic cells exposed to radiation acquire different types of DNA lesions (e.g., single-strand breaks (SSB), double-strand breaks (DSB), mismatches, modified bases etc.). This genotoxic effect induced by radiation leads to the activation of DNA damage response (DDR) pathways. DDR can be defined as the sum of functions (sensors, transducers, effectors) that orchestrate DNA damage sensing and signal transduction, triggering either DNA repair, cell survival or cell death. DDR represents a prominent example of ‘DNA self-awareness’ or ‘chemical intelligence’ [1], since this evolutionarily conserved mechanism maintains genome integrity by releasing qualitative/quantitative information on a wide range of DNA lesions and activate proper responses (lesion-specific DNA repair enzymes/pathways) [1]. Of particular importance, DDR deregulation is linked to common human diseases such as cancer [2,3].
During evolution, plants have acquired a range of genes encoding products that either participate in one or more DNA repair pathways with distinct spatial/temporal expression profiles, or contribute to complex signaling pathways that mediate DNA damage sensing and activation of DDR. The conserved features of DDR might facilitate interdisciplinary studies that cross the traditional boundaries between animal and plant biology, with the aim of expanding the collection of DNA damage biomarkers currently used in environmental and biomedical settings. We suggest the potential application of a ‘plant radiation biodosimeter’ as a potential tool for exploiting plant DDR genes selected as radiation exposure monitoring (REM) biomarkers for assessing radiation risk in environments. In all cases of radiation exposure or even radiomimetic drugs, complex DNA damage is expected to be induced triggering various DNA repair pathways [4]. The main pathways for DSB repair are homologous recombination (HR) and the less precise non-homologous end joining (NHEJ). Recent advances in the discovery of new repair factors suggest differences between the human and plant systems and more insights needed. For example, a new element of the NHEJ, human PAXX (PAralog of XRCC4 and XLF, also called C9orf142) has been recently identified as a new XRCC4 superfamily member with its crystal structure to resemble that of XRCC4. PAXX has been shown to interact directly with the DSB-repair protein Ku and to be recruited at DNA-damage sites in cells [5]. As recently discussed in [6], in most cases of genotoxic stress where DSBs occur, emphasis is given on the repair of DSBs and especially at the chromatic level. Recent discoveries of new factors (like PAXX) which operate at the level of chromatin, emphasizes the concept of a dominant role of chromatin structure in the regulation of cellular DDR regulation [6].
In addition, other features of an efficient biological system/platform used as a radiation biodosimeter would be to be able to follow DNA damage related chromatin changes like nucleosome remodeling, variant histone exchange, non-histone chromatin protein mobility alteration and histone tail post-translational modification as reviewed in [7].
Herein, we describe the design of an in silico method to identify and rescue the best candidate genes for the plant radiation biodosimeter by conducting a comparative analysis of genes implicated in DDR both in animals and plants. Towards this end, the DDR-related genes of human and Arabidopsis thaliana, representing the animal and plant kingdom, respectively, were compared.
2. Plant Radiation Biodosimeter
The application of radiation exposure biomarkers is essential for evaluating cytotoxicity and genotoxicity in the human tissues in radiation oncology, as well as for biodosimetry purposes in the case of nuclear catastrophe and accidental radiation exposure. Recent extensive comparative studies on different biodosimetry approaches on the same irradiated cells underlines the necessity of employing a multiparametric approach to accomplish an accurate dose and risk estimation [8].
Plants, unlike mammals, exhibit an inherent radioresistance, a feature considered as a limiting factor for their application as ‘radiation biodosimeters’. There are many similarities between plants and humans in response to radiation. The ATR and ATM protein kinases are recognized as key players in a variety of responses to DNA damaging agents [9]. The Arabidopsis thaliana genome includes both ATR and ATM orthologs, and plants with null alleles of these genes are viable. Arabidopsis ATR/ATM mutants display hypersensitivity to γ-irradiation similarly to humans [10]. Radiation sensitivity is often associated with reduced ability to efficiently repair DSBs and/or activate DDR as reported by Borràs-Fresneda et al. [11] who compared for example radiation response in radiosensitive and radioresistant lymphocytes.
The molecular bases of natural radiotolerance have been investigated in the IR-resistant fungus Ustilago maydis which relies on the presence of a highly efficient machinery homologous recombinational (HR) repair of DNA damage, and particularly on the activity of the BRH2 gene, homolog of the human BRCA2 gene [12]. Recently, the γ-rays responsive transcriptome of the radioresistant basidiomycetous fungus Cryptococcus neoformans has been identified by Jung et al. [13] who found a novel transcription factor containing a basic leucine zipper domain, named BDR1 (bZIP TF for DNA damage response), able to modulate the expression of DNA repair genes. The BDR1 gene expression was in turn regulated by the highly conserved DDR protein kinase RAD53 [13]. Animals and plants display different levels of radiosensitivity, with a radiotolerance range of 0.001–1 and 1–100 Gy, respectively [14]. Plants have been exposed to IR, which is part of the natural background radiation, throughout evolution, with the consequent enhancement of DNA repair mechanisms necessary to cope with genotoxic stress. It has been reported that radioresistance positively correlates with genome size, since polyploidy facilitates protection against DNA damage [14,15]. Coniferous trees (e.g., pine trees) are very radiosensitive and they show severe damage leading to mortality when exposed to doses >17 Gy. On the contrary, deciduous trees (e.g., birch, alder and aspen) shed their irradiated foliage on the ground and thus can withstand radiation doses up to 90 Gy. The most radiotolerant plants are the herbaceous (weeds) and pasture plants (e.g., grasses and legumes) which are able to withstand doses up to 870 Gy [16,17].
2.1. Bioinformatics Approaches for the Identification of Candidate Genes for the Plant Radiation Dosimeter
The basic idea was to discover through in silico analysis those genes that could serve as reliable biomarkers or a ‘global tool’ for the development of a ‘non-mammalian radiation dosimetry’. Using meta-analysis and bioinformatics procedures, Nikitaki et al. [18,19] have recently identified unique human gene biomarkers specific for different types of cellular stresses (e.g., IR, replication or oxidative stress), pointing out the potential application, in terms of experimental exploitation and technical advancement, of the selected gene products in the detection of harmful environmental stresses. The investigation described herein has been expanded to develop an in silico-plant-based platform, useful for REM in a non-mammalian and relatively inexpensive model, such as plants (‘the plant radiation dosimeter’). From a biophysical point of view, IR and non-IR exhibit substantially different DNA damage patterns, thus inducing different DDR pathways dominated by distinct genes/proteins [20]. The expected differences in radiation-induced damage at the protein/lipid level could also account for different profiles of gene induction. To this end, we focused on those genes encoding products that serve as ‘exclusive’ biomarkers for the in planta detection of radiation exposure and identification of radiation quality. These universal biomarkers could be expressed in different plant populations/communities specifically linked to different geographical areas. As for radiation quality, attention was given to genes responsive to X-rays, γ-rays and non-IR (UV-A, UV-B or UV-C). In silico searches were performed on a plethora of plant species based on Gene Ontology (GO) terms. These GO terms served as filters in plant databases and, for each plant species, the corresponding gene lists were connected to the model plant Arabidopsis thaliana, based on their orthologous counterpart. Based on a protein-protein interaction network, we detected the most important orthologues in terms of functionality (nodes). The final sorting was performed according to orthologue multiplicity (number of orthologues in different species). The detailed procedure is described below.
2.2. Screening Strategy and Final Selection of Candidate Genes
2.2.1. Selection of Gene Ontology Terms
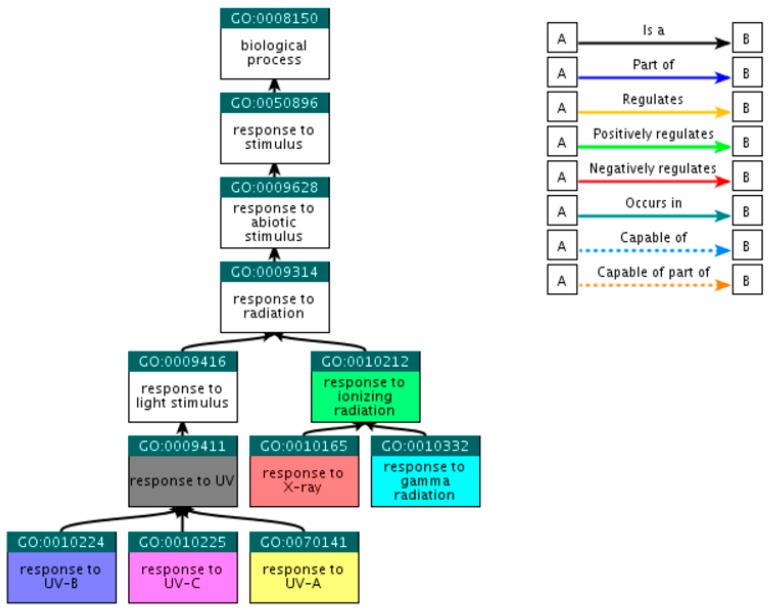
Beginning with the broad GO term ‘response to radiation’ through the QuickGO platform (http://www.ebi.ac.uk/QuickGO) [21], child GO terms that fall within the selected criteria were explored. Since QuickGO provides only the direct descendants of each term, the search for direct descendants for the resultant GO terms was repeated until all child terms of the initial input were collected. At this step, 55 GO terms were retrieved, among which only 7 were eligible as sufficient, i.e., there was no need of including further child terms (Table 1). The hierarchical relations of the selected GO terms are presented at Figure 1, including the initial GO term ‘response to radiation’; however, this term was not included in the final selection.
Table 1.
Selected Gene Ontology (GO) terms, their description and the component of the electromagnetic spectrum they represent.
| GO Term | Annotation | Category |
|---|---|---|
| GO:0010212 | response to ionizing radiation | Ionizing Radiation |
| GO:0010165 | response to X-ray | X-ray |
| GO:0010332 | response to gamma radiation | γ-ray |
| GO:0009411 | response to UV | UV |
| GO:0070141 | response to UV-A | UV-A |
| GO:0010224 | response to UV-B | UV-B |
| GO:0010225 | response to UV-C | UV-C |
Figure 1.
Ancestor tree showing the hierarchical relations of the seven selected GO terms. The 48 rest child terms, referred in the text, are not presented.
2.2.2. Orthologous Genes
In order to detect reliable and comprehensive biomarkers of assessing radiation exposure, all the available plant species from the Ensembl Plants annotation system [22] were investigated for genes orthologous to the reference plant A. thaliana. For each GO term and for every available plant (a total of 39, including the model plant), the counterpart of each orthologue was found in A. thaliana. At this step, 273 lists of genes were derived, that is, the product of the 39 plants with the seven selected GO terms. For each GO term, the corresponding 39 lists were unified, ending up with seven sets of genes with a total number of 410 different genes.
2.2.3. Exclusion of Common Genes
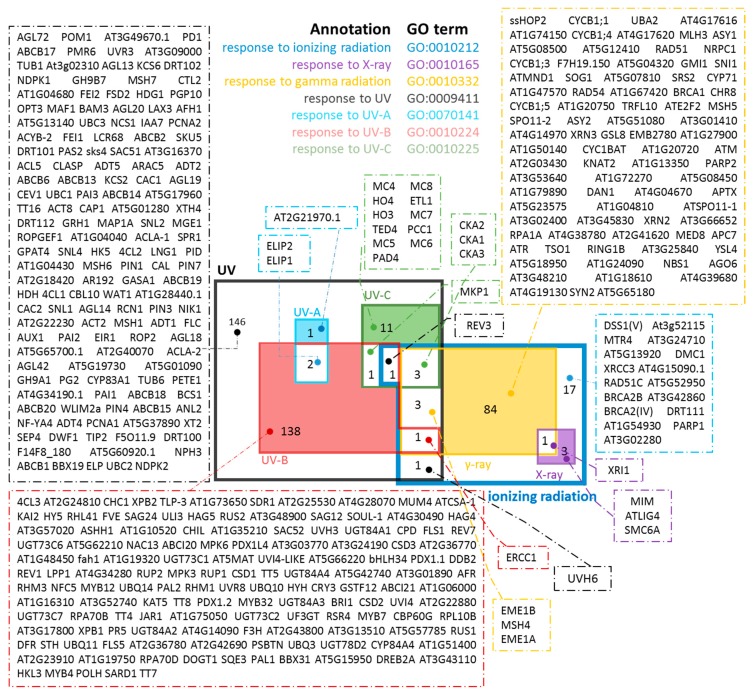
Given that in this study biodosimeters specific for radiation-quality are sought, genes categorized into more than one of the desired radiation subsets (X-ray, γ-ray, UV-A, UV-B and UV-C) had to be excluded from the subsequent steps of the analysis. Using the Draw Venn application [23], the corresponding lists of intersections among all the possible combinations of the sets were provided (Figure 2).
Figure 2.
Preliminary screening of suitable genes. Venn diagram of retrieved orthologous genes from all different plant species and for specific GO (Gene Ontology) terms. The colored regions are the suggested gene pools used for further screening, as described in Figure 3.
2.2.4. Protein-Protein Interactions Network
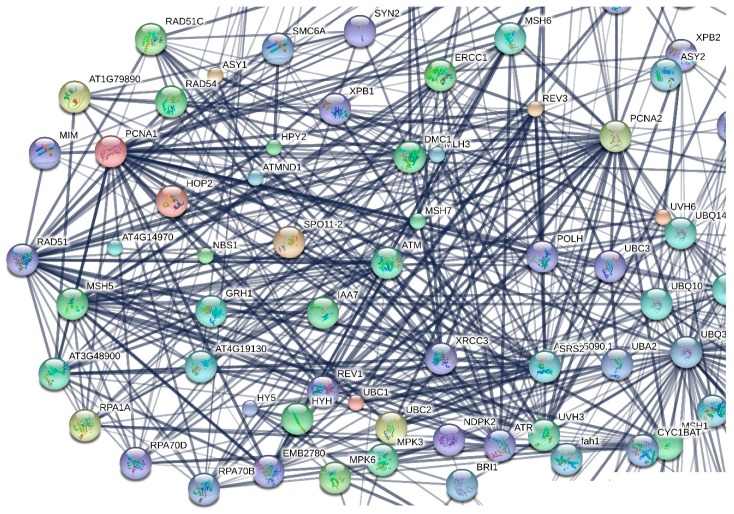
The initial screening resulted into an overall number of candidate genes that was too large to be informative. In order to reduce this number, STRING v10 (http://string-db.org) [24] was utilized. STRING V.10.0 is a database and a browser that for a given set of genes provides protein-protein interaction networks, allowing the user to set the criteria of the interaction prediction methods.
The resulting protein interaction network (a detail of which is presented in Figure 3) revealed genes/gene products that serve as nodes of dense cliques and then those genes were selected as the most critical ones for the plant functionality. The selected genes are presented in Table 2.
Figure 3.
Detail of the protein-protein interaction network, created using STRING V.10.0, where the 237 A. thaliana selected by the previous step genes were set as input. The network was rearranged in order to better identify some of the key genes.
Table 2.
Selected genes the products of which would serve as ‘exclusive’/highly specific biomarkers for the identification of radiation quality in planta, and therefore as specific indicators of the geographical region in which the ‘biosensor plant’ lives. These genes encode products that appeared as nodes of dense cliques of a protein-protein interaction network, created using STRING v10 (Figure 3). Genes are listed, along with a number, indicative of the multiplicity of their orthologues across the plant species under study. The symbol ‘#’ indicates the number of species.
| γ-rays | # | X-rays | # | UV-A | # | UV-B | # | UV-C | # |
|---|---|---|---|---|---|---|---|---|---|
| RAD54 | 30 | ATLIG4 | 28 | AT2G21970.1 | 2 | UVH3 | 32 | TED4 | 3 |
| AT4G14970 | 30 | MIM | 7 | RPA70B | 7 | MC4 | 2 | ||
| RPA1A | 27 | SMC6A | 6 | RPA70D | 5 | MC7 | 2 | ||
| MSH5 | 23 | XPB1 | 4 | MC5 | 2 | ||||
| RAD51 | 18 | XPB2 | 4 | MC6 | 2 | ||||
| AT3G48900 | 1 | MC8 | 2 | ||||||
| HO4 | 1 | ||||||||
| HO3 | 1 | ||||||||
| ETL1 | 1 | ||||||||
| PCC1 | 1 | ||||||||
| PAD4 | 1 |
Abbreviations: ETL, Enhancer Trap Locus; LIG, Ligase; MIM, Missing In Metastases; MC, MicroCystin; MSH, MutS-Homolog; PAD, Protein Arginine Aeiminase; PCC, Pathogen and Circadian Controlled; RAD, RAdiation Sensitive; RPA, Replication Protein A; SMC, Structural Maintenance of Chromosome; UVH, UltraViolet Hypersensitive; XP, Xeroderma Pigmentosum; TED, reversal of the DET phenotype.
2.2.5. Ranking Based on Multiplicity
To further reduce the number of suggested genes, candidates highlighted in the previous step were ranked according to their multiplicity of incidence among the plant species under study. Genes that have orthologues in most of the plant species under investigation (Table 2) were used for establishing the plant radiation dosimetry.
2.2.6. Human Orthologues of the Resulting Genes
The candidate genes retrieved through in silico analyses could be also experimentally validated. Towards this end, first, their distinct expression profiles in relation to the different regions of the electromagnetic spectrum need to be estimated in order to determine radiation-specific gene up-regulation. In this way, only those genes induced at a certain wavelength range would be included in the plant radiation dosimeter. Experimental validation would allow to elucidate the function of genes that are annotated to a GO term. For instance, a given gene which is annotated to the GO term ‘response to UV-B’ could likely respond to UV-C as well. Conversely, if a given gene is not annotated to a specific GO term does not necessarily mean that the gene is not classified under this term, but it has rather not been verified yet. Last but not least, in order to provide a better understanding of the ‘plant biodosimeter genes’, we searched for orthologues of the proposed plant genes (Table 2) the products of which could serve as biodosimeters in human (Table 3). In addition, we examined whether the human orthologous genes are annotated to any of the initial GO terms (Table 1), that is, we examined whether the human orthologues shown in Table 3 have been also characterized as genes involved in the response to radiation. To this end, for the genes in the second column of Table 3, the RefSeq [25] accession code of their corresponding encoded proteins was identified (Table 3, third column). For every protein, OrthoGroups were found in OrthoMCL-DB [26] (Table 3, fourth column) and scanned for orthologues in H. sapiens. The human proteins, along with their Ensembl protein identifiers (i.e., ENSP) [26], are presented in the fifth column of Table 3. The corresponding gene names (according to HUGO gene nomenclature (HGNC) [27]), were assigned to the retrieved proteins (Table 3, sixth column). Notably, among these proteins, there are also several key DSB repair proteins like RAD54, RAD51, LIG4 (DNA LIGASE 4), as well as proteins participating in HR, MMR (e.g., MSH5 or MutS-HOMOLOG 5), NER (ERCC5, ERCC3; or EXCISION REPAIR CROSS-COMPLEMENTING 5 and 3, respectively) and DDR (RPA1 or REPLICATION PROTEIN A1), further highlighting the pivotal role of the DNA damage repair components in optimized radiation biodosimetry.
Table 3.
Human orthologues of the resulting genes, proposed as exclusive biomarkers for the detection of the exposure to the several types of the electromagnetic spectrum.
| Type of Radiation | Arabidopsis thaliana | Ortho Group | Homo Sapiens | ||||
|---|---|---|---|---|---|---|---|
| TAIR | Gene Name | RefSeq | ENSP | HGNC | GO | ||
| γ-rays | RAD54 | AT3G19210 | NP_188552 | OG5_127098 | ENSP00000336606 | RAD54B | GO:0010212 |
| ENSP00000396113 | RAD54L | GO:0010212 | |||||
| AT4G14970 | NP_193233 | OG5_132711 | ENSP00000287647 | FANCD2 | GO:0010332 | ||
| RPA1A | AT2G06510 | NP_973433 | OG5_127539 | ENSP00000254719 | RPA1 | -- | |
| MSH5 | AT3G20475 | NP_188683 | OG5_129379 | ENSP00000364894 | MSH5 | -- | |
| ENSP00000387668 | MSH5 | ||||||
| ENSP00000394619 | MSH5 | ||||||
| ENSP00000394649 | MSH5 | ||||||
| ENSP00000406868 | MSH5 | ||||||
| ENSP00000407047 | MSH5 | ||||||
| ENSP00000409207 | MSH5 | ||||||
| RAD51 | AT1G07745 | NP_172254 | OG5_132909 | ENSP00000378090 | RAD51D | GO:0010212 | |
| X-rays | ATLIG4 | AT5G57160 | NP_568851 | OG5_130132 | ENSP00000402030 | LIG4 | GO:0010212 GO:0010165 GO:0010332 |
| MIM | AT5G61460 | NP_200954 | OG5_127751 | ENSP00000370672 | SMC6 | GO:0010165 | |
| SMC6A | AT5G07660 | NP_196383 | OG5_127751 | ENSP00000370672 | SMC6 | -- | |
| UV-A | SEP2 | AT2G21970 | NP_565524 | OG5_178242 | no | -- | -- |
| UV-B | UVH3 | AT3G28030 | NP_566830 | OG5_128675 | ENSP00000347978 | ERCC5 | GO:0009411 GO:0010225 |
| RPA70B | AT5G08020 | NP_196419 | OG5_127539 | ENSP00000254719 | RPA1 | -- | |
| RPA70D | AT5G61000 | NP_200908 | OG5_127539 | ENSP00000254719 | RPA1 | -- | |
| XPB2 | AT5G41360 | NP_568591 | OG5_127208 | ENSP00000285398 | ERCC3 | GO:0009411 | |
| XPB1 | AT5G41370 | NP_568592 | OG5_127208 | ENSP00000285398 | ERCC3 | -- | |
| GEN2 | AT3G48900 | NP_001118795 | OG5_174560 | no | -- | -- | |
| UV-C | TED4 | AT2G26670 | NP_001118392 | OG5_140322 | no | -- | -- |
| MC4 | AT1G79340 | NP_178052 | OG5_147205 | no | -- | -- | |
| MC8 | AT1G16420 | NP_173092 | OG5_134790 | no | -- | -- | |
| MC7 | AT1G79310 | NP_178049 | 0 | -- | -- | ||
| MC5 | AT1G79330 | NP_178051 | OG5_147205 | no | -- | -- | |
| MC6 | AT1G79320 | NP_178050 | 0 | -- | -- | ||
| HO4 | AT1G58300 | NP_176126 | OG5_140322 | no | -- | -- | |
| HO3 | AT1G69720 | NP_177130 | OG5_140322 | no | -- | -- | |
| ETL1 | AT2G02090 | NP_178318 | OG5_129286 | ENSP00000351947 | SMARCAD1 | -- | |
| PCC1 | AT3G22231 | NP_566702 | OG5_144902 | no | -- | -- | |
| PAD4 | AT3G52430 | NP_190811 | OG5_190312 | no | -- | -- | |
3. Comparison between Arabidopsis thaliana and Homo sapiens DNA Repair Mechanisms
Given that DDR pathways are the principle molecular pathways triggered following exposure to IR and non-IR, both in mammals and plants, the DNA repair mechanisms were analyzed comparatively in the model plant and animal species, Arabidopsis thaliana and human, respectively.
3.1. Comparative Analysis Strategy
3.1.1. Selection of Gene Ontology Terms
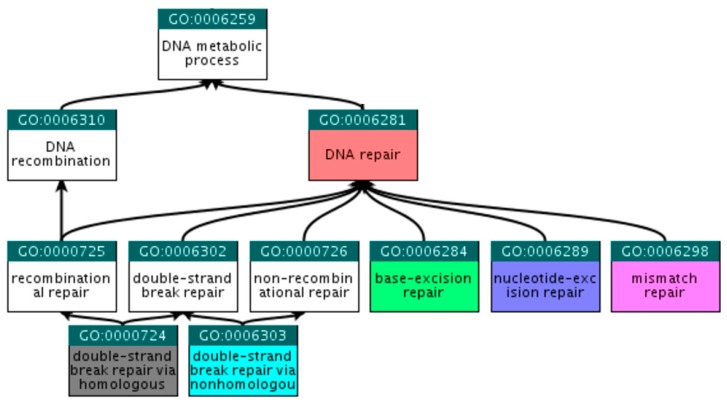
At the QuickGO (http://www.ebi.ac.uk/QuickGO) [21] platform, child terms that fall within the selected criteria were explored, beginning with the broad term ‘DNA repair’. Following steps similar to those described in Section 2.2.1, we ended up with the six GO terms presented in Table 4 and Figure 4.
Table 4.
Gene Ontology (GO) terms describing DNA repair pathways.
| GO Term | Annotation | Abbreviation |
|---|---|---|
| GO:0006281 | DNA repair | DNA repair |
| GO:0006284 | base-excision repair | BER |
| GO:0006289 | nucleotide-excision repair | NER |
| GO:0006298 | mismatch repair | MMR |
| GO:0000724 | double-strand break repair via homologous recombination | HR |
| GO:0006303 | double-strand break repair via non-homologous end joining | NHEJ |
Figure 4.
Ancestor tree showing the hierarchical relations of the six selected GO terms.
3.1.2. Human DNA Repair Genes
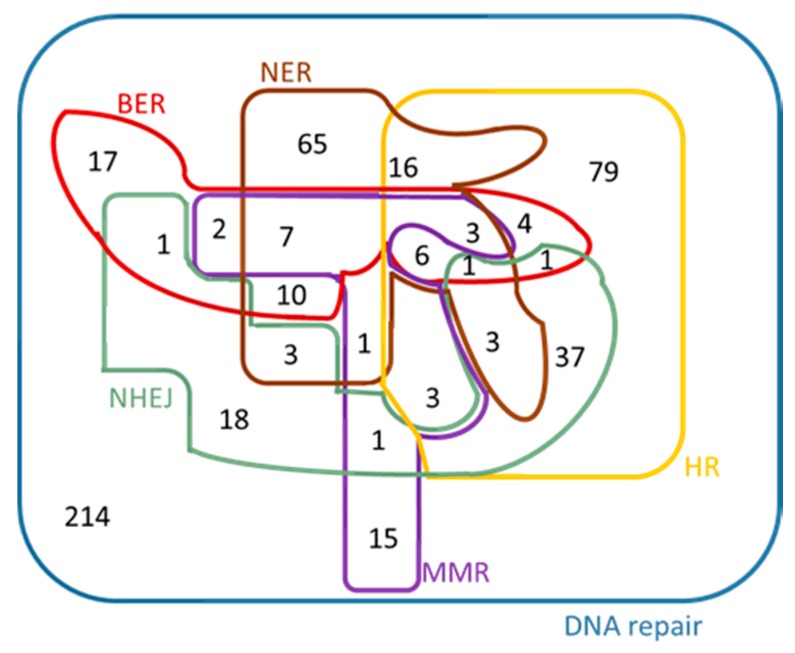
From Ensembl [28], Biomart, Ensembl Genes 83, the dataset ‘Homo sapiens genes (GRCh38.p5)’ was chosen. For each search a separate GO term listed in Table 4 was used as ‘Filter’. By choosing ’Ensembl Gene ID’ and ‘HGNC symbol’ under ‘Attributes – Features’, six ‘.txt’ files were created for Homo sapiens. The Venn diagram of the initial genes is presented in Figure 5. As it was expected, all the five DNA repair mechanisms are sub-sets of DNA repair, given that these GO terms are child terms of DNA repair (Figure 4). This Venn diagram was created manually, because it exceeds the maximum number of elements supported for automated creation by Draw Venn; however the sub-sets were determined by using Draw Venn [23]. The contents of the Venn diagram are found in the Supplementary Information Table S1.
Figure 5.
Venn diagram of the Homo sapiens genes that were found under each of the GO terms listed in Table 4.
3.1.3. Arabidopsis thaliana DNA Repair Genes
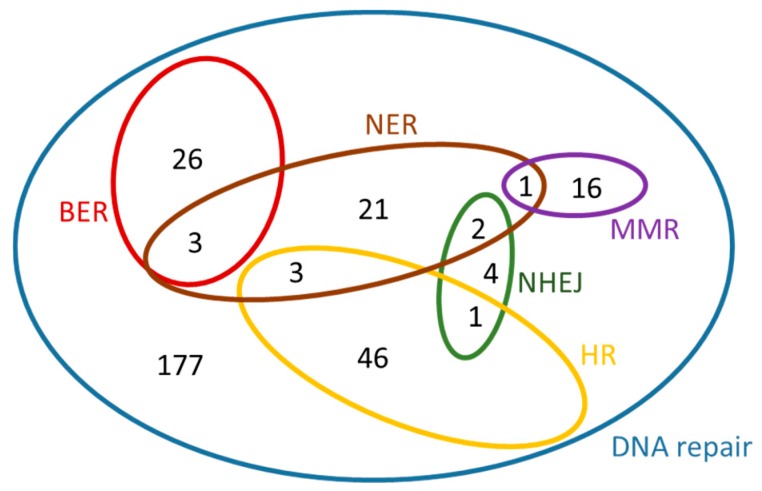
In a similar manner, for A. thaliana, the dataset ‘Arabidopsis thaliana genes (TAIR10 (2010-09-TAIR10))’ was chosen from Ensembl Plants [22], Biomart, Plant Mart. For each search, a separate GO term listed in Table 4 was used as ‘Filter’. By selecting ‘Gene stable ID’, ‘Gene name’ and ‘RefSeq protein ID’ under ‘Attributes – Features’, six ‘.txt’ files were generated for Arabidopsis thaliana. A Venn diagram for these genes is presented in Figure 6. This diagram was created manually, because it exceeds the maximum number of elements supported for automated creation by the software used, however the sub-sets were determined using the Draw Venn application [22]. The contents of this diagram can be found in the Table S2 in Supplementary Information.
Figure 6.
Venn diagram of the Arabidopsis thaliana genes that were found under each of the GO terms listed in Table 4.
3.1.4. Identification of Orthologies between Homo sapiens and Arabidopsis thaliana DNA Repair Genes
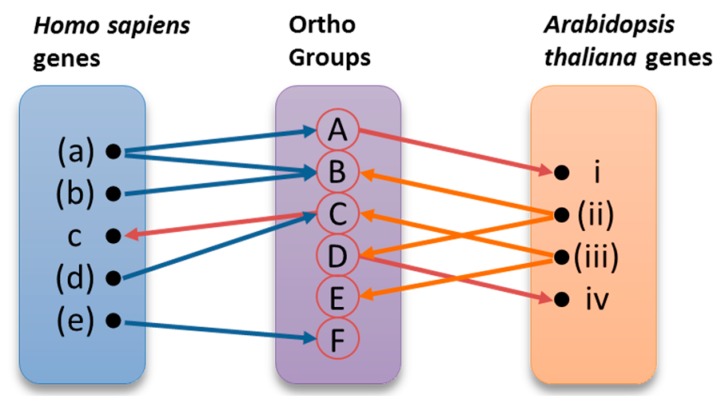
The derived twelve sets of genes were used as input to OrthoMCL-DB (http://orthomcl.cbil.upenn.edu) [29], providing for each gene a group of orthologues in the species under study. The results for the two species were collected and stored in a relational database. Data were combined in order to demonstrate known orthologies, propose new orthologies and, most importantly, suggest roles in DNA repair for orthologous genes. The orthologues pairing process, as well as the assignment of new roles to genes, are illustrated in Figure 7. Those of the initial genes that have been identified as annotated under the specific Gene Ontology (GO) terms are shown in parentheses. The same genes are represented in bold in the Supplementary Information (Tables S5–10), where the analytical results of this procedure can be found. Each one of these genes was used as input to Ortho MCL-DB, resulting to one or more Ortho Groups containing orthologous genes across several organisms. Ortho Groups A, B, and C contain both human and Arabidopsis genes. Ortho Groups D and E include only plant genes, while Ortho Group F contains only human genes. The overall procedure can be better described using the following example. As shown in Figure 7, the H. sapiens gene ‘(a)’ belongs to A and B Ortho Groups. The Arabidopsis gene ‘i’ was identified in group A, by virtue of orthology, without having been previously annotated under the initial GO term. Ortho Group B contains also the human gene ‘(a)’ and A. thaliana gene ‘(ii)’. For this reason, the previously characterized genes ‘(a)’ and ‘(ii)’ were paired as orthologues. These kinds of pairs are presented in Table 5 and Table 6. By using the A. thaliana gene ‘(ii)’ as query in OrthoMCL-BD, we identified the orthogroup D, which also contains the not yet annotated A. thaliana gene ‘iv’. On the other hand, H. sapiens gene ‘(e)’ belongs to group F, but since group F does not contain any A. thaliana gene, gene ‘(e)’ was eventually excluded from the results.
Figure 7.
Comparative analysis strategy. The orthologues pairing process and the assignment of new roles to candidate genes are represented graphically. The genes (a), (b), (d), (e), (ii), and (iii) are already characterized, while c and i are new genes.
Table 5.
DNA repair groups of orthologous genes between Homo sapiens and Arabidopsis thaliana. This table contains only the initial genes from the two organisms that have been already characterized under the GO term ‘DNA repair’ and not the genes that have arisen from this analysis.
| Homo sapiens | Arabidopsis thaliana | Homo sapiens | Arabidopsis thaliana | Homo sapiens | Arabidopsis thaliana | Homo sapiens | Arabidopsis thaliana |
|---|---|---|---|---|---|---|---|
| ERCC6 | CHR8 | GINS4 | SLD5 | GTF2H4 | AT4G17020 | ALKBH3 | ALKBH2 |
| NSMCE2 | MMS21 | UNG | ATUNG | DCLRE1A | SNM1 | XRCC1 | XRCC1 |
| RNASEH2A | AT2G25100 | APTX | BHLH140 | XRCC5 | KU80 | XRCC2 | XRCC2 |
| TOP3A | TOP3A | ALKBH1 | AT1G11780 | NSMCE1 | emb1379 | XRCC4 | XRCC4 |
| RECQL RECQL5 WRN BLM |
RECQL4A MED34 |
RAD23A RAD23B |
RAD23A RAD23B RAD23C RAD23D |
RPA1 | RPA1A RPA1B RPA1C RPA1D |
ERCC5 | UVH3 |
| OGG1 | OGG1 | ||||||
| ERCC6L2 | SWI2 | ||||||
| ERCC1 | ERCC1 | ||||||
| KAT5 | HAM2 HAM1 |
DDB1 | DDB1A DDB1B |
KIF22 | AT5G02370 | ASF1A | ASF1A ASF1B |
| MRE11A | MRE11 | ||||||
| RAD51C | RAD51C | XRCC3 | XRCC3 | DDB2 | DDB2 | GEN1 | GEN1 |
| EXO1 | AT1G18090 | MPG | MAG | RAD1 | AT4G17760 | REV3L | REV3 |
| APEX2 | ARP APE1L APE2 |
UBE2V1 | UEV1A UEV1B UEV1C |
RPS3 | RPS3A RPS3B RPS3C |
UBE2A UBE2B |
UBC1 UBC2 UBC3 |
| MLH1 | MLH1 | SHFM1 | ATDSS1(V) | POLD3 | POLD3 | XPC | ATRAD4 |
| MTOR | TOR | SSRP1 | SSRP1 | XRCC6 | KU70 | ZSWIM7 | AT4G33925 |
| CUL4ACUL4B | CUL4 | DMAP1 | SWC4 | TDP1 | TDP1 | ERCC4 | UVH1 |
| MCM8 | MCM8 | NSMCE4A | NSE4A | ERCC2 | UVH6 | ||
| RRM2B | RNR2A TSO2 |
SLX1A SLX1B |
AT2G30350 | DMC1 RAD51 |
DMC1 RAD51 |
ERCC3 | XPB2 XPB1 |
| CDC5L | CDC5 | ACTL6A | ARP4 | PARP2 | PARP2 | BRCA2 | BRCA2B |
| PMS2 | PMS1 | PNKP | ZDP | GINS2 | GINS2 | POLL | AT1G10520 |
| SUPT16H | SPT16 | ATR | ATR | POLH | POLH | FANCL | AT5G65740 |
| MSH6 | MSH6 MSH7 |
RAD9A RAD9B |
RAD9 | PCNA | PCNA PCNA2 |
LIG1 | LIG1 AT1G49250 |
| NEIL2 | FPG1 | CHAF1A | FAS1 | CDC45 | CDC45 | MSH2 | MSH2 |
| NTHL1 | NTH1 NTH2 |
POLR21 | NRPB9A NRPB9B |
GTF2H2 GTF2H2C |
ATGTF2H2 | RPA2 | RPA2A RPA2B |
| MUTYH | MYH | GTF2H1 | TFB1-1 | INO80 | INO80 | CHAF1B | FAS2 |
| PRPF19 | PRP19A | LIG4 | LIG4 | MSH3 | MSH3 | SMC5 | SMC5 |
| FEN1 | FEN1 | RFC1 | RFC1 | GTF2H3 | AT1G18340 | SMARCAD1 | ETL1 |
| RAD17 | RAD17 | PARP1 | PARP1 |
Table 6.
Sets of retrieved orthologous genes between Homo sapiens (Hs) and Arabidopsis thaliana (At) suggested to be implicated in the five main DNA repair mechanisms. For GO terms refer to Table 4. BER, base excision repair; NER, nucleotide excision repair; MMR, mismatch repair; HR, homologous recombination; NHEJ, non-homologous end joining.
| BER | NER | MMR | HR | NHEJ | |||||
|---|---|---|---|---|---|---|---|---|---|
| Hs | At | Hs | At | Hs | At | Hs | At | Hs | At |
| APEX2 | ARP APE1L APE2 |
ERCC3 | XPB2 XPB1 |
MSH6 | MSH7 MSH6 |
WRN BLM |
RECQL4A | PARP2 PARP1 |
PARP2 |
| NTHL1 | NTH2 NTH1 |
RAD23B RAD23A |
RAD23A RAD23B RAD23C RAD23D |
RNASEH2A | AT2G25100 | RAD51 DMC1 |
RAD51 | XRCC6 | KU70 |
| UNG | ATUNG | ERCC2 | UVH6 | MLH1 | MLH1 | FIGNL1 | AT3G27120 | XRCC5 | KU80 |
| FEN1 | FEN1 | GTF2H2 GTF2H2C |
ATGTF2H2 | PCNA | PCNA PCNA2 |
RAD54L RAD54B |
CHR25 | XRCC1 | XRCC1 |
| MRE11A | MRE11 | POLR2I | NRPB9A NRPB9B |
MSH2 | MSH2 | MTOR | TOR | ||
| OGG1 | OGG1 | XPC | ATRAD4 | PMS2 | PMS1 | ERCC4 | UVH1 | ||
| MUTYH | MYH | ERCC5 | UVH3 | MSH5 | MSH5 | ATR | ATR | ||
| MPG | MAG | GTF2H4 | AT4G17020 | MSH4 | MSH4 | GINS2 | GINS2 | ||
| NEIL2 | FPG1 | GTF2H3 | AT1G18340 | MSH3 | MSH3 | SMC5 | SMC5 | ||
| DDB1 | DDB1A DDB1B |
MLH3 | MLH3 | ERCC1 | ERCC1 | ||||
| GTF2H1 | TFB1-1 TFB1-3 |
GINS4 | SLD5 | ||||||
| LIG4 | LIG4 | MCM8 | MCM8 | ||||||
| POLL | AT1G10520 | MUS81 | MUS81 | ||||||
| CDC45 | CDC45 | ||||||||
| NSMCE1 | emb1379 | ||||||||
| SHFM1 | ATDSS1(V) | ||||||||
| POLD3 | POLD3 | ||||||||
| XRCC3 | XRCC3 | ||||||||
| NSMCE2 | MMS21 | ||||||||
| BRCA2 | BRCA2A BRCA2B |
||||||||
| NBN | NBS1 | ||||||||
| XRCC2 | XRCC2 | ||||||||
| ZSWIM7 | AT4G33925 | ||||||||
| RAD51B | RAD51B | ||||||||
Abbreviations: APE, Apurinic/Apyrimidinic Endonuclease; ARP, Apurinic Endonuclease-Redox protein; AT, Arabidopsis thaliana; ATR, Ataxia Telangiectasia and Rad3 Related; BLM, Bloom Syndrome RecQ like Helicase; BRCA, Breast Cancer Protein; CHR, Chromatin Remodeller; CDC, Cell Division Cycle; DDB, DNA Damage Binding Protein; DMC, Disrupted Meiotic cDNA; DSS1, Deleted in Split-Hand/Split Foot Syndrome; ERCC, Excision Repair Cross-Complementing Protein; FEN, Flap Endonuclease; FIGNL, Fidgetin-Like Protein; FPG, Formamidopyrimidine-DNA Glycosylase; GTF2H, synonimous of ERCC3; GINS, Go-Ichi-Ni-San; LIG, Ligase; MAG, Myelin-Associated Glycoprotein; MCM, Minichromosome Maintenance; MLH, MutL-homolog; MMS, Methyl Methanesulfonate Sensitive; MPG, 3-Methyladenine-DNA Glycosylase; MRE, Meiotic Recombination 11 homolog; MSH, MutS-homolog; MTOR, Mammalian Target of Rapamycin; MUS81, Structure-Specific Endonuclease Subunit; MUTYH and MYH, MuY DNA Glycosylase; NBN, Nibrin; NBS, Nijmegen Breakage Syndrome; NEIL, eukaryotic homolog of Escherichia coli endonuclese VIII (Nei); NRP, Nuclear RNA Polymerase; NSMCE, Non-smc Element 2 Mms21 homolog; NTHL, human homolog of NTH1; NTH, endonuclease III from E. coli; OGG1, 8-Oxoguanine DNA Glycosylase; PARP, Poly (ADP-ribose) Polymerase; PCNA, Proliferating Cell Nuclear Antigen; PMS, Postmeiotic Segregation Increased; RAD, Radiation Sensitive; REC, Recombinant; SLD, Synthetic Lethality with dpb11; SHF, MutS; SMC, Structural Maintenance of Chromosome; TFB, TATA-Binding Protein; TOR, Target of Rapamycin; UNG, Uracil DNA Glycosylase; UVH, Ultraviolet Hypersensitive; XRCC, X-Ray Repair Cross-Complementing; ZSWIM, Zinc Finger SWIM-Domain Containing Protein; WRN, Werner.
Of note, protein names instead of gene names were used at this step. Therefore, for Homo sapiens, the ENSP (Ensembl protein ID) [7] and for Arabidopsis thaliana, the RefSeq (Reference Sequence) [8] nomenclature was used, respectively. For instance, the corresponding proteins of the gene OGG1 (according to HGNC [9]) are ENSP00000305584 and NP_173590, in Human and Arabidopsis thaliana, respectively. Those DNA repair genes classified in orthologous groups are presented in Table 5, which contains condensed information presented in Table S5. The sets of orthologous genes between Homo sapiens (Hs) and Arabidopsis thaliana (At) retrieved for the five main DNA repair mechanisms are presented in Table 6.
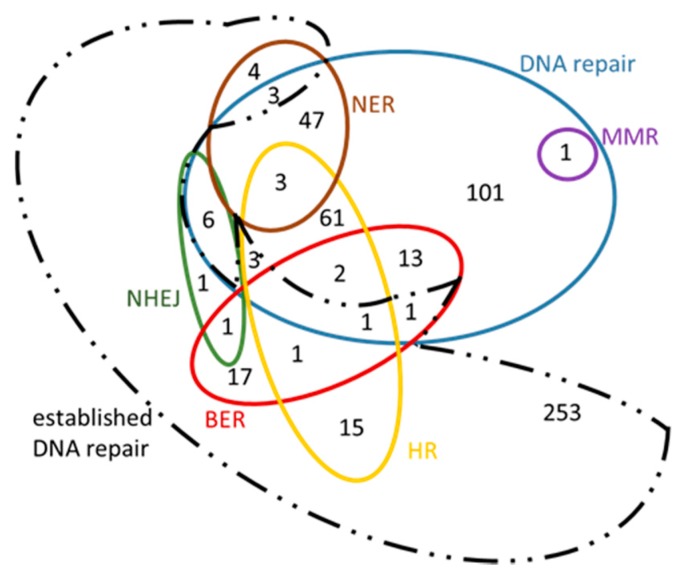
3.2. ‘New Genes’ Emerging from Comparative Analysis
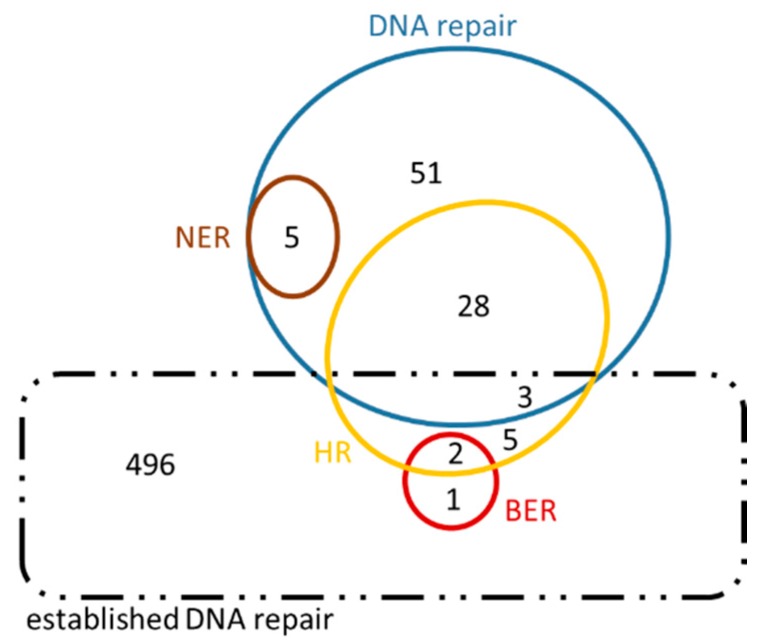
The sets of orthologous genes between H. sapiens and A. thaliana involved in the five main DNA repair mechanisms, retrieved as previously described, are reported in Table 6. Genes already known to participate in these mechanisms (termed as ‘old genes’) were included in Table 6. The bioinformatic analysis revealed a significant number of ‘new’ genes (e.g., ‘c’, ‘i' and ‘iv’ genes described in Figure 7). Analytical results are available in Supplementary Information (Tables S5–10), where detailed lists of both ‘old’ and ‘new’ genes are provided. Possible relations are presented in the form of Venn diagrams (Figure 8 and Figure 9, Supplementary Information: Tables S3 and 4). Given that the newly retrieved genes (Table 7, columns 8 and 9) have not yet been characterized and assigned to those specific GO terms, these genes are considered as novel candidate players in DNA repair. These results imply that there are unexplored areas in A. thaliana for further research and discoveries. As shown in Table 7, 300 DNA repair genes are already known, whereas 243 ‘entirely new’ genes are suggested (see also Figure 9). Of those, 87 and 8 candidates are involved in the DSBs repair pathways HR and NHEJ, respectively. These ‘new genes’ could possibly play an auxiliary or parallel role in DSB repair, as recently discovered in the case of backup NHEJ [30]. Therefore, these genes could be described as ‘genes in new roles’.
Figure 8.
Venn diagram of Homo sapiens genes in their putative ‘new roles’. This diagram also contains an intersection of the genes in ‘new roles’ with ‘established DNA repair’ genes, in order to highlight the great number of genes arisen from this analysis that have not been previously characterized under the inclusive term ‘DNA repair’ (dashed black line).
Figure 9.
Venn diagram of Arabidopsis thaliana genes in their ‘new roles’. This diagram also contains the intersection with established DNA repair genes in order to highlight the great number of genes emerged from this analysis that have not been previously characterized under the inclusive term ‘DNA repair’ (dashed black line).
Table 7.
Quantitative results obtained from the comparative analysis of DNA repair mechanisms in plants and humans. In the first column, the various ‘lemmas’ (annotations) for DNA repair and its subpathways are presented, accompanied with the corresponding GO terms. The 2nd column is referred to the number of the already characterized Homo sapiens (Hs) genes. The 3rd refers to those genes of the 2nd column that have orthologues (one or more) in Arabidopsis thaliana (At). The 4th column is just the percentage (those that have orthologues/all genes). Correspondingly, 5th–7th columns refer to Arabidopsis thaliana. Underlined (8th–9th column) are the ‘new’ genes identified from our analysis for Hsa and Atha respectively. BER, base excision repair. NER, nucleotide excision repair. MMR, mismatch repair. HR, homologous recombination. NHEJ, non-homologous end joining.
| 1. Mechanism (GO Term) |
2. # Hs Genes | 3. # Hs Genes That Have Orthologues in At | 4. % | 5. # At Genes | 6. # At Genes That Have Orthologues in Hs | 7. % | 8. # Suggested Genes Arisen from Our Analysis for Hs | 9. # Suggested Genes Arisen from Our Analysis for At |
|---|---|---|---|---|---|---|---|---|
|
DNA repair
GO:0006281 |
507 | 259 | 51.1 | 300 | 185 | 61.7 | 86 | 243 |
|
BER
GO:0006284 |
52 | 32 | 61.5 | 29 | 12 | 41.4 | 3 | 36 |
|
NER
GO:0006289 |
124 | 59 | 47.6 | 30 | 21 | 70.0 | 5 | 57 |
|
MMR
GO:0006298 |
43 | 14 | 32.6 | 17 | 12 | 70.6 | 0 | 1 |
|
HR
GO:0000724 |
162 | 76 | 46.9 | 50 | 37 | 74.0 | 38 | 87 |
|
NHEJ
GO:0006303 |
73 | 9 | 12.3 | 7 | 4 | 57.1 | 0 | 8 |
| Total number of ‘DNA repair’ genes associated with “new roles” arisen from this analysis (see Figure 8 and Figure 9) | 95 | 281 | ||||||
| Total number of “entirely new” genes that have arisen from this analysis after the comparison with the “previously established” DNA repair genes | 84 | 234 | ||||||
Our results are presented in Venn diagrams (Figure 8 and Figure 9), in a concise manner, avoiding the repetition of the common genes among the mechanisms. For the sake of completeness, we have included in these diagrams the intersection with the initial DNA repair genes, referred to as ‘established DNA repair genes’ (black dashed line in Figure 8 and Figure 9). In these Venn diagrams, only the genes of Supplementary Information: Tables S5–S10 that are not in bold, i.e., those genes that have arisen from the present analysis, are included. Thus, it is not surprising that some genes are found outside the DNA repair set (blue line Figure 8 and Figure 9) and they therefore fall into the ‘established DNA repair’ group.
4. An In Vitro Approach Monitoring Key DNA Repair Genes
Germline variants in the human BRCA1 gene are associated with familial breast and ovarian cancers [31]. The human BARD1 (BRCA1-associated RING domain protein 1) is essential for the sequestration of BRCA1 at DNA damage sites [32]. Moreover, the DDR-related protein PARP1 (Poly(ADP-ribose) polymerase-1) [33] was shown to mediate the function of BRCA1 in DDR [32]. Herein, the DNA-CL (crosslinks) comet assay, an assay modified for detection of DNA interstrand cross-links, was used to assess the effect of the Arabidopsis thaliana genes BRCA1, BARD1 and PARP1 on DNA damage repair. To this end, the alkaline/neutral (A/N) protocol of comet assay described in Angelis et al. [34], with the additional enzyme treatment step, was employed. Isolated nuclei from chopped Arabidopsis thaliana seedlings were embedded into a 0.7% agarose gel and lysed for 1 h. Then, the lysed nuclei were enzymatically treated. In particular, they were equilibrated for 20 min in a restriction endonuclease Sal1 buffer and then 50 μL of Sal1 solution (1U/ml) were added. Each gel was then spread on microscopic slides, covered with Parafilm and incubated in a sterile moist chamber for 50 min at 37 °C. The restriction enzyme digestion was stopped using TE buffer. Following enzyme treatment, the slides were dipped for 20 min into a DNA-unwinding solution (0.3 M NaOH; 10 mM EDTA), neutralized for 5 min in 1× TBE buffer and electrophoresed in the same buffer at 1 Volt/cm for 5 min.
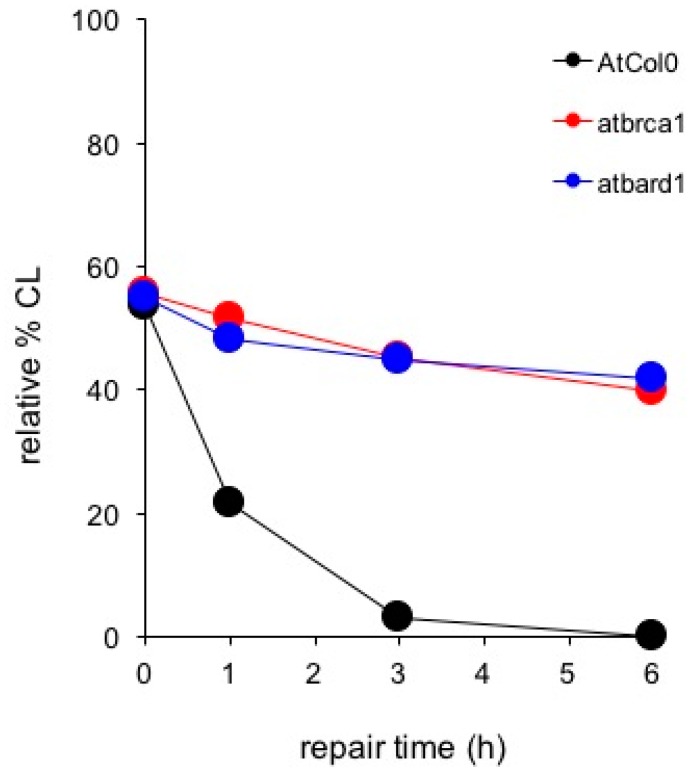
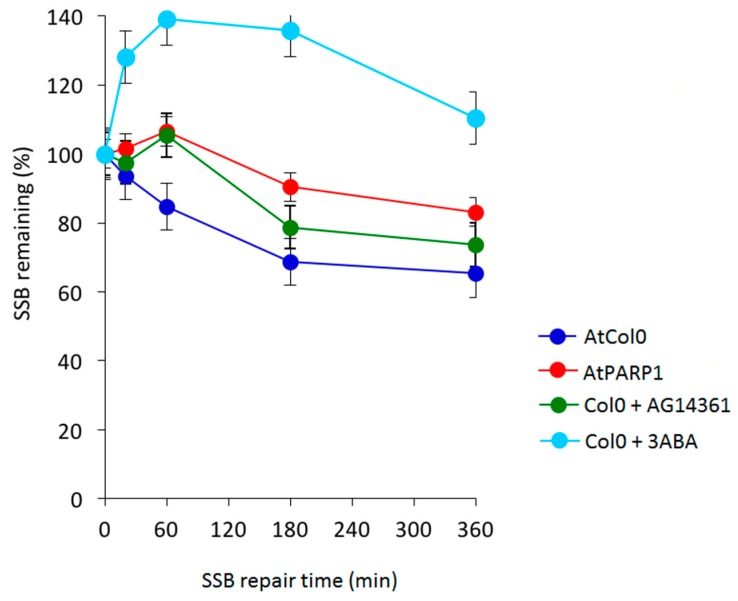
As shown in Figure 10, comparison of DDR kinetics revealed similarly reduced ability of the AtBRCA1 and AtBARD1 mutants to remove CL. Despite of the fact that wild type AtCol0 removed all CL after 3 h of DDR, residual DNA damage was observed in mutant AtBRCA1. The half-life of CL in AtBRCA1 and AtBARD1 (t1/2 = 10 h) is approximately 10 times longer compared to Col0 (t1/2 = 1.2 h). Therefore, AtBRCA1 and AtBARD1 can efficiently repair DNA interstrand cross-linked adducts generated by mitomycin C. On Figure 11 is shown the effect of SSB accumulation during the early stages of base excision repair (BER) recovery in Arabidopsis plants due to PARP1 impairment either by a knockout mutation leading to AtPARP1 or by two PARP1 inhibitors, the selective PARP1 inhibitor AG14361, developed by Pfizer for the sensitization of human breast cancer cells prior to irradiation treatment, or the non-specific PARP inhibitor 3-aminobenzamide (3-ABA). Of interest, the PARP1-mediated signaling is conserved among kingdoms, since the selective AG14361 inhibitor of HsPARP1 is also effective in Arabidopsis and exhibits the same repair kinetic behavior as the knockout AtPARP1 mutation, contrary to 3-ABA. The above observations lead to the suggestion that the genes AtBRCA1, AtBARD1 and AtPARP1 play an equally important role in DNA damage repair in plants, like Arabidopsis thaliana, as in animals and humans.
Figure 10.
Comet assay depicting DNA damage repair in wild-type Arabidopsis thaliana (AtCol0) and the knockout DNA repair mutants atbrca1 and atbard1 following treatment with 200 μM Mitomycin C and post-treatment recovery for 1, 3 and 6 h.
Figure 11.
Effect of the knockdown mutation of AtPARP1 and of specific and broad spectrum inhibitors of PARP1 on SSB repair kinetics obtained by an A/N comet assay. SSBs generated following 1 hr treatment with 2 mM MMS (methyl methanesulfonate) in AtPARP1 and in Arabidopsis Col1, in the presence of 3 mM 3-aminobenzamide (3-ABA) and 10 μM HsPARP1 specific AG14361 inhibitors.
5. Conclusions
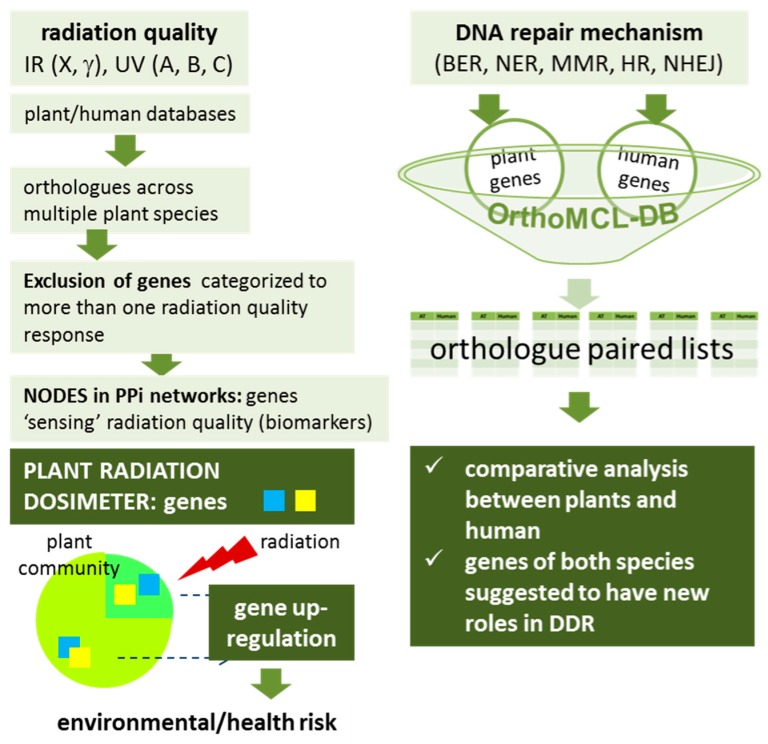
The systematic bioinformatic approach employed in this study to select candidate genes for the ‘plant radiation dosimeter’ (Figure 12) revealed that, despite the fact that the last common ancestor of human and A. thaliana is traced approximately 1.5 billion years ago [35], fundamental mechanisms underlying the maintenance of genome integrity, as well as their associated genes, are conserved between animals and plants. In particular, we have identified plant genes with human counterparts that can be used as ‘signature radiation genes’ in order to allow direct comparisons on the primary mechanisms governing the DNA damage repair response to different types of radiation (ionizing and non-ionizing) across the tree of life. The expression patterns (up- or down-regulation) of the genes which have been classified in each part of the electromagnetic spectrum should be evaluated experimentally after exposure of the plant only to a certain component of the electromagnetic spectrum. It is expected that some of the proposed genes could actually be exploited as biomarkers of exposure to electromagnetic radiation, for assessing radiation risk in environment. The main conclusion is that the results from the in silico analysis performed herein are expected to provide the foundation for future research efforts in order to design a radiation biodosimeter. Apart from the REM biomarkers, a large number of putative genes suggested to participate in DDR was also identified in human and Arabidopsis thaliana. Our studies support the further development of a plant-based radiation biodosimeter. We believe, that the importance of using a low-cost non-animal system to monitor and estimate radiation risk is high since it helps in establishing a reliable methodology avoiding all the ethical issues associated in most cases with the use of animals or human cells. In addition, this plant-based platform maybe used in other cases for the screening of specialized drugs targeting for example DNA repair like PARP inhibitors, predict response to inhibitors of the DNA damage sensors ATM and ATR, and inhibitors of nonhomologous end joining etc. As recently discussed in Stover et al. [36], producing and validating reliable biomarkers will help boost the efficiency of DNA repair targeted therapies and exploit their role(s) on cancer treatment. Also in this case, plants may prove as a first-step screening tool on all of the above cases.
Figure 12.
Schematic representation of the in silico methodology designed to select candidate genes for the plant radiation dosimeter as well as for the comparison of human and plant DNA repair machinery. BER, base excision repair. NER, nucleotide excision repair. MMR, mismatch repair. HR, homologous recombination. NHEJ, non-homologous end joining. DDR, DNA damage response. IR, ionizing radiation, UV, ultraviolet radiation. PPi, protein-protein interaction. Please see recent work by Pateras et al. [20], for analytical description of all DDR pathways.
Acknowledgments
Sponsorship from COST Action CM1201: ‘Biomimetic Radical Chemistry’ and LTC17047 hCOMET is gratefully acknowledged. This work was also supported by funds provided by the EU grant MC-CIG-303514 and DAAD grant “DNA Damage and Repair and Their Relevance to Carcinogenesis” (No. 57339330). K.J.A. and M.H. would like to acknowledge support by the Czech Science Foundation (16-01137S).
Abbreviations
| DDR | DNA damage response |
| SSB | single-strand breaks |
| DSB | double-strand breaks |
| NER | nucleotide excision repair |
| BER | base excision repair |
| MMR | Mismatch repair |
| HR | homologous recombination |
| NHEJ | non-homologous end joining |
| UV | ultraviolet radiation |
| IR | ionizing radiation |
| PAXX | PAralog of XRCC4 and XLF |
| REM | radiation exposure monitoring |
Supplementary Materials
The following are available online at http://www.mdpi.com/2072-6694/9/6/65/s1. Table S1: Human DNA repair genes. Contents of the Venn diagram presented in Figure 5 in the main text, Table S2: Arabidopsis thaliana DNA repair genes. Contents of the Venn diagram presented in Figure 6 in the main text, Table S3: Homo sapiens genes that stemmed from the present analysis (i.e., contents of the Venn diagram shown in Figure 8 in the main text), Table S4: Arabidopsis thaliana genes that stemmed from the present analysis (i.e., contents of the Venn diagram shown in Figure 9 in the main text), Table S5. DNA repair genes in human and Arabidopsis thaliana, grouped according to orthology, Table S6: BER genes in human and Arabidopsis thaliana, grouped according to orthology, Table S7: NER genes in human and Arabidopsis thaliana, grouped according to orthology, Table S8: MMR genes in human and Arabidopsis thaliana, grouped according to orthology, Table S9. HR genes in human and Arabidopsis thaliana, grouped according to orthology, Table S10: NHEJ genes in human and Arabidopsis thaliana, grouped according to orthology.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Elledge S.J. The DNA damage response--Self-awareness for DNA: The 2015 albert lasker basic medical research award. JAMA. 2015;314:1111–1112. doi: 10.1001/jama.2015.10387. [DOI] [PubMed] [Google Scholar]
- 2.Bouwman P., Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat. Rev. Cancer. 2012;12:587–598. doi: 10.1038/nrc3342. [DOI] [PubMed] [Google Scholar]
- 3.Jeggo P.A., Pearl L.H., Carr A.M. DNA repair, genome stability and cancer: A historical perspective. Nat. Rev. Cancer. 2016;16:35–42. doi: 10.1038/nrc.2015.4. [DOI] [PubMed] [Google Scholar]
- 4.Georgakilas A.G., O’Neill P., Stewart R.D. Induction and repair of clustered DNA lesions: What do we know so far? Radiat. Res. 2013;180:100–109. doi: 10.1667/RR3041.1. [DOI] [PubMed] [Google Scholar]
- 5.Ochi T., Blackford A.N., Coates J., Jhujh S., Mehmood S., Tamura N., Travers J., Wu Q., Draviam V.M., Robinson C.V., et al. DNA repair. Paxx, a paralog of xrcc4 and xlf, interacts with ku to promote DNA double-strand break repair. Science. 2015;347:185–188. doi: 10.1126/science.1261971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raschella G., Melino G., Malewicz M. New factors in mammalian DNA repair-the chromatin connection. Oncogene. 2017 doi: 10.1038/onc.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polo S.E., Jackson S.P. Dynamics of DNA damage response proteins at DNA breaks: A focus on protein modifications. Genes Dev. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeegers D., Venkatesan S., Koh S.W., Low G.K., Srivastava P., Sundaram N., Sethu S., Banerjee B., Jayapal M., Belyakov O., et al. Biomarkers of ionizing radiation exposure: A multiparametric approach. Genome Integr. 2017;8:6. doi: 10.4103/2041-9414.198911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiloh Y., Ziv Y. The atm protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013;14:197–210. doi: 10.1038/nrm3546. [DOI] [PubMed] [Google Scholar]
- 10.Culligan K.M., Robertson C.E., Foreman J., Doerner P., Britt A.B. Atr and atm play both distinct and additive roles in response to ionizing radiation. Plant J. Cell Mol. Biol. 2006;48:947–961. doi: 10.1111/j.1365-313X.2006.02931.x. [DOI] [PubMed] [Google Scholar]
- 11.Borras-Fresneda M., Barquinero J.F., Gomolka M., Hornhardt S., Rossler U., Armengol G., Barrios L. Differences in DNA repair capacity, cell death and transcriptional response after irradiation between a radiosensitive and a radioresistant cell line. Sci. Rep. 2016;6:27043. doi: 10.1038/srep27043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu E.Y., Kojic M., Holloman W.K., Lue N.F. Brh2 and rad51 promote telomere maintenance in ustilago maydis, a new model system of DNA repair proteins at telomeres. DNA Repair. 2013;12:472–479. doi: 10.1016/j.dnarep.2013.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung K.W., Yang D.H., Kim M.K., Seo H.S., Lim S., Bahn Y.S. Unraveling fungal radiation resistance regulatory networks through the genome-wide transcriptome and genetic analyses of cryptococcus neoformans. mBio. 2016;7 doi: 10.1128/mBio.01483-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L.H., Pfister T.D., Parchment R.E., Kummar S., Rubinstein L., Evrard Y.A., Gutierrez M.E., Murgo A.J., Tomaszewski J.E., Doroshow J.H., et al. Monitoring drug-induced gammah2ax as a pharmacodynamic biomarker in individual circulating tumor cells. Clin. Cancer Res. 2010;16:1073–1084. doi: 10.1158/1078-0432.CCR-09-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoenfelder K.P., Fox D.T. The expanding implications of polyploidy. J. Cell Biol. 2015;209:485–491. doi: 10.1083/jcb.201502016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geras’kin S.A. Ecological effects of exposure to enhanced levels of ionizing radiation. J. Environ. Radioact. 2016;162–163:347–357. doi: 10.1016/j.jenvrad.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 17.Bin P., Zheng Y. Association between gamma-h2ax and DNA double-strand breaks. Wei Sheng Yan Jiu. 2007;36:520–522. [PubMed] [Google Scholar]
- 18.Nikitaki Z., Hellweg C., Georgakilas A.G., Ravanat J.L. Stress-induced DNA damage biomarkers: Applications and limitations. Front. Chem. 2015;3:35–50. doi: 10.3389/fchem.2015.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikitaki Z., Mavragani I.V., Laskaratou D.A., Gika V., Moskvin V.P., Theofilatos K., Vougas K., Stewart R.D., Georgakilas A.G. Systemic mechanisms and effects of ionizing radiation: A new ‘old’ paradigm of how the bystanders and distant can become the players. Semin. Cancer Biol. 2016;37–38:77–95. doi: 10.1016/j.semcancer.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Pateras I.S., Havaki S., Nikitopoulou X., Vougas K., Townsend P.A., Panayiotidis M.I., Georgakilas A.G., Gorgoulis V.G. The DNA damage response and immune signaling alliance: Is it good or bad? Nature decides when and where. Pharmacol. Ther. 2015;154:36–56. doi: 10.1016/j.pharmthera.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 21.Binns D., Dimmer E., Huntley R., Barrell D., O'Donovan C., Apweiler R. Quickgo: A web-based tool for gene ontology searching. Bioinformatics. 2009;25:3045–3046. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolser D., Staines D.M., Pritchard E., Kersey P. Ensembl plants: Integrating tools for visualizing, mining, and analyzing plant genomics data. Methods Mol. Biol. 2016;1374:115–140. doi: 10.1007/978-1-4939-3167-5_6. [DOI] [PubMed] [Google Scholar]
- 23.Bioinformatics & Evolutionary Genomics Calculate and Draw Custom Venn Diagrams. [(accessed on 6 October 2016)]; Available online: http://bioinformatics.psb.ugent.be/webtools/Venn/
- 24.Lobachevsky P.N., Martin R.F. Plasmid DNA breakage by decay of DNA-associated auger emitters: Experiments with 123i/125i-iodohoechst 33258. Int. J. Radiat. Biol. 2004;80:915–920. doi: 10.1080/09553000400017754. [DOI] [PubMed] [Google Scholar]
- 25.Pruitt K.D., Tatusova T., Maglott D.R. Ncbi reference sequence (refseq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hubbard T., Barker D., Birney E., Cameron G., Chen Y., Clark L., Cox T., Cuff J., Curwen V., Down T., et al. The ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Povey S., Lovering R., Bruford E., Wright M., Lush M., Wain H. The hugo gene nomenclature committee (hgnc) Hum. Genet. 2001;109:678–680. doi: 10.1007/s00439-001-0615-0. [DOI] [PubMed] [Google Scholar]
- 28.Lobachevsky P.N., White J., Leung M., Skene C., White J., Martin R.F. Plasmid breakage by (125)i-labelled DNA ligands: Effect of DNA-iodine atom distance on breakage efficiency. Int. J. Radiat. Biol. 2008;84:991–1000. doi: 10.1080/09553000802478091. [DOI] [PubMed] [Google Scholar]
- 29.Chen F., Mackey A.J., Stoeckert C.J., Jr., Roos D.S. Orthomcl-db: Querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iliakis G., Murmann T., Soni A. Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombination repair: Implications for the formation of chromosome translocations. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015;793:166–175. doi: 10.1016/j.mrgentox.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 31.Welcsh P.L., King M.C. Brca1 and brca2 and the genetics of breast and ovarian cancer. Hum. Mol. Genet. 2001;10:705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]
- 32.Li M., Yu X. Function of brca1 in the DNA damage response is mediated by adp-ribosylation. Cancer Cell. 2013;23:693–704. doi: 10.1016/j.ccr.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dantzer F., Ame J.C., Schreiber V., Nakamura J., Menissier-de Murcia J., de Murcia G. Poly(adp-ribose) polymerase-1 activation during DNA damage and repair. Methods Enzymol. 2006;409:493–510. doi: 10.1016/S0076-6879(05)09029-4. [DOI] [PubMed] [Google Scholar]
- 34.Angelis K.J., Dusinska M., Collins A.R. Single cell gel electrophoresis: Detection of DNA damage at different levels of sensitivity. Electrophoresis. 1999;20:2133–2138. doi: 10.1002/(SICI)1522-2683(19990701)20:10<2133::AID-ELPS2133>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 35.Hedges S.B., Dudley J., Kumar S. Timetree: A public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- 36.Stover E.H., Konstantinopoulos P.A., Matulonis U.A., Swisher E.M. Biomarkers of response and resistance to DNA repair targeted therapies. Clin. Cancer Res. 2016;22:5651–5660. doi: 10.1158/1078-0432.CCR-16-0247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.