ABSTRACT
Oral antibiotics such as metronidazole, vancomycin and fidaxomicin are therapies of choice for Clostridium difficile infection. Several important mechanisms for C. difficile antibiotic resistance have been described, including the acquisition of antibiotic resistance genes via the transfer of mobile genetic elements, selective pressure in vivo resulting in gene mutations, altered expression of redox-active proteins, iron metabolism, and DNA repair, as well as via biofilm formation. This update summarizes new information published since 2010 on phenotypic and genotypic resistance mechanisms in C. difficile and addresses susceptibility test methods and other strategies to counter antibiotic resistance of C. difficile.
KEYWORDS: Clostridium difficile, antibiotics, drug resistance, testing, biofilm
INTRODUCTION
Clostridium difficile infection (CDI) leads to approximately 453,000 cases and 29,000 deaths yearly in the United States as reported by the Centers for Disease Control and Prevention (CDC) in 2015 (1) and has become the most common health care-associated infection in the United States and the most frequent hospital-acquired intestinal infection in Europe and worldwide (2). The prevalence of C. difficile outbreaks caused by ribotype 027 since the early 2000s has resulted in higher morbidity and mortality along with increasing medical costs throughout the world (3, 4).
CDI is typically caused by the exposure of the normal intestinal microbiota to antibiotics that are not active against C. difficile, which disrupts this flora and allows for proliferation of C. difficile (5). Many antibiotics are associated with CDI; ampicillin, amoxicillin, cephalosporins, clindamycin, and fluoroquinolones continue to be associated with the highest risk for CDI (6) (Table 1). The usual treatment for primary and recurrent CDI requires the use of antibiotics with activities against C. difficile, and includes metronidazole, vancomycin, and fidaxomicin (6–10). The choice of antibiotic treatment is dependent on the severity of CDI as per the recommendations of the Infectious Diseases Society of America (IDSA) and the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) (9, 11, 12). The emergence and spread of C. difficile isolates resistant to multiple antibiotics, especially among the hypervirulent C. difficile ribotype 027 strains, are now becoming an increasing problem for the treatment of CDI (13, 14). Finally, the spores formed by C. difficile also may allow it to survive antimicrobial therapy and thus lead to treatment failure.
TABLE 1.
Examples of current and future antibiotics useful for Clostridium difficile infections
| Antibiotic | Target | Putative resistance mechanism(s) | Reference(s) |
|---|---|---|---|
| Metronidazole | Bacterial DNA, causing DNA breakage and destabilization of the DNA helix | Alterations in some metabolic pathways, biofilm formation | 13, 42, 43, 49, 50, 52, 53 |
| Vancomycin | d-Ala-d-Ala subunit of the precursor UDP-N-acetylmuramyl pentapeptide of peptidoglycan | Mutations in peptidoglycan biosynthesis-required proteins, biofilm formation | 33 |
| Fidaxomicin | Bacterial RNA polymerase | Mutations in rpoB | 33 |
| Rifamycins | β-Subunit of DNA-dependent RNA polymerase | Mutations in rpoB | 44 |
| Ramoplanin | Inhibiting peptidoglycan biosynthesis | Not reported | |
| Fusidic acid | Inhibiting protein synthesis by binding elongation factor G on the ribosome | Mutations in fusA | 69 |
| Nitazoxanide | Pyruvate, ferredoxin oxidoreductase | Not reported | |
| Tigecycline | 30S ribosomal subunit | Not reported | |
| Cadazolid | Bacterial protein synthesis and DNA synthesis | Not reported | |
| Surotomycin | Disrupting the membrane potential | Not reported | |
| Ridinilazole (SMT19969) | Inhibits DNA synthesis | Not reported | |
| CRS3123 (REP3123) | Methionyl-tRNA synthetase (MetRS) inhibitor | Not reported |
CURRENT STATUS OF ANTIMICROBIAL RESISTANCE OF CLOSTRIDIUM DIFFICILE
Antibiotic use is thought to be the most important risk factor for CDI (6). However, C. difficile is a spore-forming organism; spores may survive antimicrobial therapy and may germinate and cause relapse of CDI after the cessation of therapy. C. difficile is known to be resistant to multiple antibiotics, such as aminoglycosides, lincomycin, tetracyclines, erythromycin, clindamycin, penicillins, cephalosporins, and fluoroquinolones, which are commonly used in the treatment of bacterial infections in clinical settings (15, 16). Recent statistics based on 30 antimicrobial susceptibility studies of C. difficile clinical isolates published between 2012 and 2015 reveal that resistance to clindamycin (8.3% to 100%), cephalosporins (51%), erythromycin (13% to 100%), and fluoroquinolones (47%) is commonly seen in C. difficile clinical isolates based on CLSI or EUCAST breakpoints (16). Clindamycin, cephalosporins, and fluoroquinolones are known to promote CDI (15–17). Among cephalosporins and fluoroquinolones, resistance to the second-generation cephalosporins (cefotetan and cefoxitin) and fluoroquinolones (ciprofloxacin) is very common (79% and 99% of the strains tested, respectively); while a certain percentage of C. difficile shows resistance to third-generation cephalosporins (ceftriaxone and cefotaxime; 38% of the strains tested) and broad-spectrum fluoroquinolones (moxifloxacin and gatifloxacin; 34% of the strains tested) (16).
Multiple studies on the antimicrobial resistance of C. difficile isolates from North America, Europe, and Asia in the last 15 years have demonstrated that the rates of moxifloxacin resistance of C. difficile isolates varied from 2% to 87%, and the rates of clindamycin resistance ranged from 15% to 97% (13). Almost 30% of ribotype 027 strains were resistant to multiple drugs, including clindamycin, moxifloxacin, and rifampin in North America, using the CLSI breakpoints for susceptibility testing of anaerobic bacteria (13). In a retrospective study of the antibiotic resistance pattern in the United States, approximately 98% of ribotype 027 strains were resistant to moxifloxacin; moreover, almost half of these isolates possessed high-level resistance based on the CLSI breakpoint (18). C. difficile strains of ribotype 078 (another hypervirulent genotype) isolated from humans and piglets in the Netherlands with active CDI showed resistance to ciprofloxacin, erythromycin, imipenem, and moxifloxacin according to the CLSI breakpoints (19). Worldwide surveillance also indicated the emergence of C. difficile strains resistant to multiple antibiotics in the past decade (16, 20–22).
The resistance of C. difficile to commonly used antibiotics for bacterial infections not only contributes to the occurrence/recurrence of CDI but also plays an important role in driving epidemiological changes and the emergence of new strain types (16). A representative example is the emergence and global spread of hypervirulent C. difficile 027/BI/NAP1 strains, which are thought to have a certain correlation with the widespread and frequent use of fluoroquinolones (14, 16). Antibiotic resistance to C. difficile also leads to suboptimal clinical outcomes and may even lead to treatment failures of CDI. When uncommon antibiotics are chosen for the treatment of CDI, collateral damage to microbiota may occur and should not be ignored.
Metronidazole and vancomycin remain the first line of antibiotics used for the treatment of CDI (6, 9). While still effective for most cases of CDI, C. difficile isolates with significantly reduced susceptibility to these antibiotics have been isolated, especially those with resistance to metronidazole (23, 24). The number of failed-treatment CDI cases following metronidazole therapy has increased remarkably in the past decade (6). C. difficile resistant to metronidazole has been reported in different regions of the world. A pan-European longitudinal surveillance of antibiotic resistance among prevalent C. difficile ribotypes showed that 0.11% of the strains investigated were resistant to metronidazole based on the CLSI breakpoint (susceptible, <8 μg/ml) (25). The metronidazole resistance in C. difficile has also been determined in Iran, as 5.3% of the clinical strains tested between November 2010 and October 2011 were resistant to metronidazole based on the CLSI breakpoint (23). In China, 15.6% of the clinical isolates recovered from June 2012 to September 2015 were revealed to be resistant to metronidazole according to the CLSI breakpoint, and the investigation even found one nontoxigenic metronidazole-resistant isolate with an MIC of >256 μg/ml (26). A national survey of the molecular epidemiology of C. difficile in Israel found that approximately 18.3% (38/208) of the strains tested were resistant to metronidazole based on the EUCAST breakpoint (susceptible epidemiological cut-off value, <2 μg/ml) (24).
The percentage of C. difficile strains with the reduced susceptibility to metronidazole has been gradually increasing (16). A surveillance study of the antimicrobial susceptibility of C. difficile isolates in the United States showed the rate of metronidazole resistance was 3.6% in 2011 based on the EUCAST breakpoint (21). Goudarzi et al. in 2013 tested the antimicrobial susceptibility of 75 C. difficile isolates from 390 CDI patients in Iran and found 5.3% of the isolates were resistant to metronidazole based on the CLSI breakpoint (23). The rates of C. difficile clinical isolates resistant to metronidazole were reported to be 0.11% (based on the CLSI breakpoint), 13.3% (based on the CLSI breakpoint), and 18% (based on the EUCAST breakpoint) in Europe in 2011 to 2012, in the United States (Texas) in 2007 to 2011, and in Israel in 2014, respectively (24, 25, 27). A recent epidemiological study showed that a total of 64 (15.6%) isolates, including one nontoxigenic isolate, were resistant to metronidazole with high MIC values (26). Some studies indicate that metronidazole resistance in C. difficile is heterogeneous (28). Moura et al. found that the use of subinhibitory concentrations of metronidazole had a role in selecting and maintaining colonies with increased minor inhibitory concentrations (29), suggesting that metronidazole heteroresistance should be a matter of concern in clinics. Metronidazole heteroresistant C. difficile can obviously result in therapeutic failure of CDI, which may not be predicted by antimicrobial susceptibility testing (AST) results.
Resistance of C. difficile to vancomycin also has been reported. In the study by Goudarzi et al., the percentage of C. difficile clinical isolates resistant to vancomycin was 8.0% based on the CLSI breakpoint (23). The rate of vancomycin-nonsusceptible C. difficile clinical isolates, including 57 ribotype 027 isolates, was 47% in Israel based on the EUCAST breakpoint (24). There are also other studies reporting C. difficile strains with vancomycin resistance. A recent longitudinal surveillance study from Europe indicated that 2.29% of C. difficile strains were intermediately resistant to vancomycin based on the EUCAST breakpoint with MICs of 4 mg/liter in the Czech Republic, Ireland, Latvia, and Poland, and 0.87% were resistant to vancomycin with MICs of >8 mg/liter in Italy and Spain (25). A US-based national sentinel surveillance study also found 17.9% of C. difficile isolates were resistant to vancomycin based on the EUCAST breakpoint (21). Even though vancomycin resistance is unlikely to affect primary treatment efficacy for CDI because of high levels of luminal in the gut (over 1,000 mg/liter in feces after oral administration) (30), these data obviously suggest a potentially serious problem for vancomycin therapy of CDI in the future. Another alarming threat is the development and dissemination of hypervirulent antibiotic-resistant C. difficile (13, 14). It has been reported that the ribotype 027 strain with reduced susceptibilities to vancomycin and metronidazole has disseminated across Israel and is now the most common clinical strain isolated (24).
In addition to metronidazole and vancomycin, C. difficile also develops resistance to other therapeutic options, such as rifamycins, fidaxomicin, tetracyclines, and chloramphenicol. In a Pan-European longitudinal surveillance of antibiotic resistance among prevalent C. difficile ribotypes, C. difficile clinical isolates resistant to rifampin (a member of rifamycin class) have been detected in 17 of the total 22 countries investigated, and the percentage of rifampin-resistant strains is over 57% (resistant strain defined as that with an MIC of >16 μg/ml because there are no CLSI or EUCAST breakpoints for rifampin currently available) in some countries, such as Italy, the Czech Republic, Denmark, and Hungary (25). The rifampin resistance problem is less severe in North America, only 7.9% of 316 tested C. difficile clinical isolates from patients in North America were resistant to rifampin (resistant strain defined as those with an MIC of >32 μg/ml) (13). In addition to those in Europe and North America, rifampin-resistant C. difficile isolates have also been detected in Asia (31, 32). Although reduced susceptibility to fidaxomicin is rare for C. difficile, mutants with decreased susceptibility to fidaxomicin could be easily developed under the selective pressure of fidaxomicin use (33), which possibly increases the risk of the occurrence of resistant strains. So far, there has been only one C. difficile isolate from a recurrence case showing an MIC of 16 μg/ml in a fidaxomicin clinical trial (34). Even though the percentages of tetracycline-resistant C. difficile isolates in different countries varied from 2.4% to 41.67% (35), it is also a potentially serious situation that should be considered in association with CDI given that tigecycline is now proposed to be an alternative antibiotic for the treatment of patients with severe or severe complicated CDI (6). Resistance to chloramphenicol is rare in C. difficile and only 3.7% of isolates (resistant strain defined as that with an MIC of >32 μg/ml) have been reported to be resistant to this antibiotic in Europe (25).
KNOWN ANTIMICROBIAL RESISTANCE MECHANISMS OF CLOSTRIDIUM DIFFICILE
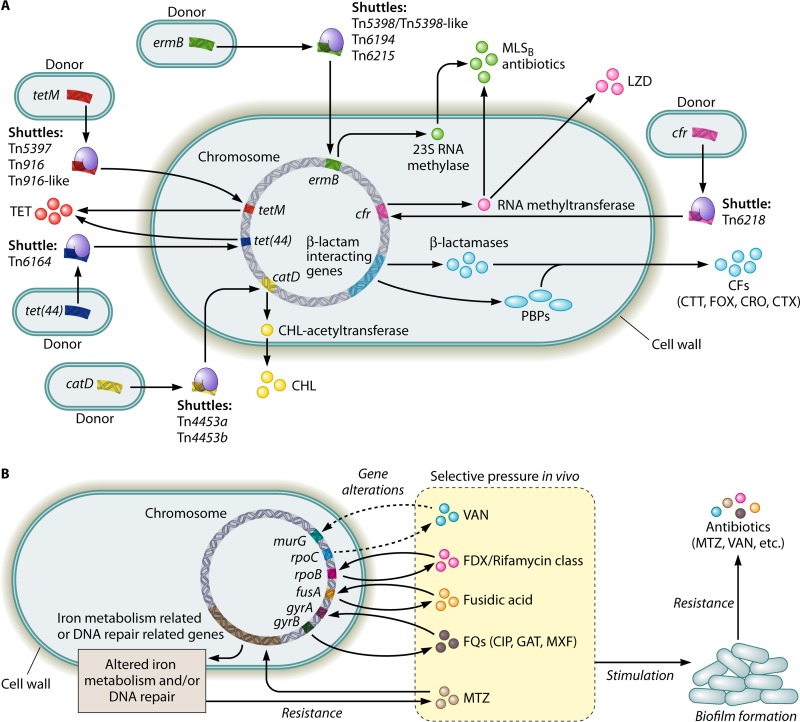
C. difficile has developed multiple mechanisms for antimicrobial resistance. Factors contributing to this development of antimicrobial resistance in C. difficile (Fig. 1) include the resistance-associated genes harbored in the bacterial chromosome, mobile genetic elements (MGEs), alterations in the antibiotic targets of antibiotics and/or in metabolic pathways in C. difficile, and biofilm formation. The C. difficile genome harbors a variety of resistance genes responsible for the resistance to different classes of antibiotics. Analysis of the C. difficile 630 genome has identified genes encoding β-lactamase-like proteins and penicillin-binding proteins (PBPs), both of which are proposed to mediate the resistance to the β-lactam antibiotics such as penicillin and cephalosporins (16).
FIG 1.
Diagram illustrating the known factors contributing to the development of antibiotic resistance in Clostridium difficile. (A) Intra- or interspecies transfers of mobile genetic elements via conjugation, transduction, and/or transformation (e.g., transposons) or the natural occurrence of gene mutations (e.g., β-lactamase genes) facilitate C. difficile in obtaining antibiotic resistance genes. (B) Selective pressure in vivo leads to alterations in the antibiotic targets and/or in the metabolic pathways in C. difficile, which on one hand, directly causes antibiotic resistance, while on the other hand, may stimulate biofilm formation. Biofilm formation via different mechanisms (e.g., C. difficile Cwp84, flagella, and the LuxS system) further promotes the development of antibiotic resistance in C. difficile. CFs, cephalosporins; CHL, chloramphenicol; CIP, ciprofloxacin; CRO, ceftriaxone; CTT, cefotetan; CTX, cefotaxime; FOX, cefoxitin; FQs, fluoroquinolones; GAT, gatifloxacin; LZD, linezolid; MLSB, macrolide-lincosamide-streptogramin B; MTZ, metronidazole; MXF, moxifloxacin; PBPs, penicillin-binding proteins; TET, tetracycline; VAN, vancomycin.
Conjugation, transduction, and/or transformation of MGEs, especially transposons among C. difficile strains and/or between C. difficile and the other bacterial species, are important mechanisms for C. difficile to acquire antimicrobial resistance genes (16). A large proportion of the C. difficile genome (approximately 11%) is made up of MGEs. Resistance to the antibiotics of the MLSB (macrolide-lincosamide-streptogramin B) family in C. difficile is mediated by at least four kinds of transposons, including Tn5398, Tn5398-like derivatives, Tn6194, and Tn6215. Transposons may also mediate the transfer of the ermB gene which encodes a 23S RNA methylase and induces the resistance to the MLSB family of antibiotics, including clindamycin and erythromycin (16, 36). Tn5398 and Tn6215 can integrate the C. difficile genome through the exchange of large genomic fragments. Tn5398 could integrate into the recipient chromosome either by homologous recombination or by using a site-specific recombination of the recipient. This element is found to be able to transfer from C. difficile to Staphylococcus aureus and to Bacillus subtilis. Tn6215 can be transferred to recipient cells via a conjugation-like mechanism, but is also able to be transduced by phage phiC2. Tn6194 likely integrates into the C. difficile genome at different sites and is also able to transfer between C. difficile strains as well from C. difficile to Enterococcus faecalis (37). Besides those four transposons, a novel Tn916-like transposon, which is similar to Tn6218, is also involved in resistance to the MLSB antibiotics in C. difficile. This element participates in the transfer of the chloramphenicol-florfenicol resistance gene (cfr) (38), which encodes an RNA methyltransferase that functions by modifying the bacterial 23S rRNA and is also found to have a role in the resistance to MLSB antibiotics, especially when the erm genes are absent (16). In addition, cfr also confers resistance to linezolid, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A (39).
Resistance to tetracycline in C. difficile is thought to be associated with transposons Tn5397, Tn916 or Tn916-like family, and Tn6164. These elements are found to be able to transfer the tet class of genes, including tet(M), tet(44), and tet(W) (16, 36), and therefore render C. difficile resistant to tetracycline. The tet(M) gene is the predominant class in C. difficile, and is responsible for tetracycline resistance and is usually carried on Tn5397, Tn916, or Tn916-like family transposons (16). The mechanism by which C. difficile acquires the tet(M) gene remains unclear. A proposed model is that C. difficile acquires this gene via a genetic transfer from some other pathogenic bacteria containing tet(M), such as Bifidobacterium longum. The tet(W) gene, thought to have the second largest host range ranking behind tet(M), was found to be copresent in tetracycline-resistant C. difficile isolated from both pigs and humans (40). Despite its infrequency, tet(44) is also proposed to have a role in resistance to tetracycline, and this gene was found to be carried by Tn6164 in some RT078 isolates. The presence of Tn6164 is likely to have a possible correlation with the higher virulence of RT078 strains (16). The transposons are also involved in the resistance to chloramphenicol in C. difficile. Two mobile transposons, Tn4453a and Tn4453b, are able to transfer the catP gene, which encodes a chloramphenicol acetyltransferase enzyme that is responsible for the chloramphenicol resistance (25, 41).
Alterations in the antibiotic targets and/or in the metabolic pathways in C. difficile represent another mechanism mediating antibiotic resistance in this microorganism. Importantly, this mechanism is thought to mediate the resistance to metronidazole and vancomycin in C. difficile, though the exact mechanism is not completely understood (16, 27). Current data suggest that the metronidazole resistance is likely due to several alterations in yet-to-be defined metabolic pathways, such as those involving the activity of nitroreductases, iron uptake, and DNA repair (42, 43), while the vancomycin resistance might be due to amino acid changes in peptidoglycan biosynthesis-associated proteins such as MurG (33). Multiple factors may induce such alterations in the antibiotic targets and/or in the metabolic pathways in C. difficile, although selective pressure from exposure to antibiotics in the environment is the most important one. For example, the selective pressure in vivo from the use of rifamycin antibiotics as alternative CDI therapies is able to mediate mutations in the β subunit of the rpoB gene, which encodes a bacterial RNA polymerase (36). These types of alterations are proposed to induce resistance to the rifamycin class of antibiotics, in particular, to rifampin and rifaximin, in C. difficile (44). The alterations of rpoB might also be involved in the reduced susceptibility of C. difficile to fidaxomicin (33). A similar mechanism is also found in the resistance to fusidic acid, as fusidic acid-resistant C. difficile strains carry fusA mutations. The selective pressure in vivo is also supposed to be the incentive for the acquisition of fluoroquinolones resistance. When the environmental concentration of fluoroquinolones is not able to inhibit C. difficile, the pathogen might acquire amino acidic substitutions harbored in two DNA gyrase subunits, GyrA and/or GyrB. Alterations in the quinolone-resistance determining region of either GyrA or GyrB might mediate the resistance to fluoroquinolones in C. difficile (16).
Biofilm formation has been proposed to be another important factor contributing to antimicrobial resistance of C. difficile by forming a multilayered structured biofilm that is composed of a thick multicomponent biofilm matrix containing proteins, DNA, and polysaccharides (45). The formation of this C. difficile biofilm is mainly driven by intrinsic C. difficile mechanisms, such as Cwp84, flagella, and LuxS, but the selective pressure from the exposure to antibiotics in the environment also has been shown to stimulate biofilm formation (45, 46). It is known that biofilms can protect pathogenic bacteria from unfavorable environmental stresses such as antibiotics and therefore contribute to survival and virulence (45). Pathogenic bacteria existing in a biofilm are known to increase resistance to antibiotics from 10- to even 1,000-fold in comparison to planktonic cells. In C. difficile, biofilm formation is proposed to play a role in both metronidazole resistance and vancomycin resistance (45, 46). Specific details on how clostridial biofilms contribute to the acquisition of the antimicrobial resistance of C. difficile are poorly understood. A hypothesis is that the biofilm matrix and the physiological state together contribute to the antimicrobial resistance seen with clostridial biofilms. Although the biofilm matrix can act as a protective barrier, it may induce antimicrobial tolerance by altering the physiological state of C. difficile contained within the biofilm, such as bacteria in a dormant state, which are then more resistant to the antibiotics (45). Further studies are necessary for understanding the mechanisms by which C. difficile biofilms contribute to antibiotic resistance.
ROLE OF ANTIMICROBIAL SUSCEPTIBILITY TESTING OF CLOSTRIDIUM DIFFICILE
In the near future, clinical microbiology laboratories will need to rapidly perform antimicrobial susceptibility testing (AST) to determine antimicrobial resistance profiles of C. difficile isolates recovered from patients and present easy-to-understand AST results to physicians. Also, AST is frequently used to monitor resistance patterns in the epidemiology of CDI. With dynamic changes of CDI epidemiology, the US CDC, European Centre for Disease Prevention and Control (ECDC), and other national CDC should establish a surveillance network to track CDI in real time. Obviously, AST for C. difficile is more difficult to perform than AST for aerobes for many reasons; these include slow growth and the need for strict maintenance of anaerobic conditions (47).
There are numerous AST methods available for C. difficile that have been used in clinical microbiology and public health laboratories. Most of the well-accepted methods focus on phenotypic characteristics of C. difficile, including agar dilution, broth dilution, and MIC gradient diffusion (13, 48–50). Phenotypic methods are classified into quantitative and qualitative ones. Molecular assays, including whole-genome sequencing and proteomics to detect resistance determinants or single nucleotide polymorphisms, have also shown promise in gene-based AST (51).
The agar dilution assay, also known as agar incorporation, is recommended as a gold standard AST for C. difficile by the Clinical and Laboratory Standards Institute (CLSI) (52). A detailed test method, interpretive categories, and breakpoints have been described in the CLSI document M100 (52). There have been plenty of C. difficile studies employing the agar dilution assay to test antimicrobial susceptibility (18, 21, 53, 54). In addition, the agar dilution assay is always chosen as a reference method to which other AST methods are compared (55).
The agar dilution assay is useful for clinical labs and for non-patient care testing, such as in epidemiological studies, with some advantages. First, it is an accurate and high-throughput assay. Second, the choice of antibiotics tested is flexible and can be changed according to clinical or investigational needs. Third, this assay is easily established and inexpensive as well as suitable for large numbers of isolates. However, it is both labor and time consuming and requires skilled and experienced microbiology technologists to properly perform the test. Moreover, the agar dilution test is usually batched and does not readily allow testing of individual isolates one by one, which is often needed in clinical laboratories to meet clinical needs.
Some studies have used broth microdilution for C. difficile AST, which was recommended by the CLSI (52, 56–58). For example, broth microdilution has been used to evaluate antimicrobial susceptibilities of the fluoroquinolone finafloxacin (57) and a novel lipopeptide antibiotic (CB-183,315) (56). The results from these studies indicated that the broth microdilution method has a good reproducibility of 100% and an agreement of 90 to 95% in comparison to agar dilution (56, 57). These studies showed that the broth microdilution method is easier to perform and more convenient for clinical use than agar dilution. Furthermore, multiple antibiotics are measured at one time with low costs. This method is still labor and time consuming like the agar dilution assay. Despite the lack of standardization, the advantages of broth microdilution make it a useful AST tool in today's surveillance and public health laboratories.
The Etest strip (bioMérieux, Durham, NC) is one of the gradient diffusion assays available for clinical laboratories and for non-patient care testing. This assay has been widely used to identify antibiotic susceptibility profiles for patient care in clinical sets and for surveillance of antibiotic resistance in molecular epidemiology (13, 29, 59). A comparative study of performance for 238 C. difficile isolates in a Swedish university hospital has demonstrated high categorical agreement between the Etest and agar dilution (60). Although there were significant differences in MICs between the two methods, the results did not lead to any discrepancy in susceptible-intermediate-resistant categorization. In another study, the MIC value in 80% of isolates tested by the Etest was lower than those tested by agar dilution. The same results were observed in other studies (29, 61, 62). Nevertheless, the results have no effect on the categorization of most antimicrobials, including vancomycin, fusidic acid, clindamycin, tetracycline, moxifloxacin, gatifloxacin, teicoplanin, rifampin, and others, with the exact breakpoints. In the case of metronidazole, the Etest MIC results did not correlate with those of the agar dilution assay, especially when the MIC value was close to the metronidazole breakpoint. The high MIC of metronidazole should be confirmed by the agar dilution assay. The accuracy of metronidazole susceptibility testing usually depends on the anaerobic condition and medium quality. The Etest is a convenient easy-to-use assay by which multiple antimicrobials can be measured on a plate at the same time. The Etest delivers quantitative results with exact MIC values. The disadvantage of high cost still significantly hinders the extensive use of the Etest in clinical laboratory and epidemiological surveillance.
Although disk diffusion testing generally is not recommended by CLSI, it has recently become an attractive alternative for C. difficile AST in epidemiological studies. Wong et al. in 1999 performed a prospective susceptibility study of 100 C. difficile strains for vancomycin and metronidazole using the Etest and disk diffusion test; the MIC value by the Etest was correlated with the zone size of inhibition determined by the disk diffusion assay. The study indicated that correlation coefficients were too low to accurately predict the MIC value of C. difficile using the disk diffusion test (70). Erikstrup et al. showed that the same MIC results were obtained when they tested 211 C. difficile isolates using Etest and disk diffusion (49). The zone diameter breakpoints for vancomycin, moxifloxacin, and metronidazole were reported to be ≥19 mm, ≥20 mm, and ≥23 mm, respectively, with no very major errors. Less than 2.0% of major errors were found in a tolerable range, including a 1.4% error for metronidazole, 0.5% for vancomycin, and 1.8% for moxifloxacin. An excellent agreement was found between MIC results when the Etest and disk diffusion were used, which can be an alternative for C. difficile AST (49). Based on the above-mentioned zone diameter breakpoints, the disk diffusion test was used to assess 2,717 C. difficile isolates with reduced susceptibility to metronidazole and vancomycin in Denmark (63). Similar conclusions were also drawn in a recent comparative study between agar dilution and disk diffusion by Fraga et al. in Brazil in 2016 (53).
Disk diffusion also has been used for testing for susceptibility to rifaximin (60), tetracycline, erythromycin, penicillin G, and chloramphenicol. Especially, disk diffusion (5-μg disk) was recommended as an easy rapid assay to distinguish metronidazole-heteroresistant strains. There is still a debate about whether disk diffusion is qualified for C. difficile AST without the exact zone diameter breakpoints determined by both CLSI and EUCAST. The recent studies indicate that there remain ongoing interests for the disk diffusion assay with its assets, including low cost, flexible antibiotic selection, and adaptability for changes of interpretive breakpoints.
The phenotypic tests are well recognized as traditional methods for C. difficile AST as mentioned above. It takes almost 1 week to get the final results after isolation, purification, and susceptibility testing of bacteria. Patients may have more than one C. difficile strain in their stool specimens. Microorganisms are not isolated from patient specimens in most clinical laboratories; therefore, phenotypic AST results are not provided. The delayed feedback to clinicians potentially results in therapy failure because no appropriate treatment can be chosen. Gene-based analysis can be an alternative to MIC testing for C. difficile in clinical laboratories. Antimicrobial resistance has been correlated with mobile genetic elements ermB (MLSB antibiotics resistance gene), tet(M) (tetracycline resistance gene), gyrA or gyrB, catD, and so on (16, 63, 64). Whole-genome sequencing and proteomics have also been applied to study C. difficile resistance with promising performance, and many targeted genes/proteins associated with metronidazole have been found (42, 65). More biomarkers related with antimicrobial resistance will be disclosed on the basis of whole-genome sequencing and proteomics. With the cost decreasing in recent years, these technologies will be widely used as gene-based analyses for AST in clinical microbiological laboratories and in epidemiological surveillance.
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has been successfully used to identify antibiotic resistance through the detection of enzymatic activity, bacterial extracts, specific proteins, and cell wall components in methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus spp., and others (66). Gene mutants are also sequenced using MALDI-TOF MS combined with PCR amplification. MALDI-TOF MS approaches have been used for the recognition of C. difficile ribotypes (67). A rapid identification of C. difficile combined with the chromID C. difficile chromogenic agar (68) has not yet been applied in the detection of antibiotic resistance in C. difficile (67).
CONCLUDING REMARKS
The utilization of antimicrobial agents is a double-edged sword in terms of C. difficile. Infections caused by this pathogen are somewhat unique in that their incidence increases with increased utilization of certain antibiotics; yet these infections are typically treated with other antibiotics that are active against C. difficile. Currently available antibiotics for treating CDI are becoming limited due to the increasing resistance in this pathogen. Understanding the resistance mechanisms of C. difficile is one of the key issues in the strategy for preventing CDI. In addition to the proper use of antimicrobial agents and avoidance of the over use of these agents, antimicrobial resistance of C. difficile should be monitored over time. Continued research on the resistance mechanisms of C. difficile are needed along with the development of new antimicrobial agents effective against C. difficile. Alternative therapies, as well as novel therapies, for CDI also should be pursued.
ACKNOWLEDGMENTS
This work was supported in part by grants from the Key Research and Development Program of Zhejiang (2015C03048) and the National Institutes of Health (K01-DK092352, R21-AI113470 and P30-CA008748).
REFERENCES
- 1.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. 2015. Burden of Clostridium difficile infection in the United States. N Engl J Med 372:825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Donoghue C, Kyne L. 2011. Update on Clostridium difficile infection. Curr Opin Gastrenterol 27:38–47. doi: 10.1097/MOG.0b013e3283411634. [DOI] [PubMed] [Google Scholar]
- 3.Kwon JH, Olsen MA, Dubberke ER. 2015. The morbidity, mortality, and costs associated with Clostridium difficile infection. Infect Dis Clin North Am 29:123–134. doi: 10.1016/j.idc.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 4.McGlone SM, Bailey RR, Zimmer SM, Popovich MJ, Tian Y, Ufberg P, Muder RR, Lee BY. 2012. The economic burden of Clostridium difficile. Clin Microbiol Infect 18:282–289. doi: 10.1111/j.1469-0691.2011.03571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun X, Hirota SA. 2015. The roles of host and pathogen factors and the innate immune response in the pathogenesis of Clostridium difficile infection. Mol Immunol 63:193–202. doi: 10.1016/j.molimm.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leffler DA, Lamont JT. 2015. Clostridium difficile infection. N Engl J Med 372:1539–1548. doi: 10.1056/NEJMra1403772. [DOI] [PubMed] [Google Scholar]
- 7.Ofosu A. 2016. Clostridium difficile infection: a review of current and emerging therapies. Ann Gastroenterol 29:147–154. doi: 10.20524/aog.2016.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bagdasarian N, Rao K, Malani PN. 2015. Diagnosis and treatment of Clostridium difficile in adults: a systematic review. JAMA 313:398–408. doi: 10.1001/jama.2014.17103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH, Society for Healthcare Epidemiology of America, Infectious Diseases Society of America. 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect Control Hosp Epidemiol 31:431–455. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 10.Gerding DN, File TM Jr, McDonald LC. 2016. Diagnosis and treatment of Clostridium difficile infection. Infect Dis Clin Pract (Baltim Md) 24:3–10. doi: 10.1097/IPC.0000000000000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bauer MP, Kuijper EJ, van Dissel JT, European Society of Clinical Microbiology and Infectious Diseases. 2009. European Society of Clinical Microbiology and Infectious Diseases (ESCMID): treatment guidance document for Clostridium difficile infection (CDI). Clin Microbiol Infect 15:1067–1079. doi: 10.1111/j.1469-0691.2009.03099.x. [DOI] [PubMed] [Google Scholar]
- 12.Debast SB, Bauer MP, Kuijper EJ, European Society of Clinical Microbiology and Infectious Diseases. 2014. European Society of Clinical Microbiology and Infectious Diseases: update of the treatment guidance document for Clostridium difficile infection. Clin Microbiol Infect 20 Suppl 2:S1–S26. doi: 10.1111/1469-0691.12418. [DOI] [PubMed] [Google Scholar]
- 13.Tenover FC, Tickler IA, Persing DH. 2012. Antimicrobial-resistant strains of Clostridium difficile from North America. Antimicrob Agents Chemother 56:2929–2932. doi: 10.1128/AAC.00220-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, Connor TR, Harris SR, Fairley D, Bamford KB. 2013. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet 45:109–113. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johanesen PA, Mackin KE, Hutton ML, Awad MM, Larcombe S, Amy JM, Lyras D. 2015. Disruption of the gut microbiome: Clostridium difficile infection and the threat of antibiotic resistance. Genes 6:1347–1360. doi: 10.3390/genes6041347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spigaglia P. 2016. Recent advances in the understanding of antibiotic resistance in Clostridium difficile infection. Ther Adv Infect Dis 3:23–42. doi: 10.1177/2049936115622891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slimings C, Riley TV. 2014. Antibiotics and hospital-acquired Clostridium difficile infection: update of systematic review and meta-analysis. J Antimicrob Chemother 69:881–891. doi: 10.1093/jac/dkt477. [DOI] [PubMed] [Google Scholar]
- 18.Wieczorkiewicz JT, Lopansri BK, Cheknis A, Osmolski JR, Hecht DW, Gerding DN, Johnson S. 2015. Fluoroquinolone and macrolide exposure predict Clostridium difficile infection with the highly fluoroquinolone-and macrolide-resistant epidemic C. difficile strain BI/NAP1/027. Antimicrob Agents Chemother 60:418–423. doi: 10.1128/AAC.01820-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keessen EC, Hensgens MP, Spigaglia P, Barbanti F, Sanders IM, Kuijper EJ, Lipman LJ. 2013. Antimicrobial susceptibility profiles of human and piglet Clostridium difficile PCR-ribotype 078. Antimicrob Resist Infect Control 2:14. doi: 10.1186/2047-2994-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freeman J, Baines SD, Jabes D, Wilcox MH. 2005. Comparison of the efficacy of ramoplanin and vancomycin in both in vitro and in vivo models of clindamycin-induced Clostridium difficile infection. J Antimicrob Chemother 56:717–725. doi: 10.1093/jac/dki321. [DOI] [PubMed] [Google Scholar]
- 21.Snydman D, McDermott L, Jacobus N, Thorpe C, Stone S, Jenkins S, Goldstein E, Patel R, Forbes B, Mirrett S, Johnson S, Gerding DN. 2015. U.S.-based national sentinel surveillance study for the epidemiology of Clostridium difficile-associated diarrheal isolates and their susceptibility to fidaxomicin. Antimicrob Agents Chemother 59:6437–6443. doi: 10.1128/AAC.00845-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spigaglia P, Barbanti F, Mastrantonio P, European Study Group on Clostridium difficile (ESGCD). 2011. Multidrug resistance in European Clostridium difficile clinical isolates. J Antimicrob Chemother 66:2227–2234. doi: 10.1093/jac/dkr292. [DOI] [PubMed] [Google Scholar]
- 23.Goudarzi M, Goudarzi H, Alebouyeh M, Azimi Rad M, Shayegan Mehr FS, Zali MR, Aslani MM. 2013. Antimicrobial susceptibility of Clostridium difficile clinical isolates in Iran. Iran Red Crescent Med J 15:704–711. doi: 10.5812/ircmj.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adler A, Miller-Roll T, Bradenstein R, Block C, Mendelson B, Parizade M, Paitan Y, Schwartz D, Peled N, Carmeli Y. 2015. A national survey of the molecular epidemiology of Clostridium difficile in Israel: the dissemination of the ribotype 027 strain with reduced susceptibility to vancomycin and metronidazole. Diagn Microbiol Infect Dis 83:21–24. doi: 10.1016/j.diagmicrobio.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 25.Freeman J, Vernon J, Morris K, Nicholson S, Todhunter S, Longshaw C, Wilcox MH, Pan-European Longitudinal Surveillance of Antibiotic Resistance among Prevalent Clostridium difficile Ribotypes' Study Group. 2015. Pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes. Clin Microbiol Infect 21:248.e9–248.e16. doi: 10.1016/j.cmi.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 26.Jin D, Luo Y, Huang C, Cai J, Ye J, Zheng Y, Wang L, Zhao P, Liu A, Fang W, Wang X, Xia S, Jiang J, Tang YW. 2017. Molecular epidemiology of Clostridium difficile infection in hospitalized patients in eastern China. J Clin Microbiol 55:801–810. doi: 10.1128/JCM.01898-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Norman KN, Scott HM, Harvey RB, Norby B, Hume ME. 2014. Comparison of antimicrobial susceptibility among Clostridium difficile isolated from an integrated human and swine population in Texas. Foodborne Pathog Dis 11:257–264. doi: 10.1089/fpd.2013.1648. [DOI] [PubMed] [Google Scholar]
- 28.Peláez T, Cercenado E, Alcalá L, Marin M, Martín-López A, Martínez-Alarcón J, Catalán P, Sánchez-Somolinos M, Bouza E. 2008. Metronidazole resistance in Clostridium difficile is heterogeneous. J Clin Microbiol 46:3028–3032. doi: 10.1128/JCM.00524-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moura I, Spigaglia P, Barbanti F, Mastrantonio P. 2013. Analysis of metronidazole susceptibility in different Clostridium difficile PCR ribotypes. J Antimicrob Chemother 68:362–365. doi: 10.1093/jac/dks420. [DOI] [PubMed] [Google Scholar]
- 30.Baines SD, Wilcox MH. 2015. Antimicrobial resistance and reduced susceptibility in Clostridium difficile: potential consequences for induction, treatment, and recurrence of C. difficile infection. Antibiotics (Basel) 4:267–298. doi: 10.3390/antibiotics4030267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J, Kang JO, Pai H, Choi TY. 2012. Association between PCR ribotypes and antimicrobial susceptibility among Clostridium difficile isolates from healthcare-associated infections in South Korea. Int J Antimicrob Agents 40:24–29. doi: 10.1016/j.ijantimicag.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 32.Dong D, Zhang L, Chen X, Jiang C, Yu B, Wang X, Peng Y. 2013. Antimicrobial susceptibility and resistance mechanisms of clinical Clostridium difficile from a Chinese tertiary hospital. Int J Antimicrob Agents 41:80–84. doi: 10.1016/j.ijantimicag.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 33.Leeds JA, Sachdeva M, Mullin S, Barnes SW, Ruzin A. 2013. In vitro selection, via serial passage, of Clostridium difficile mutants with reduced susceptibility to fidaxomicin or vancomycin. J Antimicrob Chemother 69:41–44. doi: 10.1093/jac/dkt302. [DOI] [PubMed] [Google Scholar]
- 34.Goldstein EJ, Babakhani F, Citron DM. 2012. Antimicrobial activities of fidaxomicin. Clin Infect Dis 55:S143–S148. doi: 10.1093/cid/cis339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linkevicius M, Sandegren L, Andersson DI. 2016. Potential of tetracycline resistance proteins to evolve tigecycline resistance. Antimicrob Agents Chemother 60:789–796. doi: 10.1128/AAC.02465-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsutsumi LS, Owusu YB, Hurdle JG, Sun D. 2014. Progress in the discovery of treatments for C. difficile infection: a clinical and medicinal chemistry review. Cur Top Med Chem 14:152–175. doi: 10.2174/1568026613666131113154753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wasels F, Monot M, Spigaglia P, Barbanti F, Ma L, Bouchier C, Dupuy B, Mastrantonio P. 2014. Inter-and intraspecies transfer of a Clostridium difficile conjugative transposon conferring resistance to MLSB. Microb Drug Resist 20:555–560. doi: 10.1089/mdr.2014.0015. [DOI] [PubMed] [Google Scholar]
- 38.Dingle KE, Elliott B, Robinson E, Griffiths D, Eyre DW, Stoesser N, Vaughan A, Golubchik T, Fawley WN, Wilcox MH. 2014. Evolutionary history of the Clostridium difficile pathogenicity locus. Genome Biol Evol 6:36–52. doi: 10.1093/gbe/evt204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen J, Wang Y, Schwarz S. 2013. Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J Antimicrob Chemother 68:1697–1706. doi: 10.1093/jac/dkt092. [DOI] [PubMed] [Google Scholar]
- 40.Fry PR, Thakur S, Gebreyes W. 2012. Antimicrobial resistance, toxinotype and genotypic profiling of Clostridium difficile of swine origin. J Clin Microbiol 50:2366–2372. doi: 10.1128/JCM.06581-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kali A, Charles MVP, Srirangaraj S. 2015. Cadazolid: a new hope in the treatment of Clostridium difficile infection. Australas Med J 8:253–262. doi: 10.4066/AMJ.2015.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chong PM, Lynch T, McCorrister S, Kibsey P, Miller M, Gravel D, Westmacott GR, Mulvey MR, Canadian Nosocomial Infection Surveillance Program (CNISP). 2014. Proteomic analysis of a NAP1 Clostridium difficile clinical isolate resistant to metronidazole. PLoS One 9:e82622. doi: 10.1371/journal.pone.0082622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moura I, Monot M, Tani C, Spigaglia P, Barbanti F, Norais N, Dupuy B, Bouza E, Mastrantonio P. 2014. Multidisciplinary analysis of a nontoxigenic Clostridium difficile strain with stable resistance to metronidazole. Antimicrob Agents Chemother 58:4957–4960. doi: 10.1128/AAC.02350-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Connor JR, Galang MA, Sambol SP, Hecht DW, Vedantam G, Gerding DN, Johnson S. 2008. Rifampin and rifaximin resistance in clinical isolates of Clostridium difficile. Antimicrob Agents Chemother 52:2813–2817. doi: 10.1128/AAC.00342-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ðapa T, Leuzzi R, Baban ST, Ng YK, Adamo R, Kuehne SA, Scarselli M, Minton NP, Serruto D, Unnikrishnan M. 2013. Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J Bacteriol 195:545–555. doi: 10.1128/JB.01980-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vuotto C, Moura I, Barbanti F, Donelli G, Spigaglia P. 2016. Subinhibitory concentrations of metronidazole increase biofilm formation in Clostridium difficile strains. Pathog Dis 74:ftv114. doi: 10.1093/femspd/ftv114. [DOI] [PubMed] [Google Scholar]
- 47.Jorgensen JH, Pfaller M, Carroll KC (ed). 2015. Manual of clinical microbiology, 11th ed, vol 1 ASM press, Washington, DC. [Google Scholar]
- 48.Brook I, Wexler HM, Goldstein EJ. 2013. Antianaerobic antimicrobials: spectrum and susceptibility testing. Clin Microbiol Rev 26:526–546. doi: 10.1128/CMR.00086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Erikstrup LT, Danielsen TK, Hall V, Olsen KE, Kristensen B, Kahlmeter G, Fuursted K, Justesen US. 2012. Antimicrobial susceptibility testing of Clostridium difficile using EUCAST epidemiological cut-off values and disk diffusion correlates. Clin Microbiol Infect 18:E266–272. doi: 10.1111/j.1469-0691.2012.03907.x. [DOI] [PubMed] [Google Scholar]
- 50.Pirs T, Avbersek J, Zdovc I, Krt B, Andlovic A, Lejko-Zupanc T, Rupnik M, Ocepek M. 2013. Antimicrobial susceptibility of animal and human isolates of Clostridium difficile by broth microdilution. J Med Microbiol 62:1478–1485. doi: 10.1099/jmm.0.058875-0. [DOI] [PubMed] [Google Scholar]
- 51.Fluit AC, Visser MR, Schmitz FJ. 2001. Molecular detection of antimicrobial resistance. Clin Microbiol Rev 14:836–871, table of contents. doi: 10.1128/CMR.14.4.836-871.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.CLSI. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. CLSI M100-S24. CLSI, Wayne, PA. [Google Scholar]
- 53.Fraga EG, Nicodemo AC, Sampaio JL. 2016. Antimicrobial susceptibility of Brazilian Clostridium difficile strains determined by agar dilution and disk diffusion. Braz J Infect Dis 20:476–481. doi: 10.1016/j.bjid.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiuff C, Brown DJ, Mather H, Banks AL, Eastaway A, Coia JE. 2011. The epidemiology of Clostridium difficile in Scotland. J Infect 62:271–279. doi: 10.1016/j.jinf.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 55.Rennie RP, Turnbull L, Brosnikoff C, Cloke J. 2012. First comprehensive evaluation of the M.I.C. evaluator device compared to Etest and CLSI broth microdilution for MIC testing of aerobic Gram-positive and Gram-negative bacterial species. J Clin Microbiol 50:1147–1152. doi: 10.1128/JCM.05395-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Citron DM, Goldstein EJ. 2011. Reproducibility of broth microdilution and comparison to agar dilution for testing CB-183,315 against clinical isolates of Clostridium difficile. Diagn Microbiol Infect Dis 70:554–556. doi: 10.1016/j.diagmicrobio.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 57.Genzel GH, Stubbings W, Stingu CS, Labischinski H, Schaumann R. 2014. Activity of the investigational fluoroquinolone finafloxacin and seven other antimicrobial agents against 114 obligately anaerobic bacteria. Int J Antimicrob Agents 44:420–423. doi: 10.1016/j.ijantimicag.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 58.Lim SC, Foster NF, Riley TV. 2016. Susceptibility of Clostridium difficile to the food preservatives sodium nitrite, sodium nitrate and sodium metabisulphite. Anaerobe 37:67–71. doi: 10.1016/j.anaerobe.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 59.Spigaglia P, Drigo I, Barbanti F, Mastrantonio P, Bano L, Bacchin C, Puiatti C, Tonon E, Berto G, Agnoletti F. 2015. Antibiotic resistance patterns and PCR-ribotyping of Clostridium difficile strains isolated from swine and dogs in Italy. Anaerobe 31:42–46. doi: 10.1016/j.anaerobe.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 60.Huhulescu S, Sagel U, Fiedler A, Pecavar V, Blaschitz M, Wewalka G, Allerberger F, Indra A. 2011. Rifaximin disc diffusion test for in vitro susceptibility testing of Clostridium difficile. J Med Microbiol 60:1206–1212. doi: 10.1099/jmm.0.028571-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baines SD, O'Connor R, Freeman J, Fawley WN, Harmanus C, Mastrantonio P, Kuijper EJ, Wilcox MH. 2008. Emergence of reduced susceptibility to metronidazole in Clostridium difficile. J Antimicrob Chemother 62:1046–1052. doi: 10.1093/jac/dkn313. [DOI] [PubMed] [Google Scholar]
- 62.Poilane I, Cruaud P, Torlotin JC, Collignon A. 2000. Comparison of the E test to the reference agar dilution method for antibiotic susceptibility testing of Clostridium difficile. Clin Microbiol Infect 6:155–156. doi: 10.1046/j.1469-0691.2000.00034-4.x. [DOI] [PubMed] [Google Scholar]
- 63.Holt HM, Danielsen TK, Justesen US. 2015. Routine disc diffusion antimicrobial susceptibility testing of Clostridium difficile and association with PCR ribotype 027. Eur J Clin Microbiol Infect Dis 34:2243–2246. doi: 10.1007/s10096-015-2475-x. [DOI] [PubMed] [Google Scholar]
- 64.Brouwer MS, Warburton PJ, Roberts AP, Mullany P, Allan E. 2011. Genetic organisation, mobility and predicted functions of genes on integrated, mobile genetic elements in sequenced strains of Clostridium difficile. PLoS One 6:e23014. doi: 10.1371/journal.pone.0023014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lynch T, Chong P, Zhang J, Hizon R, Du T, Graham MR, Beniac DR, Booth TF, Kibsey P, Miller M, Gravel D, Mulvey MR, Canadian Nosocomial Infection Surveillance Program (CNISP). 2013. Characterization of a stable, metronidazole-resistant Clostridium difficile clinical isolate. PLoS One 8:e53757. doi: 10.1371/journal.pone.0053757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hrabak J, Chudackova E, Walkova R. 2013. Matrix-assisted laser desorption ionization-time of flight (maldi-tof) mass spectrometry for detection of antibiotic resistance mechanisms: from research to routine diagnosis. Clin Microbiol Rev 26:103–114. doi: 10.1128/CMR.00058-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reil M, Erhard M, Kuijper EJ, Kist M, Zaiss H, Witte W, Gruber H, Borgmann S. 2011. Recognition of Clostridium difficile PCR-ribotypes 001, 027 and 126/078 using an extended MALDI-TOF MS system. Eur J Clin Microbiol Infect Dis 30:1431–1436. doi: 10.1007/s10096-011-1238-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen JH, Cheng VC, Wong OY, Wong SC, So SY, Yam WC, Yuen KY. 11 January 2016. The importance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for correct identification of Clostridium difficile isolated from chromID C. difficile chromogenic agar. J Microbiol Immunol Infect 2016:S1684–1182(16)00003-7. doi: 10.1016/j.jmii.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 69.Norén T, Åkerlund T, Wullt M, Burman L, Unemo M. 2007. Mutations in fusA associated with posttherapy fusidic acid resistance in Clostridium difficile. Antimicrob Agents Chemother 51:1840–1843. doi: 10.1128/AAC.01283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wong SS, Woo PC, Luk WK, Yuen KY. 1999. Susceptibility testing of Clostridium difficile against metronidazole and vancomycin by disk diffusion and Etest. Diagn Microbiol Infect Dis 34:1–6. [DOI] [PubMed] [Google Scholar]