ABSTRACT
Infections of the central nervous system (CNS) are often acute, with significant morbidity and mortality. Routine diagnosis of such infections is limited in developing countries and requires modern equipment in advanced laboratories that may be unavailable to a number of patients in sub-Saharan Africa. We developed a TaqMan array card (TAC) that detects multiple pathogens simultaneously from cerebrospinal fluid. The 21-pathogen CNS multiple-pathogen TAC (CNS-TAC) assay includes two parasites (Balamuthia mandrillaris and Acanthamoeba), six bacterial pathogens (Streptococcus pneumoniae, Haemophilus influenzae, Neisseria meningitidis, Mycoplasma pneumoniae, Mycobacterium tuberculosis, and Bartonella), and 13 viruses (parechovirus, dengue virus, Nipah virus, varicella-zoster virus, mumps virus, measles virus, lyssavirus, herpes simplex viruses 1 and 2, Epstein-Barr virus, enterovirus, cytomegalovirus, and chikungunya virus). The card also includes human RNase P as a nucleic acid extraction control and an internal manufacturer control, GAPDH (glyceraldehyde-3-phosphate dehydrogenase). This CNS-TAC assay can test up to eight samples for all 21 agents within 2.5 h following nucleic acid extraction. The assay was validated for linearity, limit of detection, sensitivity, and specificity by using either live viruses (dengue, mumps, and measles viruses) or nucleic acid material (Nipah and chikungunya viruses). Of 120 samples tested by individual real-time PCR, 35 were positive for eight different targets, whereas the CNS-TAC assay detected 37 positive samples across nine different targets. The CNS-TAC assays showed 85.6% sensitivity and 96.7% specificity. Therefore, the CNS-TAC assay may be useful for outbreak investigation and surveillance of suspected neurological disease.
KEYWORDS: central nervous system, TaqMan PCR, meningitis, encephalitis
INTRODUCTION
Infections of the central nervous system (CNS) such as meningitis, encephalitis, or meningoencephalitis may present as an acute illness with significant mortality and extended sequelae (1). Because these infections are often difficult to diagnose in the laboratory, clinical diagnoses often rely upon modern noninvasive techniques, including computerized tomography scans and in some cases magnetic resonance imaging. Access to these advanced clinical diagnostic techniques is limited to large metropolitan hospitals rarely found in developing countries. Even with neuroimaging, the determination of etiologic agents causing meningitis and encephalitis remains complicated, requiring confirmation using other laboratory tests (2, 3). Laboratory tests can identify a number of etiological agents responsible for bacterial and viral meningitis. Timely identification of these agents continues to be challenging in developing countries, where physicians frequently resort to empirical treatment with little or no benefit.
Advances in molecular diagnostic technology have fostered the development of multiple pathogen detection systems based on PCR. The advent of real-time PCR and multiplexing technologies has facilitated the detection of multiple targets from a single clinical sample (4–7). Although multiplex PCR is susceptible to reduced efficiency and sensitivity due to competition for PCR reagents by the different targets, specificity may also be affected if closely related targets are not selected and validated carefully. These problems can be overcome by using the TaqMan array card (TAC), which utilizes microfluidic technology and single-plex PCRs configured in a 384-well array format. The TAC has previously been used to detect pathogens responsible for respiratory (8, 9), enteric (10), and neonatal (11) infections as well as other acute febrile illnesses (12). Even though a number of multiplex pathogen detection assays, e.g., BioFire, SeeGene, and Fast Track diagnostics, are commercially available, there are numerous advantages to the TAC, including ease of use, low risk of contamination attributable to the sealed format, the ability to modify or replace individual targets without additional optimization, and a small sample volume requirement compared to using multiple single agent real-time PCR assays (13, 14).
In this study, we evaluated a CNS multiple-pathogen TAC (CNS-TAC) assay for 21 etiologies and validated the CNS-TAC assay results alongside individual real-time PCR (IRTP) assays for nine pathogens (four viruses, four bacteria, and one parasite). The purpose of this evaluation was to determine the sensitivity and specificity of the CNS-TAC assay compared to IRTP assays in detecting multiple pathogens from clinical samples. We likewise propose the use of this tool in outbreak settings, providing reduced turnaround times resulting in timely and agent appropriate interventions. In addition, the method will be used to improve our understanding of the epidemiology of the CNS.
RESULTS
Analytical performance (plasmid controls).
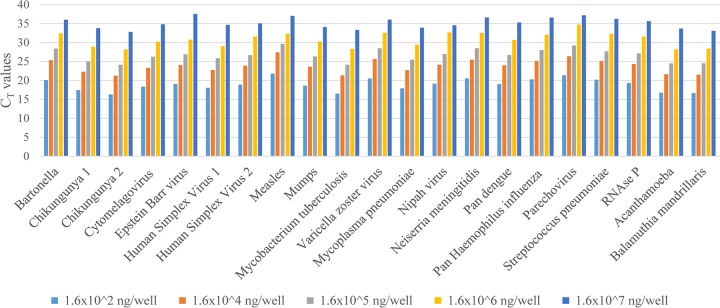
All assays exhibited a linear relationship between threshold cycle (CT) values and the concentrations of nucleic acids. CNS-TAC assays demonstrated linearity with R2 values ranging between 0.987 and 0.998, except for the measles assay which had a R2 value of 0.920. The PCR efficiency for detection of all targets in the plasmid ranged from 98.7 to 99.7%, whereas measles virus had an efficiency of 92.0% (Table 1).
TABLE 1.
Analytical performance of the CNS-TAC assaya
| Target | Linearity |
Accuracy (sensitivity %) |
Reproducibility (CV%) |
|||
|---|---|---|---|---|---|---|
| Linearity (R2) | Efficiency (%) | High concn | Low concn | High concn | Low concn | |
| Bartonella | 0.994 | 99.4 | 100 | 80.0 | 1.6 | 2.3 |
| Chikungunya 1 virus | 0.991 | 99.1 | 100 | 100 | 1.3 | 5.7 |
| Chikungunya 2 virus | 0.994 | 99.4 | 100 | 100 | 1.8 | 5.5 |
| CMV | 0.994 | 99.4 | 100 | 100 | 1.5 | 4.1 |
| EBV | 0.987 | 98.7 | 100 | 100 | 1.5 | 6.8 |
| HSV 1 | 0.987 | 98.7 | 96.7 | 100 | 2.3 | 4 |
| HSV 2 | 0.993 | 99.3 | 100 | 100 | 0.9 | 4 |
| Measles virus | 0.92 | 92.0 | 100 | 55.0 | 8.6 | 6.7 |
| Mumps virus | 0.997 | 99.7 | 100 | 100 | 1.1 | 2 |
| Mycobacterium tuberculosis | 0.993 | 99.3 | 96.7 | 100 | 1.0 | 5.1 |
| VZV | 0.993 | 99.3 | 100 | 95.0 | 2.4 | 2.7 |
| Mycoplasma pneumoniae | 0.993 | 99.3 | 100 | 100 | 1.0 | 2.9 |
| Nipah virus | 0.980 | 98.0 | 100 | 100 | 1.4 | 1 |
| Neisseria meningitidis | 0.994 | 99.4 | 100 | 100 | 1.8 | 4 |
| Pan-dengue virus | 0.993 | 99.3 | 100 | 100 | 2.1 | 1.7 |
| Haemophilus influenza | 0.993 | 99.3 | 100 | 100 | 1.2 | 5.2 |
| Parechovirus | 0.995 | 99.5 | 96.6 | 85.0 | 1.9 | 5.6 |
| Streptococcus pneumoniae | 0.992 | 99.2 | 100 | 95.0 | 1.2 | 7.4 |
| RNase P | 0.993 | 99.3 | 100 | 95.0 | 1.8 | 1.6 |
| Acanthamoeba | 0.990 | 99.0 | 100 | 100 | 1.8 | 4.1 |
| Balamuthia mandrillaris | 0.990 | 99.0 | 100 | 100 | 1.7 | 5.4 |
High concentration = 1.6 × 10−2 ng/well or 5.3 × 106 copies per well; low concentration = 1.6 × 10−7 ng/well or 54 copies per well. CV, coefficient of variance.
The lower limit of detection (LOD) for all targets was 1.6 × 10−7 ng/well, an equivalent of 54 copies per well (Fig. 1). At a concentration of 1.6 × 10−2 ng/well, the assay sensitivity ranged from 80 to 100% for the plasmid targets and was 55% for the measles virus nucleic material. The specificity for all of the positive controls was 100%. The assays showed an accuracy range of 96.7 to 100% at a concentration of 1.6 × 10−2 ng/well. The variation in reproducibility of the CT values for the 21 targets ranged from 0.9 to 2.2% for the high-concentration control, and it was 8.6% for the measles assay. The variation in the reproducibility of the low-concentration control material ranged from 1.0 to 7.5% for the plasmid targets, and it was 6.7% for the measles assay (Table 1).
FIG 1.
Test of linearity for all targets in the TAC. Dilutions from 1.6 × 10−2 to 1.6 × 10−7 are shown as bars on the x axis.
Clinical performance.
We tested by CNS-TAC assay 120 specimens, 35 of which were initially positive upon IRTP analysis to validate the assays. The specimens were positive for nine targets: 4% (5/120) cytomegalovirus (CMV), 4% (5/120) Neisseria meningitidis, 8% (10/120) Epstein-Barr virus (EBV), 3% (3/120) varicella-zoster virus (VZV), 5% (6/120) Streptococcus pneumoniae, 2% (2/120) mumps virus, 6% (7/120) Mycobacterium tuberculosis, 2% (2/120) Acanthamoeba, and 2% (2/120) Haemophilus influenzae. The specificity for all nine targets across 120 samples ranged from 87.5 to 100%. Streptococcus pneumoniae had the lowest specificity (87.5%), whereas CMV, VZV, mumps virus, and Haemophilus influenzae all had specificities of 100%. Compared to IRTP, the overall sensitivity of the CNS-TAC assay ranged from 33.3% for VZV to 100% for Neisseria meningitidis (Table 2). Low sensitivities (<50%) were observed in cases where there were small sample sizes of positive targets as seen with the mumps virus and VZV assays (Table 2). Although Acanthamoeba was not detected by IRTP in any of the samples, the CNS-TAC assay detected this target in two samples. Further attempts to detect this by IRTP failed to yield positive results.
TABLE 2.
Sensitivity of CNS-TAC assays compared to IRTP assays using a CT of 40 as a cutoffa
| Target and parameter | IRTP assay | TAC assay | % Sensitivity (95% CI) |
|---|---|---|---|
| CMV | |||
| No. positive | 7 | 5 | 100 (59.0–100) |
| Mean CT ± SD | 35.5 ± 2.8 | 33.0 ± 2.3 | |
| Median CT (range) | 36 (30.7–38.9) | 33.2 (29.9–36) | |
| Neisseria meningitidis | |||
| No. positive | 5 | 5 | 100 (47.8–100) |
| Mean CT ± SD | 24.2 ± 1.9 | 23.16 ± 1.6 | |
| Median CT (range) | 24.4 (21.5–26.7) | 23 (21.1–24.9) | |
| EBV | |||
| No. positive | 8 | 10 | 80.0 (44.4–97.5) |
| Mean CT ± SD | 34.9 ± 2.0 | 32.2 ± 1.1 | |
| Median CT (range) | 35.5 (31.5–37.0) | 32.1 (29.8–34.0) | |
| VZV | |||
| No. positive | 1 | 3 | 33.3 (0.8–90.6) |
| Mean CT ± SD | 30.6 | 28.3 ± 3.5 | |
| Median CT (range) | 29.0 (24.4–31.3) | ||
| Streptococcus pneumoniae | |||
| No. positive | 5 | 6 | 83.3 (35.9–99.6) |
| Mean CT ± SD | 23.0 ± 3.2 | 22.4 ± 4.0 | |
| Median CT (range) | 22.8 (18.8–27.5) | 22.3 (17.6–27.6) | |
| Mumps virus | |||
| No. positive | 1 | 2 | 50.0 (1.3–98.7) |
| Mean CT ± SD | 31.8 | 27.3 ± 6.2 | |
| Median CT (range) | 27.3 (22.9–31.8) | ||
| Mycobacterium tuberculosis | |||
| No. positive | 6 | 7 | 85.7 (42.1–99.6) |
| Mean CT ± SD | 33.4 ± 2.5 | 32.2 ± 4.0 | |
| Median CT (range) | 33.1 (29.9–37.7) | 34.0 (24.8–36.8) | |
| Haemophilus influenzae | |||
| No. positive | 2 | 2 | 100 (47.8–100) |
| Mean CT ± SD | 22.1 ± 6.6 | 23.2 ± 5.6 | |
| Median CT (range) | 22.1 (17.4–26.8) | 23.2 (19.2–27.2) | |
| Acanthamoeba | |||
| No. positive | 0 | 2 | |
| Mean CT ± SD | 34.9 ± 3.2 | ||
| Median CT (range) | 34.1 (33.9–38.4) |
IRTP, individual real-time PCR; TAC, TaqMan array card; CI, confidence interval.
DISCUSSION
We describe our evaluation of an in-house-developed CNS-TAC assay that can be used to test cerebrospinal fluid (CSF) for infections associated with meningitis and encephalitis. Infections of the CNS comprise a number of serious and often fatal infections, and yet such infections often pose challenges in diagnosis (15). Many of the pathogens associated with CNS infections are detected by culture, microscopy, or antigen detection techniques. PCR is generally more reliable at detecting pathogens in the CSF, with substantially higher sensitivity than other diagnostic methods such as culture and enzyme-linked immunosorbent assay if the samples are collected at the appropriate time during the infection (16–18). PCR-based approaches for detecting multiple pathogens in a single array not only increase the number of pathogens that can be detected but also reduce the overall amount of time needed to rule out multiple pathogens. Therefore, we utilized previously published real-time PCR assays incorporated into a CNS-TAC assay. With improvements in sensitivity and ease of use, such multipathogen TAC assays have been used in the detection of both human respiratory and enteric pathogens (8–10). All CNS-TAC assays had LODs similar to what has been described elsewhere for respiratory and enteric pathogens (8, 10). Although this CNS-TAC method was designed for East Africa, many of the pathogens evaluated here cause CNS infections worldwide, and therefore the card is suitable for broader use. In this study, we designed and evaluated a CNS-TAC method that was able to detect 13 viruses, 6 bacteria, and 2 parasites. Clinical evaluation was against 120 patient samples; 35 of these specimens were found to be positive for eight pathogens using IRTP. The patient samples used in clinical validation were collected from subjects presenting at either Mbagathi District Hospital or Siaya District Hospital for patient care.
Analysis of the clinical validation demonstrated an average sensitivity of 79% across the TAC. This excluded the two Acanthamoeba positive specimens that failed to amplify by IRTP, suggesting that these were false-positive reactions. However, this average sensitivity was skewed by the VZV assay, which had a suboptimal sensitivity of 33.3%; eliminating these results from the calculation yields an average sensitivity of 85.6%. The low sensitivity for VZV and mumps virus may be attributed to the low numbers of positive samples tested. A parasite with global distribution, Balamuthia mandrillaris, was not detected by the assay in this card. This could have been due to the small sample size tested or to the absence of this pathogen in patients from the two geographical regions sampled in Kenya. Additional positive samples for these targets are needed to accurately determine the sensitivity of these assays. Alternatively, we advise the exclusion of these targets with low sensitivities from cards designed for future studies. Acanthamoeba spp. and Balamuthia mandrillaris are free-living amoebas that can potentially cause infections in humans and have been implicated in CNS infections worldwide (19). However, the prevalence of these pathogens is not well documented in sub-Saharan Africa. On the other hand, Plasmodium falciparum infection can present in a severe form of cerebral malaria, with a mortality rate of 10 to 25%, and is most common in sub-Saharan Africa (20, 21). Similarly, several studies have implicated cryptococcal meningitis as the major cause of meningitis among HIV-infected individuals in sub-Saharan Africa (22–24). Therefore, we recommend replacement of the Acanthamoeba and Balamuthia mandrillaris targets with Plasmodium falciparum and cryptococcal meningitis, which are more prevalent in sub-Saharan Africa, in future versions of these cards. The average specificity for CNS-TAC assay for the eight targets was 96.7%. A subset of samples failed to amplify RNase P in CNS-TAC (13%) and IRTP (6%) analyses. Usually, this would suggest inappropriate specimen collection, sample degradation, or the complete absence of human DNA in some CSF samples. However, the concentration of RNase P should reflect the concentration of white blood cells in the specimen, since CSF is usually free of human DNA. This suggests that a different marker should be used in the future as a control for specimen integrity for CSF.
The CNS-TAC assay detected seven targets that were not detected by the IRTP assays. These additional detections failed to yield positive results using IRTP despite numerous attempts, and as such, are possibly false positives that would negatively impact the specificity of the TAC assays since the IRTP method was considered the gold standard for these comparisons. These discrepancies could possibly be explained by additional freeze-thaw cycles negatively impacting nucleic acid integrity for the IRTP assay. However, the observed mean CT of 30.8 ± 4.2 indicates significant amplification and would argue against this possibility.
In our protocol, two of eight lanes were occupied on the first TAC: one of the eight lanes of the card was designated a no-template negative control, and another was designated the combined positive control. Subsequent cards tested would hold a negative control and seven specimens on a card. Up to three cards were tested per day in one ViiA-7 machine, which allowed for 20 specimens to be tested for 21 pathogens each per day. This greatly reduces the turnaround time for specimen testing compared to IRTP assays. Despite the discrepancies observed between TAC and IRTP, we think there is added value in the use of CNS-TAC as a screening assay in outbreak settings. Indeed, samples with positive IRTP or TAC results require further investigations, including gene sequencing, among other confirmatory tests. Our future plans are therefore to confirm all the TAC positives by sequencing, as well as to further validate the CNS-TAC assay using a larger sample size from patients presenting with CNS infections from other geographical sites within Kenya. This will help us better understand the utility of CNS-TAC in outbreak investigations.
MATERIALS AND METHODS
CNS-TAC design.
The CNS card includes assays for the detection of six bacterial pathogens, Streptococcus pneumoniae, Haemophilus influenzae, Neisseria meningitidis, Mycoplasma pneumoniae, Mycobacterium tuberculosis, and Bartonella (genus specific); 13 viruses, parechovirus, pan-dengue virus (detects all four serotypes), Nipah virus, VZV, mumps virus, measles virus, pan-Lyssa virus, HSV-1 and -2, EBV, enterovirus, CMV, and chikungunya virus (detection is based upon two different gene targets for all three genotypes); and two parasites, Balamuthia mandrillaris and Acanthamoeba (Fig. 2). All primers and probes were adapted from previously published assays, except for the M. tuberculosis assay, which is described for the first time here. Primers and probes for the targets were titrated individually by real-time PCR using genomic DNA, plasmid DNA, or RNA on a Bio-Rad CFX 96 platform and AgPath-ID One-Step RT-PCR master mix (Life Technologies, United Kingdom). Once titrated, the primers and probe for each assay were preloaded and dried by a ViiA7 Applied Biosystems (Foster City, CA) instrument in duplicate wells on the TAC, which included three intrinsic controls: (i) an extraction control, RNase P; (ii) measles virus RNA acting as both a target control and a RNA control; and (iii) an internal manufacturer control, glyceraldehyde-3-phosphate (GAPDH) (Table 3). All of the assays in this card apart from lyssavirus and pan-dengue virus assays were developed at the Centers for Disease Control and Prevention (CDC) and are used routinely for clinical diagnosis in the United States. The other two assays were developed at the University of Pretoria, South Africa, and the Bernhard Nocht Institute, Germany, respectively.
FIG 2.
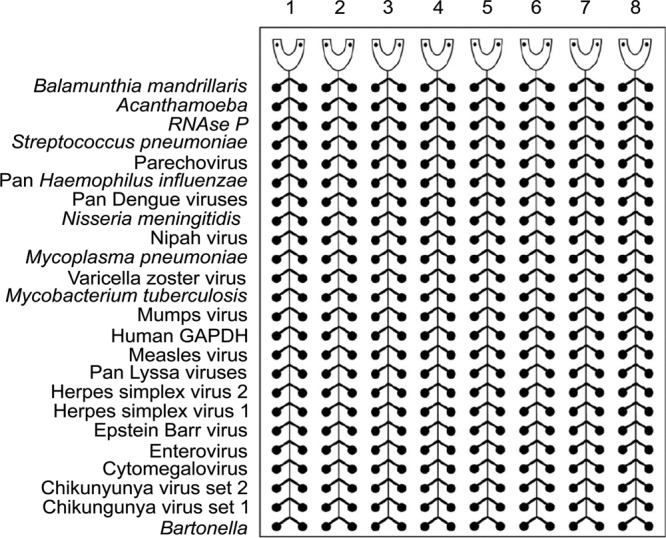
CNS-TAC layout with 22 encephalitis targets, as well two human DNA/RNA controls, GAPDH and RNase P. PCRs for all the targets, including intrinsic controls, were customized for testing in duplicates.
TABLE 3.
Oligonucleotide sequences for CNS-TAC assaysa
| Target | Gene | ID | Sequence (5′–3′) | Reference |
|---|---|---|---|---|
| CMV | UL55 | For | AGG TCT TCA AGG AAC TCA GCA AGA | 26 |
| Rev | CGG CAA TCG GTT TGT TGT AAA | |||
| Pr | FAM-ACC CCG TCA GCC ATT CTC TCG GC-BHQ 1 | |||
| EBV | BNRF1 | For | GGA ACC TGG TCA TCC TTT GC | 27 |
| Rev | ACG TGC ATG GAC CGG TTA AT | |||
| Pr | FAM-CGC AGG CAC TCG TAC TGC TCG CT-BHQ 1 | |||
| Mycobacterium tuberculosis | IS6110 | For | CCT ACT ACG ACC ACA TCA | —b |
| Rev | CCG TAA ACA CCG TAG TTG | |||
| Pr | FAM-ATG TGC TCC TTG AGT TCG CCA T-BHQ 1 | |||
| Neisseria meningitidis | sodC | For | CCA CCC GTG TGG ATC ATA ATA GA | 28 |
| Rev | GCA CAC TTA GGT GAT TTA CCT GCA T | |||
| Pr | FAM-CA TGA TGG CAC AGC AAC AAA TCC TGT TT-BHQ 1 | |||
| Streptococcus pneumoniae | lytA | For | ACG CAA TCT AGC AGA TGA AGC A | 29 |
| Rev | TCG TGC GTT TTA ATT CCA GCT | |||
| Pr | FAM-TG CCG AAA ACG CTT GAT ACA GGG AG-BHQ 1 | |||
| VZV | ORF29 | For | CAC GTA TTT TCA GTC CTC TTC AAG TG | 30 |
| Rev | TTAGACGTGGAGTTGACATCGTTT | |||
| Pr | FAM-TACCGCCCGTGGAGCGCG-BHQ 1 | |||
| Chikungunya virus 1 | NSP1 | For | AAAGGGCAAACTCAGCTTCAC | 31 |
| Rev | GCCTGGGCTCATCGTTATTC | |||
| Pr | FAM-CTGTGATACAGTGGTTTCGTGTG-BHQ 1 | |||
| Chikungunya virus 2 | NSP4 | For | TCACTCCCTGTTGGACTTGATAGA | 31 |
| Rev | TTGACGAACAGAGTTAGGAACATACC | |||
| Pr | FAM-AGGTACGCGCTTCAAGTTCGGCG-BHQ1 | |||
| Enterovirus | 5′ UTR | For | GGC CCC TGA ATG CGG CTA ATC C | 32 |
| Rev | GCG ATT GTC ACC ATWA GCA GYC A | |||
| Pr | FAM-CC GAC TAC TTT GGG WGT CCG TGT-BHQ1 | |||
| Mycoplasma pneumoniae | CARDS toxin | For | TTT GGT AGC TGG TTA CGG GAA T | 33 |
| Rev | GGT CGG CAC GAA TTT CAT ATA AG | |||
| Pr | FAM-TG TAC CAG AGC ACC CCA GAA GGG CT-BHQ1 | |||
| HSV 1 | US4 | For | TAT TGG TGC GAT GGC GAC AC | 34 |
| Rev | CTT TCC GCA TGT GGG CTC TC | |||
| Pr | FAM-CCC CGC CCC ATA CCC TAC CCG C-BHQ1 | |||
| HSV 2 | US6 | For | AGC ATC CCG ATC ACT GTG TAC TA | 34 |
| Rev | GCG ATG GTC AGG TTG TAC GT | |||
| Pr | FAM-CAG TGC TGG AAC GTG CCT GCC GC-BHQ 1 | |||
| Measles virus | N | For | TGG CAT CTG AAC TCG GTA TCA C | 35 |
| Rev | TGT CCT CAG TAG TAT GCA TTG CAA | |||
| Pr | FAM-CCGAG GAT GCA AGG CTT GTT TCA GA-BHQ1 | |||
| Mumps virus | NP | For | GTA TGA CAG CGT ACG ACC AAC CT | 36 |
| Rev | GCG ACC TTG CTG CTG GTA TT | |||
| Pr | FAM-CC GGG TCT GCT GAT CGG CGA T-BHQ 1 | |||
| Parechovirus | 5′ UTR | For | GTAACASWWGCCTCTGGGSCCAAAAG | 37 |
| Rev | GGCCCCWGRTCAGATCCAYAGT | |||
| Pr | FAM-CCTRYGGGTACCTYCWGGGCATCCTT-BHQ 1 | |||
| Bartonella | ssrA | For | GCTATGGTAATAAATGGACAATGAAATAA | 38 |
| Rev | GCTTCTGTTGCCAGGTG | |||
| Pr | FAM-ACCCCGCTTAAACCTGCGACG-BHQ1 | |||
| Haemophilus influenzae | bexA | For | TGCGGTAGTGTTAGAAAATGGTATTATG | 39 |
| Rev | GGACAAACATCACAAGCGGTTA | |||
| Pr | FAM-ACAAAGCGTATCAATACTACAACGAGACGCAAAAA-BHQ1 | |||
| Acanthamoeba | 18S rRNA | For | CCCAGATCGTTTACCGTGAA | 40 |
| Rev | TAAATATTAATGCCCCCAACTATCC | |||
| Pr | FAM-CTGCCACCGAATACATTAGCATGG-BHQ1 | |||
| Lyssavirus | N | For | GTRCTCCARTTAGCRCACAT | 41 |
| Rev | CACMGSNAAYTAYAARACNAA | |||
| Pr | FAM-CATCACACCTTGATGACAACTCACAA-BHQ1 | |||
| Balamunthia mandrillaris | 18S rRNA | For | TAA CCT GCT AAA TAG TCA TGC CAA T | 40 |
| Rev | CAA ACT TCC CTC GGC TAA TCA | |||
| Pr | FAM-AG TAC TTC TAC CAA TCC AAC CGC CA- BHQ1 | |||
| Pan-dengue virus | 3′ NCR | For | GGA TAG ACC AGA GAT CCT GCT GT | 42 |
| Rev 1 | CAT TCC ATT TTC TGG CGT TC | |||
| Rev 2 | CAA TCC ATC TTG CGG CGC TC | |||
| Pr | FAM CA GCA TCA TTC CAG GCA CAG-BHQ1 | |||
| Nipah virus | N | For | CTG GTC TCT GCA GTT ATC ACC ATC GA | 43 |
| Rev | ACG TAC TTA GCC CAT CTT CTA GTT TCA | |||
| Pr | FAM-CAG CTC CCG ACA CTG CCG AGG AT-BHQ | |||
| RNase P | RPP30 | For | AGA TTT GGA CCT GCG AGC G | 44 |
| Rev | GAG CGG CTG TCT CCA CAA GT | |||
| Pr | FAM-TTC TGA CCT GAA GGC TCT GCG CG-BHQ |
ID, oligonucleotide identity; For, forward; Rev, reverse; Pr, probe; CMV, cytomegalovirus; EBV, Epstein-Barr virus; VZV, varicella-zoster virus; HSV, herpes simplex virus; UTR, untranslated region; NCR, noncoding region; ORF, open reading frame.
—, J. Posey, unpublished data.
Design of combined positive control.
Customized combined positive controls were designed and synthesized in two different plasmids. The design and orientation of the positive-control plasmid maps are similar to that described by Kodani and Winchell (25). The forward primer sequence was placed downstream of the plasmid pUC57 T7 sequence, followed by the probe sequence and finally the reverse primer sequence. The plasmid comprised these concatenated sequences for all targets. The two plasmids were designated A and B. Plasmid A contained sequences for Bartonella, CMV, EBV, HSV-1 and -2, mumps virus, Mycobacterium tuberculosis, VZV, Mycoplasma pneumoniae, Nipah virus, Neisseria meningitidis, pan-dengue virus, Haemophilus influenzae, parechovirus, Streptococcus pneumoniae, and RNase P. Plasmid B contained sequences for chikungunya virus targets 1 and 2, Acanthamoeba, and Balamuthia mandrillaris. Measles virus RNA was spiked into plasmid B preparation as an exogenous control for the virus, as well as an RNA control. Positive-control extracts were titrated following a 10-fold dilution to determine the LOD.
Analytical validation.
The LOD, linearity, repeatability, and reproducibility were determined using 10-fold dilutions of the positive-control material using infection-free CSF as the diluent. The positive-control materials were derived from nucleic acid materials from the respective targets. Nucleic acid material from RNA viruses was transcribed into cDNA and prepared for gene cloning. Repeatability was tested using eight repeats on a single card, whereas reproducibility was tested with 10 serial dilutions of each plasmid and assayed over 5 days. The lower LOD was defined as the lowest concentration at which the target could be detected in all of the diluted samples. Analytical validation of these assays was performed at the National Center for Emerging and Zoonotic Infectious Diseases at the CDC.
Testing of CSF.
CSF samples were obtained from patients in either Mbagathi District Hospital in Nairobi or Siaya District Hospital in rural Western Kenya. Children older than 6 weeks and adults of all ages were eligible for lumbar puncture if they presented with two or more signs and symptoms of CNS infection, such as fever (≥38°C) and/or history of reported fever in the last 3 days, neck stiffness and/or bulging fontanel, headache, reduced level of consciousness, or new-onset seizures. A total of 120 samples, including 35 that were positive for any of the CDC in-house IRTP assays, were also tested using the CNS-TAC method. CDC in-house assays were designed to include targets for CMV, EBV, mumps virus, Mycobacterium tuberculosis, VZV, Neisseria meningitidis, Haemophilus influenzae, Acanthamoeba, and Streptococcus pneumoniae. In addition, 85 randomly selected samples, determined to be negative in IRTP assays, were tested by the CNS-TAC assay to determine specificity.
Nucleic acids were extracted from CSF specimens using the KingFisher ML extraction platform (Thermo Scientific, Waltham, MA) and MagMax nucleic isolation kit (Life Technologies, Carlsbad, CA). Portions (100 μl) of CSF specimens were mixed with 260 μl of lysis binding solution and added to the columns. The column was washed once with 600 μl of wash solution 1 and then twice with 450 μl of wash solution 2 according to the manufacturer's recommendations. After the wash steps, the nucleic acids were eluted with 60 μl of elution buffer. An additional 166 μl of previously PCR-positive samples was reextracted for IRTP testing using the same platform and kit. We used 433 μl of lysis binding solution to adjust for the increased sample volume. The samples were then eluted in 100 μl of elution buffer. An increased extraction volume was required for the eight IRTP assays, and this increased volume did not alter the sensitivity of the assays. The CNS-TAC assays were compared to the cognate IRTP assays on 96-well plates under the same thermocycling conditions using the same PCR master mix and 5 μl of nucleic acids as the template. Samples with a CT of ≥40 were interpreted as negative, and those with a CT of 35 to 40 were classified as indeterminate and retested. If these CT values remained within the range of 35 to 40, they were ultimately classified as weak positives.
The CNS-TAC assays were run on a ViiA-7 real-time PCR system using an AgPath-ID One-Step real-time PCR kit (Applied Biosystems, Foster City, CA). The PCR master mix for each card included 1× RT-PCR buffer, RT-PCR enzyme in a final 100-μl reaction volume. A 46-μl portion of nucleic acid extract was added to the master mix. Each run consisted of a negative control and a positive control for the first card of the day to be tested. A minimum of three cards were tested per day, with thermal cycling conditions as follows: 45°C for 10 min, 94°C for 10 min, and then 45 cycles of 94°C for 30 s and 60°C for 1 min. These clinical analyses were performed at the Centre for Global Health Research of the Kenya Medical Research Institute (KEMRI) in western Kenya.
Data analysis.
Receiver operating characteristic analysis was used to derive CT cutoffs. The CT values for CNS-TAC and IRTP assays were compared using a t test, whereas dichotomous measures of the presence or absence of extrinsic controls were compared using a Fisher exact test. Linearity was tested by fitting linear regression models of CT values against the concentrations of nucleic acids and interpreting the R2. The sensitivity of the CNS-TAC assay was also calculated against the gold standard: IRTP for nine targets. All analyses were performed using STATA v13 (StataCorp).
Ethical approval.
This study was covered under an investigational protocol reviewed by human subject review experts from the institutional review boards at the CDC (protocol 6092) and KEMRI (SSC protocol 1948). Informed written consent for survey participation, and CSF collection was obtained from all adult participants 18 years of age and older and from mature minors 13 to 17 years old. Verbal assent from minors (children 13 to 17 years old) and written consent from parents or guardians of those minors were obtained, and written consent from parents was obtained for children ≤13 years old. If a patient of any age was unable to provide consent or assent because of altered mental status, consent was obtained from the patient's responsible family member or guardian.
ACKNOWLEDGMENTS
We thank Jan Pohl, Maya Kodani, and Dean Erdman for assistance in the analytical validation. We also acknowledge the KEMRI staff who helped collect specimens used in clinical validation and laboratory testing. Work in Kenya was made possible through assistance by a number of personnel, including Janet Awando, Bryan Nyawanda, Weldon Korir, Newton Wamola, Patrick Emojoong, Jeremiah Nyaundi, Fredrick Ade, Victoria Mwende, and Jim Katieno.
This study was partly funded by the 2012 U.S. Department of Defense, Defense Threat Reduction Agency/Cooperative Biologic Engagement Program (DTRA/CBEP) funding (KE 06; Deployment of Diagnostic Tests, Including Rapid Diagnostic Tests for Diagnosis of Human and Animal Infections to District Laboratories in Kenya). Additional funding was from internal CDC-Kenya funds.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the U.S. Centers for Disease Control and Prevention.
REFERENCES
- 1.Mace SE. 2010. Central nervous system infections as a cause of an altered mental status? What is the pathogen growing in your central nervous system? Emerg Med Clin North Am 28:535–570. doi: 10.1016/j.emc.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Venkatesan A, Tunkel AR, Bloch KC, Lauring AS, Sejvar J, Bitnun A, Stahl JP, Mailles A, Drebot M, Rupprecht CE, Yoder J, Cope JR, Wilson MR, Whitley RJ, Sullivan J, Granerod J, Jones C, Eastwood K, Ward KN, Durrheim DN, Solbrig MV, Guo-Dong L, Glaser CA. 2013. Case definitions, diagnostic algorithms, and priorities in encephalitis: consensus statement of the International Encephalitis Consortium. Clin Infect Dis 57:1114–1128. doi: 10.1093/cid/cit458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitley RJ. 1990. Viral encephalitis. N Engl J Med 323:242–250. doi: 10.1056/NEJM199007263230406. [DOI] [PubMed] [Google Scholar]
- 4.Briese T, Palacios G, Kokoris M, Jabado O, Liu Z, Renwick N, Kapoor V, Casas I, Pozo F, Limberger R, Perez-Brena P, Ju J, Lipkin WI. 2005. Diagnostic system for rapid and sensitive differential detection of pathogens. Emerg Infect Dis 11:310–313. doi: 10.3201/eid1102.040492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dominguez SR, Briese T, Palacios G, Hui J, Villari J, Kapoor V, Tokarz R, Glode MP, Anderson MS, Robinson CC, Holmes KV, Lipkin WI. 2008. Multiplex MassTag-PCR for respiratory pathogens in pediatric nasopharyngeal washes negative by conventional diagnostic testing shows a high prevalence of viruses belonging to a newly recognized rhinovirus clade. J Clin Virol 43:219–222. doi: 10.1016/j.jcv.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pabbaraju K, Tokaryk KL, Wong S, Fox JD. 2008. Comparison of the Luminex xTAG respiratory viral panel with in-house nucleic acid amplification tests for diagnosis of respiratory virus infections. J Clin Microbiol 46:3056–3062. doi: 10.1128/JCM.00878-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reijans M, Dingemans G, Klaassen CH, Meis JF, Keijdener J, Mulders B, Eadie K, van Leeuwen W, van Belkum A, Horrevorts AM, Simons G. 2008. RespiFinder: a new multiparameter test to differentially identify fifteen respiratory viruses. J Clin Microbiol 46:1232–1240. doi: 10.1128/JCM.02294-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kodani M, Yang G, Conklin LM, Travis TC, Whitney CG, Anderson LJ, Schrag SJ, Taylor TH Jr, Beall BW, Breiman RF, Feikin DR, Njenga MK, Mayer LW, Oberste MS, Tondella ML, Winchell JM, Lindstrom SL, Erdman DD, Fields BS. 2011. Application of TaqMan low-density arrays for simultaneous detection of multiple respiratory pathogens. J Clin Microbiol 49:2175–2182. doi: 10.1128/JCM.02270-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinberg GA, Schnabel KC, Erdman DD, Prill MM, Iwane MK, Shelley LM, Whitaker BL, Szilagyi PG, Hall CB. 2013. Field evaluation of TaqMan Array Card (TAC) for the simultaneous detection of multiple respiratory viruses in children with acute respiratory infection. J Clin Virol 57:254–260. doi: 10.1016/j.jcv.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Platts-Mills JA, Gratz J, Mduma E, Svensen E, Amour C, Liu J, Maro A, Saidi Q, Swai N, Kumburu H, McCormick BJ, Kibiki G, Houpt ER. 2014. Association between stool enteropathogen quantity and disease in Tanzanian children using TaqMan array cards: a nested case-control study. Am J Trop Med Hyg 90:133–138. doi: 10.4269/ajtmh.13-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz MH, Waller JL, Napoliello RA, Islam MS, Wolff BJ, Burken DJ, Holden RL, Srinivasan V, Arvay M, McGee L, Oberste MS, Whitney CG, Schrag SJ, Winchell JM, Saha SK. 2013. Optimization of multiple pathogen detection using the TaqMan array card: application for a population-based study of neonatal infection. PLoS One 8:e66183. doi: 10.1371/journal.pone.0066183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Ochieng C, Wiersma S, Stroher U, Towner JS, Whitmer S, Nichol ST, Moore CC, Kersh GJ, Kato C, Sexton C, Petersen J, Massung R, Hercik C, Crump JA, Kibiki G, Maro A, Mujaga B, Gratz J, Jacob ST, Banura P, Scheld WM, Juma B, Onyango CO, Montgomery JM, Houpt E, Fields B. 2016. Development of a TaqMan array card for acute-febrile-illness outbreak investigation and surveillance of emerging pathogens, including Ebola virus. J Clin Microbiol 54:49–58. doi: 10.1128/JCM.02257-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Driscoll AJ, Karron RA, Bhat N, Thumar B, Kodani M, Fields BS, Whitney CG, Levine OS, O'Brien KL, Murdoch DR. 2014. Evaluation of fast-track diagnostics and TaqMan array card real-time PCR assays for the detection of respiratory pathogens. J Microbiol Methods 107:222–226. doi: 10.1016/j.mimet.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Kabir F, Manneh J, Lertsethtakarn P, Begum S, Gratz J, Becker SM, Operario DJ, Taniuchi M, Janaki L, Platts-Mills JA, Haverstick DM, Kabir M, Sobuz SU, Nakjarung K, Sakpaisal P, Silapong S, Bodhidatta L, Qureshi S, Kalam A, Saidi Q, Swai N, Mujaga B, Maro A, Kwambana B, Dione M, Antonio M, Kibiki G, Mason CJ, Haque R, Iqbal N, Zaidi AK, Houpt ER. 2014. Development and assessment of molecular diagnostic tests for 15 enteropathogens causing childhood diarrhoea: a multicentre study. Lancet Infect Dis 14:716–724. doi: 10.1016/S1473-3099(14)70808-4. [DOI] [PubMed] [Google Scholar]
- 15.Weber MW, Herman J, Jaffar S, Usen S, Oparaugo A, Omosigho C, Adegbola RA, Greenwood BM, Mulholland EK. 2002. Clinical predictors of bacterial meningitis in infants and young children in The Gambia. Trop Med Int Health 7:722–731. doi: 10.1046/j.1365-3156.2002.00926.x. [DOI] [PubMed] [Google Scholar]
- 16.Meyer T, Franke G, Polywka SK, Lutgehetmann M, Gbadamosi J, Magnus T, Aepfelbacher M. 2014. Improved detection of bacterial central nervous system infections by use of a broad-range PCR assay. J Clin Microbiol 52:1751–1753. doi: 10.1128/JCM.00469-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welinder-Olsson C, Dotevall L, Hogevik H, Jungnelius R, Trollfors B, Wahl M, Larsson P. 2007. Comparison of broad-range bacterial PCR and culture of cerebrospinal fluid for diagnosis of community-acquired bacterial meningitis. Clin Microbiol Infect 13:879–886. doi: 10.1111/j.1469-0691.2007.01756.x. [DOI] [PubMed] [Google Scholar]
- 18.Wu HM, Cordeiro SM, Harcourt BH, Carvalho M, Azevedo J, Oliveira TQ, Leite MC, Salgado K, Reis MG, Plikaytis BD, Clark TA, Mayer LW, Ko AI, Martin SW, Reis JN. 2013. Accuracy of real-time PCR, Gram stain, and culture for Streptococcus pneumoniae, Neisseria meningitidis, and Haemophilus influenzae meningitis diagnosis. BMC Infect Dis 13:26. doi: 10.1186/1471-2334-13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Visvesvara GS, Moura H, Schuster FL. 2007. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol Med Microbiol 50:1–26. doi: 10.1111/j.1574-695X.2007.00232.x. [DOI] [PubMed] [Google Scholar]
- 20.Mishra SK, Newton CR. 2009. Diagnosis and management of the neurological complications of falciparum malaria. Nat Rev Neurol 5:189–198. doi: 10.1038/nrneurol.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor TE, Fu WJ, Carr RA, Whitten RO, Mueller JS, Fosiko NG, Lewallen S, Liomba NG, Molyneux ME. 2004. Differentiating the pathologies of cerebral malaria by postmortem parasite counts. Nat Med 10:143–145. doi: 10.1038/nm986. [DOI] [PubMed] [Google Scholar]
- 22.Britz E, Perovic O, von Mollendorf C, von Gottberg A, Iyaloo S, Quan V, Chetty V, Sriruttan C, Ismail NA, Nanoo A, Musekiwa A, Reddy C, Viljoen K, Cohen C, Govender NP. 2016. The epidemiology of meningitis among adults in a South African province with a high HIV prevalence, 2009-2012. PLoS One 11:e0163036. doi: 10.1371/journal.pone.0163036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jarvis JN, Meintjes G, Williams A, Brown Y, Crede T, Harrison TS. 2010. Adult meningitis in a setting of high HIV and TB prevalence: findings from 4961 suspected cases. BMC Infect Dis 10:67. doi: 10.1186/1471-2334-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajasingham R, Rhein J, Klammer K, Musubire A, Nabeta H, Akampurira A, Mossel EC, Williams DA, Boxrud DJ, Crabtree MB, Miller BR, Rolfes MA, Tengsupakul S, Andama AO, Meya DB, Boulware DR. 2014. Epidemiology of meningitis in an HIV-infected Ugandan cohort. Am J Trop Med Hyg 92:274–279. doi: 10.4269/ajtmh.14-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kodani M, Winchell JM. 2012. Engineered combined-positive-control template for real-time reverse transcription-PCR in multiple-pathogen-detection assays. J Clin Microbiol 50:1057–1060. doi: 10.1128/JCM.05987-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boppana SB, Fowler KB, Pass RF, Rivera LB, Bradford RD, Lakeman FD, Britt WJ. 2005. Congenital cytomegalovirus infection: association between virus burden in infancy and hearing loss. J Pediatr 146:817–823. doi: 10.1016/j.jpeds.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 27.Ramamurthy M, Alexander M, Aaron S, Kannangai R, Ravi V, Sridharan G, Abraham AM. 2011. Comparison of a conventional polymerase chain reaction with real-time polymerase chain reaction for the detection of neurotropic viruses in cerebrospinal fluid samples. Indian J Med Microbiol 29:102–109. doi: 10.4103/0255-0857.81777. [DOI] [PubMed] [Google Scholar]
- 28.Dolan Thomas J, Hatcher CP, Satterfield DA, Theodore MJ, Bach MC, Linscott KB, Zhao X, Wang X, Mair R, Schmink S, Arnold KE, Stephens DS, Harrison LH, Hollick RA, Andrade AL, Lamaro-Cardoso J, de Lemos AP, Gritzfeld J, Gordon S, Soysal A, Bakir M, Sharma D, Jain S, Satola SW, Messonnier NE, Mayer LW. 2011. sodC-based real-time PCR for detection of Neisseria meningitidis. PLoS One 6:e19361. doi: 10.1371/journal.pone.0019361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carvalho MDGS, Tondella ML, McCaustland K, Weidlich L, McGee L, Mayer LW, Steigerwalt A, Whaley M, Facklam RR, Fields B, Carlone G, Ades EW, Dagan R, Sampson JS. 2007. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol 45:2460–2466. doi: 10.1128/JCM.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinlivan ML, Ayres K, Ran H, McElwaine S, Leedham-Green M, Scott FT, Johnson RW, Breuer J. 2007. Effect of viral load on the outcome of herpes zoster. J Clin Microbiol 45:3909–3914. doi: 10.1128/JCM.00874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanciotti RS, Kosoy OL, Laven JJ, Panella AJ, Velez JO, Lambert AJ, Campbell GL. 2007. Chikungunya virus in US travelers returning from India, 2006. Emerg Infect Dis 13:764–767. doi: 10.3201/eid1305.070015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kilpatrick DR, Yang CF, Ching K, Vincent A, Iber J, Campagnoli R, Mandelbaum M, De L, Yang SJ, Nix A, Kew OM. 2009. Rapid group-, serotype-, and vaccine strain-specific identification of poliovirus isolates by real-time reverse transcription-PCR using degenerate primers and probes containing deoxyinosine residues. J Clin Microbiol 47:1939–1941. doi: 10.1128/JCM.00702-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winchell JM, Thurman KA, Mitchell SL, Thacker WL, Fields BS. 2008. Evaluation of three real-time PCR assays for detection of Mycoplasma pneumoniae in an outbreak investigation. J Clin Microbiol 46:3116–3118. doi: 10.1128/JCM.00440-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miari VF, Wall GR, Clark DA. 2015. Evaluation of non-extracted genital swabs for real-time HSV PCR. J Med Virol 87:125–129. doi: 10.1002/jmv.23967. [DOI] [PubMed] [Google Scholar]
- 35.Hummel KB, Lowe L, Bellini WJ, Rota PA. 2006. Development of quantitative gene-specific real-time RT-PCR assays for the detection of measles virus in clinical specimens. J Virol Methods 132:166–173. doi: 10.1016/j.jviromet.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Rota JS, Rosen JB, Doll MK, McNall RJ, McGrew M, Williams N, Lopareva EN, Barskey AE, Punsalang A Jr, Rota PA, Oleszko WR, Hickman CJ, Zimmerman CM, Bellini WJ. 2013. Comparison of the sensitivity of laboratory diagnostic methods from a well-characterized outbreak of mumps in New York city in 2009. Clin Vaccine Immunol 20:391–396. doi: 10.1128/CVI.00660-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nix WA, Maher K, Johansson ES, Niklasson B, Lindberg AM, Pallansch MA, Oberste MS. 2008. Detection of all known parechoviruses by real-time PCR. J Clin Microbiol 46:2519–2524. doi: 10.1128/JCM.00277-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diaz MH, Bai Y, Malania L, Winchell JM, Kosoy MY. 2012. Development of a novel genus-specific real-time PCR assay for detection and differentiation of Bartonella species and genotypes. J Clin Microbiol 50:1645–1649. doi: 10.1128/JCM.06621-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Mair R, Hatcher C, Theodore MJ, Edmond K, Wu HM, Harcourt BH, Carvalho MDGS, Pimenta F, Nymadawa P, Altantsetseg D, Kirsch M, Satola SW, Cohn A, Messonnier NE, Mayer LW. 2011. Detection of bacterial pathogens in Mongolia meningitis surveillance with a new real-time PCR assay to detect Haemophilus influenzae. Int J Med Microbiol 301:303–309. doi: 10.1016/j.ijmm.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Qvarnstrom Y, Visvesvara GS, Sriram R, da Silva AJ. 2006. Multiplex real-time PCR assay for simultaneous detection of Acanthamoeba spp., Balamuthia mandrillaris, and Naegleria fowleri. J Clin Microbiol 44:3589–3595. doi: 10.1128/JCM.00875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coertse J, Weyer J, Nel LH, Markotter W. 2010. Improved PCR methods for detection of African rabies and rabies-related lyssaviruses. J Clin Microbiol 48:3949–3955. doi: 10.1128/JCM.01256-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drosten C, Gottig S, Schilling S, Asper M, Panning M, Schmitz H, Gunther S. 2002. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J Clin Microbiol 40:2323–2330. doi: 10.1128/JCM.40.7.2323-2330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lo MK, Lowe L, Hummel KB, Sazzad HM, Gurley ES, Hossain MJ, Luby SP, Miller DM, Comer JA, Rollin PE, Bellini WJ, Rota PA. 2012. Characterization of Nipah virus from outbreaks in Bangladesh, 2008-2010. Emerg Infect Dis 18:248–255. doi: 10.3201/eid1802.111492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo W, Yang H, Rathbun K, Pau CP, Ou CY. 2005. Detection of human immunodeficiency virus type 1 DNA in dried blood spots by a duplex real-time PCR assay. J Clin Microbiol 43:1851–1857. doi: 10.1128/JCM.43.4.1851-1857.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]