ABSTRACT
Multidrug-resistant (MDR) and extensively drug resistant (XDR) strains of Mycobacterium tuberculosis pose major problems for global health. The GeneXpert MTB/RIF (Xpert) assay rapidly detects resistance to rifampin (RIFr), but for detection of the additional resistance that defines MDR-TB (MDR tuberculosis) and XDR-TB, and for molecular epidemiology, specimen cultures and a biosafe infrastructure are generally required. We sought to determine whether the remnants of sputa prepared for the Xpert assay could be used directly to find mutations associated with drug resistance and to study molecular epidemiology, thus providing precise characterization of MDR-TB cases in countries lacking biosafety level 3 (BSL3) facilities for M. tuberculosis cultures. After sputa were processed and run on the Xpert instrument, the leftovers of the samples prepared for the Xpert assay were used for PCR amplification and sequencing or for a line probe assay to detect mutations associated with resistance to additional drugs, as well as for molecular epidemiology with spoligotyping and selective mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) typing. Of 130 sputum samples from Gabon tested with the Xpert assay, 124 yielded interpretable results; 21 (17%) of these were determined to be RIFr. Amplification and sequencing or a line probe assay of the Xpert remnants confirmed 18/21 samples as MDR, corresponding to 12/116 (9.5%) new and 6/8 (75%) previously treated TB patients. Spoligotyping and MIRU typing with hypervariable loci identified an MDR Beijing strain present in five samples. We conclude that the remnants of samples processed for the Xpert assay can be used in PCRs to find mutations associated with the resistance to the additional drugs that defines MDR and XDR-TB and to study molecular epidemiology without the need for culturing or a biosafe infrastructure.
KEYWORDS: Mycobacterium tuberculosis, Libreville, Gabon, MDR-TB, molecular epidemiology, GeneXpert, sequencing, Beijing family, drug resistance, drug sensitivity testing, multidrug resistance
INTRODUCTION
Tuberculosis (TB) is now the leading cause of death from an infectious disease worldwide, with an estimated 10.4 million new cases and 1.4 million deaths in 2015 (1). Of these, an estimated 480,000 new cases were multidrug-resistant TB (MDR-TB), caused by strains resistant to at least isoniazid (INH) and rifampin (RIF). There are effective multidrug regimens that can cure 80% of MDR-TB cases in 9 to 12 months (2–4), but the MDR-TB cases must first be detected (5). When an MDR strain is also resistant to the fluoroquinolones (FQ) and an injectable agent, the disease is termed extensively drug resistant TB (XDR-TB) and is unlikely to be cured with standard MDR treatment regimens (6).
Africa accounts for about one-quarter of the world's TB cases, but few countries in the region have detailed data on TB management and the prevalence of MDR cases (7). Gabon is a country in Central Africa with about 1.7 million inhabitants, half of whom live in the capital, Libreville. The World Health Organization (WHO) estimated that in 2014, Gabon had an annual incidence of 393 to 497 cases, with an accompanying mortality rate of 56 to 93 TB deaths per 100,000 inhabitants, making it one of the 10 highest-incidence countries (7). Between 8 and 11% of Gabonese TB patients are suspected of having MDR-TB (9), and a recent report from Lambaréné, Gabon, found that 42% of adult and 16% of pediatric TB patients were coinfected with the human immunodeficiency virus (HIV) (10).
The high incidence of TB cases and TB mortality reflects the deficiencies in Gabon's TB control program (11–13). A survey in 2006 found that only 55% of patients, whose strains were not routinely tested for drug resistance, completed the standard first-line drug regimen, and a more recent study found only 53% treatment success (7, 10–12). There is no public health laboratory in Gabon with the ability to perform phenotypic drug susceptibility testing (DST) (5), and MDR drug regimens are not administered with standardized protocols in most of the country. However, a WHO-approved MDR treatment program is currently being implemented, and therefore, identification of MDR-TB patients to receive this treatment has become a priority. The GeneXpert MTB/RIF assay (Xpert assay; Cepheid, Sunnyvale, CA, USA), was recently introduced into the country and can rapidly analyze sputum samples to detect the presence of Mycobacterium tuberculosis bacilli, as well as mutations associated with resistance to rifampin (RIFr), a marker for MDR-TB (14, 15). The Xpert assay does not, however, detect the mutations associated with the additional drug resistance that defines XDR-TB.
We report here a novel method, not requiring cultures or a biosafe infrastructure, that can detect resistance to drugs other than rifampin. The method first employs the Xpert assay to detect the presence of M. tuberculosis bacilli and RIFr; it then uses the remainder of the sputum samples prepared for the Xpert assay in PCRs to identify mutations associated with resistance to additional drugs as well as to define the lineage of circulating TB strains. This strategy was used to profile the nature and extent of drug resistance in Libreville, Gabon.
RESULTS
Use of Xpert remnants for amplification and sequencing to find mutations conferring resistance to first- and second-line drugs.
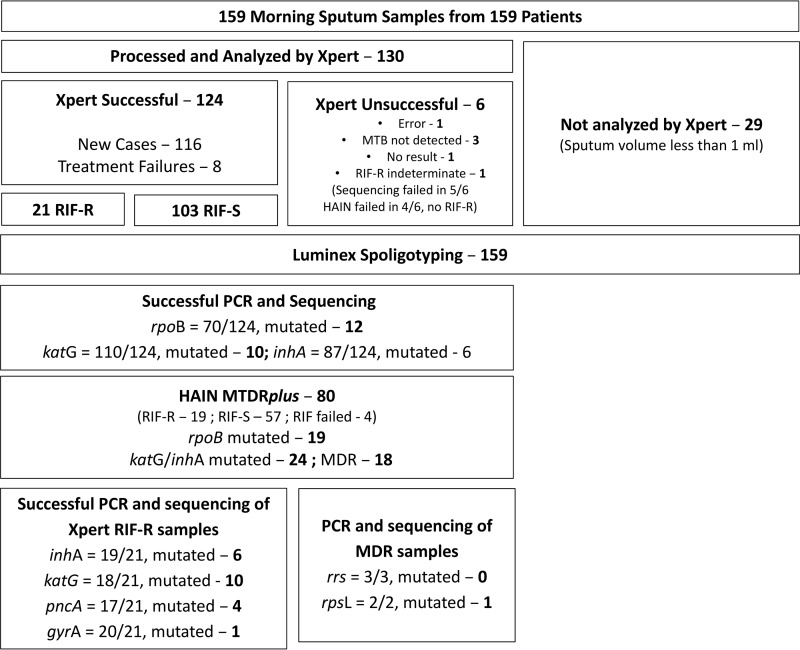
Of the 130 samples run on the Xpert instrument, 1 gave a no-result signal, 1 gave an error signal, 1 was positive for M. tuberculosis but indeterminate for RIFr, and M. tuberculosis was not detected in 3. Among the remaining 124 Xpert samples, RIFr was detected in 21 (17%), of which 12 were mutated at RpoB amino acid residue 531 or 533, and 9 were mutated at RpoB residue 516 or 522 (Table 1; Fig. 1).
TABLE 1.
Mutations determined by the GeneXpert MTB/RIF assay, the Hain MTBDRplus test, and sequencing of the individual genes, performed with leftovers of the Xpert processed sputum specimens
| Sample | Patienta |
GeneXpert result |
rpoB result |
MDR |
katG result (codon 315) |
inhA promoter result (position −15) |
Sequencing result for: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | Age (yr) | Probe(s)b | Mutation(s) detected at codon(s): | Sequencingc | Hain test | Sequencing | Hain test | Sequencing | Hain test | pncA | rrs | rpsL | gyrA | ||
| GAB-001 | M | 54 | B and C | 516 or 522 | D516Y | Mutated (probes WT4 and -5 absent) | x | S315T | S315T | WT | WT | WT | WT | ||
| GAB-065 | F | 19 | B and C | 516 or 522 | D516Y | Mutated (probe WT4 absent) | x | S315T | S315T | WT | WT | WT | WT | ||
| GAB-152 | F | 18 | B | 516 | D516Y | Mutated (probe WT4 absent) | x | S315T | S315T | WT | WT | WT | WT | ||
| GAB-076 | F | 26 | C | 522 | D516Y, S522T | Mutated (probe WT4 absent) | x | S315T | S315T | WT | WT | WT | WT | ||
| GAB-163 | F | 23 | E | 531 or 533 | NS | S531L | x | S315T | S315T | WT | WT | NS | WT | ||
| GAB-003 | M | 35 | E | 531 or 533 | S531L | S531L | x | S315T | S315T | WT | WT | WT | WT | ||
| GAB-009 | F | 37 | E | 531 or 533 | S531L | S531L | x | S315T | S315T | WT | WT | G97S | WT | ||
| GAB-182 | M | 32 | E | 531 or 533 | S531L | S531L | x | S315T | S315T | WT | WT | T135P | WT | ||
| GAB-002 | F | 50 | B | 516 | D516Y | Mutated (probe WT4 absent) | x | S315T | S315T | WT | C → T | WT | WT | K43R | D94G |
| GAB-180 | M | 19 | B | 516 | D516Y | Mutated (probe WT4 absent) | x | S315T | S315T | WT | C → T | WT | WT | ||
| GAB-014 | M | 30 | E | 531 or 533 | NS | Mutated (probe WT8 absent) | x | WT | WT | C → T | C → T | NS | WT | NS | |
| GAB-173 | M | 44 | E | 531 or 533 | S531L | S531L | x | WT | WT | C → T | C → T | WT | WT | WT | WT |
| GAB-062 | M | 18 | E | 531 or 533 | NS | S531L | x | WT | WT | C → T | C → T | WT | WT | ||
| GAB-151 | F | 42 | E | 531 or 533 | NS | S531L | x | WT | WT | C → T | C → T | WT | WT | ||
| GAB-157 | M | 25 | E | 531 or 533 | NS | S531L | x | WT | WT | C → T | C → T | WT | WT | ||
| GAB-010 | F | 28 | E | 531 or 533 | S531L | S531L | x | WT | WT | C → T | WT | S88Stop | WT | ||
| GAB-068 | F | 54 | E | 531 or 533 | NS | S531L | x | NS | S315T | WT | WT | G97S | WT | ||
| GAB-072 | M | 23 | C | 522 | NS | Mutated (probes WT3 and -4 absent) | x | NS | S315T | NS | WT | WT | WT | ||
| GAB-017 | M | 26 | E | 531 or 533 | S531L | S531L | WT | S315T | WT | WT | NS | WT | |||
| GAB-059 | F | 32 | B | 516 | NS | WT | NS | WT/S315Td | WT | WT | WT | WT | |||
| GAB-191 | ? | ? | B | 516 | NS | TUBe control absent | WT | WT | NS | C → T | NS | WT | |||
M, male; F, female; ?, unknown.
Xpert probe B can also detect less-frequent mutations affecting amino acid 513 and a deletion of amino acids 516 and 517 (31).
NS, PCR amplification was not successful.
Mixed profile; see Results.
TUB, M. tuberculosis complex specific.
FIG 1.
Flow chart showing the processing of the 159 sputum samples.
We then wanted to determine whether the leftover sputa processed in the Xpert assay could be used with other tests to obtain information on additional drug resistance. The Hain GenoType MTBDRplus assay is a line probe assay that detects rifampin resistance-associated mutations in rpoB, as well as mutations in katG (16) and inhA (17) that are associated with resistance to isoniazid (INH). The Hain assay was performed using Xpert-processed leftovers in order to confirm the Xpert rpoB results and to identify strains that were also INH resistant, thus confirming them as MDR-TB (Table 1). The Hain test found rpoB mutations in 19/21 samples that were RIFr by the Xpert assay and also detected the KatG S315T substitution in 14/21 samples and the C → T mutation at position −15 in the inhA promoter in 8 samples, with GAB-180 and GAB-002 showing both katG and inhA mutations. In one of the two rpoB-discordant samples (GAB-059), where the Hain test did not confirm the presence of an rpoB mutation found by the Xpert assay, the Hain test detected a mixed KatG profile with positive results for both the wild-type (WT) sequence and the S315T substitution, suggesting that the sample might have contained a mixed population of two strains of M. tuberculosis. In that case, the rpoB codon 516 mutation detected by the Xpert assay could have been hidden in the Hain test by the presence of a WT rpoB sequence. In the other discordant strain (GAB-191), the RIF resistance mutation detected by the Xpert assay was not found with the Hain test, but because there was no hybridization to the M. tuberculosis complex-specific (TUB) control band, the Hain test for this sample was not valid.
To confirm which strains were MDR, the Xpert leftovers were used in PCRs to amplify the rpoB, katG, and inhA genes for DNA sequencing (Fig. 1). Readable sequences were obtained from 70/124 samples (56%) for rpoB, of which 12 had mutations; from 110/124 samples (89%) for katG, of which 10 had mutations; and from 87/124 (67%) samples for inhA, of which 6 had mutations. The 12 mutated rpoB sequences contained the same mutations found with the Xpert and Hain tests (Table 1), except that for sample GAB-076, the Xpert assay detected the codon 522 mutation while the Hain test detected the codon 516 mutation, but sequencing showed that both mutations were present. Unfortunately, sequencing of the rpoB gene was not successful for either of the two samples where the Hain test did not confirm the Xpert results (GAB-059 and GAB-191). Among the 21 samples found to be RIFr by the Xpert assay, readable katG sequences were obtained from 18, of which 10 had mutations resulting in the S315T substitution, and readable inhA sequences were obtained from 19, of which 6 carried the C → T mutation at position −15 in the promoter. Despite repeated attempts, it was not possible to amplify the rpoB gene from 9 samples, or the katG or inhA gene from 3 or 2 samples, respectively.
Most of the sequences yielded the same results found by the Hain test, but a few were discordant. For two samples (GAB-002 and -180), the Hain test detected the C → T mutation at position −15 in the inhA promoter, which was not present in the sequences, but katG mutations were detected in these samples by both the Hain test and sequencing, making them MDR. Sample GAB-010 was WT for both katG and inhA by the Hain test but showed the C → T mutation at position −15 in the inhA promoter by sequencing and was therefore classified as MDR.
Based on sequencing results or a Hain test without a discordant sequence, 18 of the 21 samples (86%) determined to be RIFr by the Xpert assay would be classified as MDR. Results for the remaining three samples were as follows. For GAB-017, the Hain test found a KatG S315T substitution, but the katG sequence was WT. For GAB-191, the Hain test found the inhA promoter mutation, but the absence of a positive control invalidated the Hain test, and no inhA sequence was available. For GAB-059, the Hain test did not detect an rpoB mutation and showed both mutated and WT katG sequences, suggesting the possible presence of two strains, one WT and one MDR with rpoB and katG mutations.
To detect additional antibiotic resistance in the 21 samples determined to be RIFr by the Xpert assay, we amplified and sequenced the pncA, gyrA, rrs, and rpsL genes for detection of mutations associated with resistance to pyrazinamide (PZA), fluoroquinolones, the injectable antibiotics, and streptomycin, respectively. Readable sequences were obtained for pncA in 17/21 (81%) samples, of which 4 (24%) carried mutations (Table 1; Fig. 1). For the gyrA gene, 20/21 (95%) samples yielded readable sequences, of which 1 (5%) was mutated. The rrs gene was amplified and sequenced from the only three samples that could contain XDR-TB: the sample with the gyrA mutation, the sample for which sequencing of gyrA was unsuccessful, and a sample from a patient who had received 7 months of treatment with streptomycin (SM). All three were WT for the rrs gene, showing that there were no XDR-TB cases. The rpsL gene was sequenced from the samples of two patients: the patient treated with SM (GAB-173) had a WT rpsL sequence; but the sample with the gyrA mutation (GAB-002) also had a mutation causing a K43R substitution in RpsL, making this MDR-FQr sample also SMr.
Epidemiology of MDR-TB patients.
Epidemiological data for MDR-TB patients are shown in Table 2. Of the 18 patients confirmed to have MDR-TB strains, 9 were male and 9 were female. The mean age was 32 years (range, 18 to 54), and 2 patients were non-Gabonese. Of the 8 patients who had been treated with first-line drugs previously, 6 (75%) had samples that were MDR. One of these 6 patients also had mutations associated with resistance to FQ and SM, and 2 others had mutations associated with resistance to PZA. Of the 116 individuals with new TB cases, 12 (10.3%) had strains that were MDR. Of these 12, 6 (50%) had not started TB treatment when the samples were taken. One of their strains had a pncA mutation. The remaining 6 individuals with new MDR-TB cases had begun first-line treatment 2, 3, 4 (2 patients), 6, or 8 months prior to the time the samples were obtained.
TABLE 2.
Epidemiology and spoligotypes of sputum samples determined by the Xpert assay to be RIFr
| Sample | Patient |
Spoligotype | Additional resistancea | SIT | Clade | Medical information |
Demographic information |
Laboratory and Xpert results |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | Age (yr) | Examb | Statusc | District | Nationality | Microscopy scored | Xpert score | |||||
| GAB-157 | M | 25 | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◻◼◼◼◼◼◼◼◼◻◻◼◼◼◼◼◼◼◼◼ | INH | Orphan | C4 | N | Akanda | Gabon | 1+ | Medium | |
| GAB-065 | F | 19 | ◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◼◼◼◼◼◼◼◼◼ | INH | 1 | Beijing | D | N | Owendo | Gabon | 2+ | Medium |
| GAB-001 | M | 54 | ◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◼◼◼◼◼◼◼◼◼ | INH | 1 | Beijing | C | FR | ND | Gabon | 10 AFB | Medium |
| GAB-072 | M | 23 | ◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◼◼◼◼◼◼◼◼◼ | INH | 1 | Beijing | C6 | N | 5th | Gabon | 1+ | Very low |
| GAB-076 | F | 26 | ◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◼◼◼◼◼◼◼◼◼ | INH | 1 | Beijing | C4 | N | Akanda | Gabon | 2+ | Medium |
| GAB-180 | M | 19 | ◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◼◼◼◼◼◼◼◼◼ | INH | 1 | Beijing | C2 | N | 5th | Gabon | 1+ | High |
| GAB-003 | M | 35 | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◼◻◻◻◻◼◼◼◼◼◼◼ | INH | 50 | H3 | D | FR | 5th | Gabon | 3+ | Medium |
| GAB-163 | F | 23 | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◼◻◻◻◻◼◼◼◼◼◼◼ | INH | 50 | H3 | D | N | 6th | Gabon | 2+ | High |
| GAB-182 | M | 32 | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◼◻◻◻◻◼◼◼◼◼◼◼ | INH, PZA | 50 | H3 | C3 | FR | 2nd | Other | 1+ | Medium |
| GAB-014 | M | 30 | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◻◻◻◼◼◼◼◼◼◼ | INH | 53 | T1 | D | N | 5th | Gabon | 1+ | Medium |
| GAB-002 | F | 50 | ◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◼◼◻◻◼◼◼◼◼ | INH, FQ, SM | 260 | Beijing | C | FR | 5th | Gabon | 3+ | High |
| GAB-151 | F | 42 | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◻◼◼◼◼◼◼◼◼◻◻◻◻◼◻◻◼◼◼◼ | INH | 370 | T1 | D | N | 2nd | Gabon | 1+ | Medium |
| GAB-173 | M | 44 | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◻◼◼◼◼◼◼◼◼◻◻◻◻◼◻◻◼◼◼◼ | INH | 370 | T1 | C5 | FR | 2nd | Other | 7 AFB | High |
| GAB-068 | F | 54 | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◻◻◼◼◼◼◼◼◼◼ | INH, PZA | 1196 | C3 | N | 1st | Gabon | 1+ | Medium | |
| GAB-009 | F | 37 | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◻◻◼◼◼◼◼◼◼◼ | INH, PZA | 1196 | D | FR | Owendo | Gabon | 2+ | High | |
| GAB-010 | F | 28 | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◻◻◼◼◼◼◼◼◼◼ | INH, PZA | 1196 | D | N | 6th | Gabon | 2+ | Medium | |
| GAB-062 | M | 18 | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◻◼◼◼◼◼◼◼◼◻◻◻◻◼◼◼◼◼◼◼ | INH | 1580 | T1 | D | N | 2nd | Gabon | 1+ | Medium |
| GAB-017 | M | 26 | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◻◻◼◼◼◼◼◼◼◻◻◼◼◼◼◼◼◼◼◼ | INHe | 2697 | MANU2 | D | FR | 2nd | Gabon | 2+ | Very low |
| GAB-152 | F | 18 | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◻◻◼◼◼◼◼◼◼◻◻◼◼◼◼◼◼◼◼◼ | INH | 2697 | MANU2 | C8 | N | 1st | Gabon | 7 AFB | Medium |
| GAB-059 | F | 32 | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◻◻◼◼◼◼◼◼◼◻◻◼◼◼◼◼◼◼◼◼ | INHf | 2697 | MANU2 | D | N | 5th | Gabon | 1+ | Low |
| GAB-191 | ND | ND | ◻◻◻◻◻◼◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◼◻◻◻◼◻◼◼◼◼◼◼◼◼◼ | Orphan | ND | ND | ND | Gabon | ND | Medium | ||
INH, isoniazid; PZA, pyrazinamide; FQ, fluoroquinolones; SM, streptomycin.
C, control microscopy (with the number of months for which the individual had been under treatment with first-line drugs given in parentheses); D, initial microscopy for diagnosis; ND, no data.
N, new TB case; FR, failed or relapsed case.
3+, >10 acid-fast bacilli (AFB) per high-power field (HPF); 2+, 1 to 10 AFB per HPF; 1+, 10 to 99 AFB per 100 HPFs. If there are <10 AFB per 100 HPFs, the absolute number of AFB observed in 100 HPFs is shown.
For this strain, the Hain test found an S315T KatG substitution, but sequencing found WT katG.
This strain was RIFr by the Xpert assay but had a WT rpoR sequence by the Hain test, which also detected both WT katG and katG S315T. The Hain test detected a C → T mutation at position −15 in the inhA promoter, while sequencing found WT inhA.
Molecular epidemiology of MDR strains.
Spoligotypes were obtained for all 124 samples tested by the Xpert assay, as well as for the 29 samples with volume insufficient for the Xpert assay and the 6 samples for which Xpert analysis was unsuccessful (Fig. 1). The spoligotypes of two strains grouped with Mycobacterium africanum West African 1. The most frequent spoligotyping international type (SIT) was SIT61, found in 42 isolates (Table 3), but none of these were RIFr by any method used. In contrast, RIFr was found in 5 of 5 ST1, 3 of 16 SIT50, 3 of 6 SIT1196, 2 of 6 SIT370, 3 of 6 SIT2697, 1 of 2 SIT1580, 1 of 6 SIT53, and 1 of the 17 orphan isolates (Table 2). The single sample with SIT260, which, like ST1, belongs to the Beijing family, was also MDR and, additionally, FQr and SMr.
TABLE 3.
Clustered spoligotypesa
| SIT | Clade | No. of isolates (% of total) | Spoligotype | Octal |
|---|---|---|---|---|
| 61 | LAM10-CAM | 42 (26.42) | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◻◻◼◼◼◼◼◼◼◻◻◻◻◼◼◼◼◼◼◼ | 777777743760771 |
| 50 | H3 | 16 (10.1) | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◼◻◻◻◻◼◼◼◼◼◼◼ | 777777777720771 |
| 53 | T1 | 6 (3.8) | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◻◻◻◼◼◼◼◼◼◼ | 777777777760771 |
| 54 | MANU2 | 6 (3.8) | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◻◼◼◼◼◼◼◼◼◼ | 777777777763771 |
| 370 | T1 | 6 (3.8) | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◻◼◼◼◼◼◼◼◼◻◻◻◻◼◻◻◼◼◼◼ | 777777747760471 |
| 1196 | 6 (3.8) | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◻◻◼◼◼◼◼◼◼◼ | 777777777761771 | |
| 2697 | MANU2 | 6 (3.8) | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◻◻◼◼◼◼◼◼◼◻◻◼◼◼◼◼◼◼◼◼ | 777777743763771 |
| 1 | Beijing | 5 (3.1) | ◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◻◼◼◼◼◼◼◼◼◼ | 000000000003771 |
| 42 | LAM9 | 4 (2.5) | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◻◻◻◼◼◼◼◼◼◼◼◻◻◻◻◼◼◼◼◼◼◼ | 777777607760771 |
| 523 | MANU_ancestor | 4 (2.5) | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼ | 777777777777771 |
| 237 | 3 (1.9) | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◻◻◻◻◻◻◻◻◻◻◻◻ | 777777777700000 | |
| 378 | T1 | 3 (1.9) | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◼◼◻◼◼◼◼◼◼◼◼◻◻◻◻◼◼◼◼◼◼◼ | 777777667760771 |
| 2298 | T1 | 3 (1.9) | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◼◻◻◼◼◼◼◼◼◼◼◻◻◻◻◼◼◼◼◼◼◼ | 777777647760771 |
| 176 | LAM6 | 2 (1.3) | ◼◼◼◼◼◼◼◼◼◼◼◼◻◼◼◼◼◼◼◼◻◻◻◻◼◼◼◼◻◼◼◼◻◻◻◻◼◼◼◼◼◼◼ | 777737607560771 |
| 373 | T1 | 2 (1.3) | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◼◼◼◼◼◼◼◼◻◻◻◻◼◼◼◼◼◼◼ | 777777767760771 |
| 741 | H3 | 2 (1.3) | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◼◼◼◼◼◼◼◻◼◻◻◻◻◼◼◼◼◼◼◼ | 777777757720771 |
| 1580 | T1 | 2 (1.3) | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◻◼◼◼◼◼◼◼◼◻◻◻◻◼◼◼◼◼◼◼ | 777777747760771 |
| 1690 | MANU2 | 2 (1.3) | ◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◼◻◼◻◻◼◼◼◼◼◼◼ | 777777777762771 |
Of 159 total isolates, 120 (75.5%) belonged to clustered spoligotypes; 22 (13.8%) had unique SITs, and 17 (10.7%) were orphan strains.
The standard mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) loci are poorly discriminatory for Beijing strains, so in order to determine whether the five SIT1 Beijing MDR strains belonged to the same genotype, they were analyzed with 4 VNTR systems (VNTR systems 1982, 3232, 3820, and 4120) reported to be hypervariable for Beijing strains (18, 19). VNTR systems 1982, 3820, and 4120 yielded identical bands for the five SIT1 Beijing MDR samples (see Fig. S1 in the supplemental material), suggesting that they all belonged to the same genotype. Amplification with primers for system 3232 yielded several bands per sample and was considered uninterpretable. Sequencing of the Rv2629 gene showed that all five strains had the 191C genotype, described as a marker for the Beijing-W clade, cluster group 2 (20).
DISCUSSION
The Xpert technology has improved TB diagnosis and can rapidly detect RIFr, which, as confirmed in this study, is a reliable indicator of MDR-TB (21). However, most methods for detecting resistance to antibiotics other than rifampin require culturing of the specimen. Cultures on solid media take more than 2 weeks to turn positive, while cultures in liquid media can turn positive in 7 to 10 days (22) but pose a greater biosafety hazard. The Hain MTBDRplus test can be performed directly on sputum specimens but detects resistance to RIF and INH only. Amplification and sequencing of Xpert leftovers, the alternative method proposed here, yielded readable inhA sequences for 19, and readable katG sequences for 18, of the 21 strains determined to be RIFr by the Xpert assay. This method confirmed that 18/21 strains were unequivocally MDR. In four instances, the sequence was discordant with the Hain test result (GAB-002, -0180, -010, and -017), but two of these samples (GAB-002 and 180) were nonetheless defined as MDR because both sequencing and the Hain test detected substitutions at KatG residue 315. The other two had confirmed rpoB mutations and a mutation associated with INH resistance detected by only one technique, but for the sake of caution, these should probably be regarded as MDR despite the discordant results. The rpoB discrepancy for GAB-059, together with the presence of signals for both WT and mutated katG, suggests a mixed infection with both sensitive and resistant bacilli.
Testing for FQ resistance in MDR strains is critical, because treatment failures with FQ-containing MDR regimens are more likely when the strain is FQr, although MDR strains with particular gyrA mutations may be cured with higher FQ doses (23). The Hain MTBDRsl test can detect resistance to FQ and injectable antibiotics, and a new version, which appeared after the present study was completed, can be used directly with sputum specimens (24). It might be worthwhile to compare the performance of this test on Xpert leftovers with its results when it is used directly on sputa.
Our alternative method for detecting XDR-TB, using the leftovers from sputa processed for the Xpert assay to amplify and sequence the gyrA and rrs genes, is easy, quick, and relatively inexpensive. The sequencing can be done by a commercial service, and if the specimens are processed rapidly and promptly sent to be sequenced, the results could be available by Internet within about 7 days after the sputum specimen is obtained. Although amplification of the rpoB gene was successful for only 12 of 21 samples, it is unlikely that this low success rate was due to inhibitors in the stored Xpert leftovers, because readable sequences were obtained from 18/21 samples for katG, 19/21 samples for inhA, and 20/21 samples with gyrA. Perhaps the success rate with rpoB could be improved by optimizing the primers. The success rates of sequencing and the Hain test were roughly the same for sputa with 1+, 2+, or 3+ positivity but were lower for sputa with fewer than 1+ bacilli on microscopy and would probably be lower still for smear-negative specimens in which the Xpert assay detects M. tuberculosis and RIFr.
Xpert leftovers were also used for molecular epidemiology studies with both spoligotyping and MIRU-VNTR typing. Spoligotyping with the Luminex format showed that most of the MDR strains belonged to spoligotypes that were also present in non-MDR strains, but surprisingly, there were no MDR isolates with the most frequent spoligotype, SIT61, which was seen in 26% of all isolates examined (42/159). An alarming finding was that 6 of the 18 confirmed MDR strains belonged to SIT1 or SIT260, genotypes of the Beijing family that has been associated with many MDR-TB outbreaks (25, 26). Because MIRU-VNTR typing, even with 24 loci, lacks discriminatory power for the Beijing family, we used VNTR loci reported to be hypervariable in the Beijing lineage, which confirmed that the five SIT1 strains apparently belonged to the same genotype (18, 19). SIT1 was found only in these five MDR strains, and SIT260 was found only in one strain that was MDR, FQr, and SMr. This suggests that while most of the MDR strains could have developed drug resistance within Gabon, the SIT1 strains and the SIT260 strain may have been MDR when they were introduced into Libreville. The origin of these strains is a mystery, since reports from neighboring countries suggest that Beijing strains are not common in the region, and those found were generally not MDR (27–29). Follow-up studies are in progress to determine the extent and epidemiology of the apparent SIT1 MDR outbreak strain and to see if the MDR, FQr, SMr SIT260 Beijing strain is also spreading.
The strategy employed in this study provided important information on the urgent tuberculosis problem in Gabon, without the need for cultures or biohazard facilities. Because only 124 strains were examined in this study, and the sample was not representative of the entire country, the exact percentages of MDR-TB we found, 10% of new cases and 75% (6/8) of previously treated cases, may not accurately represent the situation for all of Gabon, but these high rates are nevertheless ominous and are likely attributable to the low success rates (only 53 to 55%) reported for first-line therapy in Gabon (10).
This method could be used to efficiently provide information on drug resistance and molecular epidemiology in other low-resource countries and can likely be used with any molecular test based on the amplification of specific genes or loci (30). Methods for phenotypic drug sensitivity testing rely on cultures and require a biosafe infrastructure to reduce the biohazards inherent in the manipulation of clinical samples, and heat inactivation of bacteria can generate aerosol risks. In contrast, the bacilli in samples processed for the Xpert assay are no longer viable (31), so they can be used in PCRs without any special infrastructure.
Whole-genome sequencing (WGS) has been proposed as a more-comprehensive and safer alternative to phenotypic drug testing and molecular epidemiology (32, 33). With advances in technology and cost reductions, WGS performed directly on clinical samples suspected to be MDR or XDR could eventually be cost-effective, even in low-resource settings. In a possible future algorithm, Xpert leftovers of RIFr samples could be sent for WGS to determine the full resistance profile, but this would require better sequencing technology and improved methods for isolating M. tuberculosis DNA from clinical specimens (34, 35). The strategy used in this study could represent an efficient interim solution for finding mutations associated with resistance to second- and third-line drugs, especially in settings where phenotypic drug sensitivity testing is not possible, and also for basic or retrospective molecular epidemiology.
MATERIALS AND METHODS
Specimens.
Between October 2014 and February 2015, 159 morning sputum samples were collected from 159 microscopy-positive patients who either were newly diagnosed with TB or were not cured after two courses of standard first-line therapy. Some patients had also received streptomycin (SM). The patients presented to one of the three main TB diagnostic laboratories in Libreville, the capital of Gabon: Nkembo Respiratory Hospital (124 samples), the National Public Health Laboratory (LNSP) (31 samples), and the Melen regional hospital (4 samples). The bacilli were seen in all samples after Ziehl-Neelsen staining and were scored as +/−, 1+, 2+, or 3+ (36).
The Ethics Committee of the Gabon LNSP approved the study (Décision 23022015-1). It was undertaken in the context of a research agreement between the Institut Pasteur of Paris, France, and the Government of Gabon. After a written or oral explanation, patients signed an informed consent to allow the use of their sputa in the study and to provide sociodemographic data and information on previous TB treatment. All sputum samples were transported to the National Public Health Laboratory and were processed for the Xpert assay.
Before the addition of the Xpert diluent, 29 sputum samples, 28 from new cases and 1 from a previously treated patient, had less than the 1-ml volume considered to be the minimum required for the Xpert assay (15). These samples were processed and, though not assayed with the Xpert test (Fig. 1), were used in PCRs for spoligotyping. After the specified volumes of the 130 processed sputum samples were run on the Xpert system, 375 μl of each of the leftover diluted samples was individually placed in a 50-ml tube containing 25 ml of autoclaved phosphate buffer (Na2HPO4·2H2O and KH2PO4 at 0.067 M [pH 6.8]) to neutralize the alkaline pH of the Xpert leftover to pH 7.0. After centrifugation at 4,500 rpm for 15 min, the pellets were resuspended in 100 μl 1× TE (10 mM Tris-HCl, 1 mM disodium EDTA [pH 8]) and were transferred to a microtube. The microtube was heated at 90°C for 30 min in a water bath and was frozen at −40°C for 1 h. After thawing, the lysate was centrifuged at 8,000 rpm for 5 min, and the supernatant was transferred to a new microtube and was stored at −40°C until it was sent to the Unité de Génétique Mycobacterienne at the Institut Pasteur in Paris, France, where the rest of the studies were performed. The 29 samples not run on the Xpert system were similarly processed.
Detection of drug resistance mutations.
The processing of the samples is shown in Fig. 1. PCR was performed with primers targeting rpoB, katG, inhA, pncA, gyrA, rrs, and rpsL (Table 4) to detect mutations conferring resistance to RIF, isoniazid (INH), pyrazinamide (PZA), the fluoroquinolones (FQ), amikacin (AMK), kanamycin (KAN), capreomycin (CPN), and SM (Table 4) (37–39). The rpoB, katG, inhA, pncA, gyrA, and rpsL genes were amplified separately in 50-μl reaction mixtures containing 1× NH4 buffer (Bioline), 10% dimethyl sulfoxide (DMSO), 1.5 mM MgCl2, 0.3 μM each primer, 200 μM deoxynucleoside triphosphates (dNTP), 1 U BioTaq DNA polymerase (Bioline), and 5 μl of the Xpert sample remnant diluted 1:10 in water. The thermocycler program was as follows: 95°C for 5 min; 45 cycles of 95°C for 1 min, the hybridization temperature (Table 4) for 1 min, and 72°C for 1 min; and a final extension at 72°C for 7 min. The rrs gene was amplified in a 30-μl reaction mixture containing 1× GC buffer (TaKaRa), 400 μM dNTP, 0.4 μM each primer, 1 U TaKaRa LA Taq (TaKaRa Bio Inc., Japan) and 5 μl of the Xpert sample remnant diluted 1:10 in water. The amplification program was as follows: 94°C for 5 min; 35 cycles of 94°C for 30 s, 59°C for 30 s, and 72°C for 2 min; and a final extension at 72°C for 5 min. Ten microliters of each PCR product was electrophoresed on a 1% agarose gel to verify the amplification. The PCR products were sequenced with the forward and reverse amplification primers (Cochin Sequencing Platform, Paris, France), and the sequencing results were aligned to the respective wild-type (WT) genes of M. tuberculosis H37Rv using Geneious (v9.04), MEGA (v6.06), or BLAST (www.ncbi.nlm.nih.gov). For all strains without readable rpoB sequences and all strains that were RIFr by the Xpert assay, the Hain GenoType MTBDRplus assay, v2.0 (Hain Lifescience, Nehren, Germany), was used to detect mutations in rpoB (31), katG, and inhA (40) according to the manufacturer's protocol, using 5 μl of the undiluted Xpert sample remnant.
TABLE 4.
Primers used to amplify and sequence the Mycobacterium tuberculosis genes associated with resistance to antibiotics
| Gene | Antibiotic | Primer name | Primer sequence (5′–3′) | Hybridization temp (°C) | Product size (bp) |
|---|---|---|---|---|---|
| rpoB | Rifampin | TR1 | TACGGTCGGCGAGCTGATCC | 53 | 411 |
| TR2 | TACGG CGTTTCGATGAACC | ||||
| katG | Isoniazid | katG1 | TGGCCGCGGCGGTCGACATT | 60 | 330 |
| katG2 | CCAGCAGGGCTCTTCGTCAG | ||||
| inhA | Isoniazid | inhA promo-1 | CCTCGCTGCCCAGAAAGGGA | 60 | 248 |
| inhA promo-2 | ATCCCCCGGTTTCCTCCGGT | ||||
| pncA | Pyrazinamide | pncA1 | ATCGCGATGGAACGTGATA | 60 | 950 |
| pncA2 | CTGTCACCGGACGGATTTG | ||||
| gyrA | Fluoroquinolones | gyrA-F | GATGACAGACACGACGTTGC | 55 | 398 |
| gyrA-R | GGGCTTCGGTGTACCTCAT | ||||
| gyrB | Fluoroquinolones | gyrB-F | CCACCGACATCGGTGGATT | 54 | 427 |
| gyrB-R | CTGCCACTTGAGTTTGTACA | ||||
| rrs | Aminoglycosides (capreomycin) | rrs-F | AAACCTCTTTCACCATCGAC | 59 | 1,329 |
| rrs-R | GTATCCATTGATGCTCGC | ||||
| rpsL | Streptomycin | rpsLfw | GGCCGACAAACAGAACGT | 51 | 501 |
| rpsLrev | GTTCACCAACTGGGTGAC |
Genotyping.
Spoligotyping (41, 42) was performed with 43 spacer probes coupled to MagPlex microspheres using the Luminex MagPix instrument (Luminex Corporation, Austin, TX, USA) and TB-SPOL Beamedex reagents (Beamedex SAS, Orsay, France). The spoligotyping PCR was carried out with 1× MasterMix (Applied Biological Materials Inc., Richmond, BC, Canada), containing the reaction buffer, 1 U Taq polymerase, 1.5 mM MgCl2, 0.2 mM dNTPs, with 0.6 μM each primer, and 5 μl of the Xpert sample remnant diluted 1:10 in water, in a 30-μl final volume. The amplification program was as follows: 95°C for 5 min; 45 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 30 s; and a final extension of 72°C for 7 min. The spoligotyping patterns obtained were sent to the SITVIT Web database (http://www.pasteur-guadeloupe.fr:8081/SITVIT_ONLINE/description.jsp) for identification of the spoligotype international types.
MIRU-VNTR typing.
Typing with four MIRU-VNTR systems (systems 1982, 3232, 3820, and 4120) reported to be hypervariable in strains of the Beijing lineage was performed with the primers described previously (18, 19) in a 50-μl final volume containing 1× PCR buffer, 1× Q-Solution, 500 nM dNTP, 1.5 mM MgCl2, 400 μM each primer, 1 U HotStarTaq DNA polymerase (Qiagen), and 5 μl of the Xpert sample remnant diluted 1:10 in water. The PCR parameters were as follows: 95°C for 15 min; 45 cycles of 94°C for 1 min, 59°C (MIRU 1982, 3820, 4102) or 60°C (MIRU 3232) for 1 min, and 72°C for 1.5 min; and a final extension of 72°C for 10 min. Eight microliters of each amplified PCR product was electrophoresed on a 1% agarose gel for 60 min at 160 V.
Supplementary Material
ACKNOWLEDGMENTS
This work is part of the NAREB (Nanotheraputics for Antibiotic Resistant Emerging Bacterial Pathogens) European Research Network (Collaborative Project), supported by the European Union's Seventh Framework Program for research, technological development, and demonstration under grant agreement 604237. This work is also part of the “Programme de recherche sur les nouvelles molécules intervenant dans la lutte contre la tuberculose,” supported by the Gabonese Republic. This study was also supported by the Chinese Academy of Sciences, Beijing, China; the Institut Pasteur of Shanghai, Shanghai, China; the Institut Pasteur of Paris, Paris, France; and the Instituto Venezolano de Investigaciones Científicas (IVIC), Caracas, Venezuela.
We declare that we have no conflicts of interest other than that C.S. was a founder of Beamedex.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.02257-16.
REFERENCES
- 1.World Health Organization. 2016. Global tuberculosis report 2016. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Kuaban C, Noeske J, Rieder HL, Ait-Khaled N, Abena Foe JL, Trebucq A. 2015. High effectiveness of a 12-month regimen for MDR-TB patients in Cameroon. Int J Tuberc Lung Dis 19:517–524. doi: 10.5588/ijtld.14.0535. [DOI] [PubMed] [Google Scholar]
- 3.Aung KJ, Van Deun A, Declercq E, Sarker MR, Das PK, Hossain MA, Rieder HL. 2014. Successful ‘9-month Bangladesh regimen’ for multidrug-resistant tuberculosis among over 500 consecutive patients. Int J Tuberc Lung Dis 18:1180–1187. doi: 10.5588/ijtld.14.0100. [DOI] [PubMed] [Google Scholar]
- 4.Van Deun A, Maug AK, Salim MA, Das PK, Sarker MR, Daru P, Rieder HL. 2010. Short, highly effective, and inexpensive standardized treatment of multidrug-resistant tuberculosis. Am J Respir Crit Care Med 182:684–692. doi: 10.1164/rccm.201001-0077OC. [DOI] [PubMed] [Google Scholar]
- 5.Dye C, Williams BG. 2000. Criteria for the control of drug-resistant tuberculosis. Proc Natl Acad Sci U S A 97:8180–8185. doi: 10.1073/pnas.140102797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonilla CA, Crossa A, Jave HO, Mitnick CD, Jamanca RB, Herrera C, Asencios L, Mendoza A, Bayona J, Zignol M, Jaramillo E. 2008. Management of extensively drug-resistant tuberculosis in Peru: cure is possible. PLoS One 3:e2957. doi: 10.1371/journal.pone.0002957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization. 2015. Global tuberculosis report 2015. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 8.Reference deleted.
- 9.Cremers AL, Janssen S, Huson MA, Bikene G, Belard S, Gerrets RP, Grobusch MP. 2013. Perceptions, health care seeking behaviour and implementation of a tuberculosis control programme in Lambaréné, Gabon. Public Health Action 3:328–332. doi: 10.5588/pha.13.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belard S, Remppis J, Bootsma S, Janssen S, Kombila DU, Beyeme JO, Rossatanga EG, Kokou C, Osbak KK, Obiang Mba RM, Kaba HM, Traore AN, Ehrhardt J, Bache EB, Flamen A, Rusch-Gerdes S, Frank M, Adegnika AA, Lell B, Niemann S, Kremsner PG, Loembe MM, Alabi AS, Grobusch MP. 2016. Tuberculosis treatment outcome and drug resistance in Lambaréné, Gabon: a prospective cohort study. Am J Trop Med Hyg doi: 10.4269/ajtmh.15-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mounguengui D, Ondounda M, Mandji Lawson JM, Fabre M, Gaudong L, Mangouka L, Magne C, Nzenze JR, L'Her P. 2012. Tuberculose multirésistante à l'hôpital d'instruction des armées de Libreville (Gabon) à propos de 16 cas. Bull Soc Pathol Exot 105:1–4. doi: 10.1007/s13149-011-0195-8. [DOI] [PubMed] [Google Scholar]
- 12.Nkoghe D, Mve MT, Nnegue S, Nkoume MO, Ba JI, Hypolite J, Leonard P, Kendjo E. 2005. Séroprévalence du VIH au sein des tuberculeux de l'hôpital de Nkembo à Libreville, Gabon. Bull Soc Pathol Exot 98:121–122. [PubMed] [Google Scholar]
- 13.Stolp SM, Huson MAM, Janssen S, Beyeme JO, Grobusch MP. 2013. Tuberculosis patients hospitalized in the Albert Schweitzer Hospital, Lambaréné, Gabon—a retrospective observational study. Clin Microbiol Infect 19:E499–E501. doi: 10.1111/1469-0691.12278. [DOI] [PubMed] [Google Scholar]
- 14.World Health Organization. 2016. Tuberculosis diagnostics: Xpert MTB/RIF assay. World Health Organization, Geneva, Switzerland: http://www.who.int/tb/publications/factsheet_xpert.pdf?ua=1. [Google Scholar]
- 15.Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, Allen J, Tahirli R, Blakemore R, Rustomjee R, Milovic A, Jones M, O'Brien SM, Persing DH, Ruesch-Gerdes S, Gotuzzo E, Rodrigues C, Alland D, Perkins MD. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med 363:1005–1015. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pym AS, Saint-Joanis B, Cole ST. 2002. Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect Immun 70:4955–4960. doi: 10.1128/IAI.70.9.4955-4960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hazbón MH, Brimacombe M, Bobadilla del Valle M, Cavatore M, Guerrero MI, Varma-Basil M, Billman-Jacobe H, Lavender C, Fyfe J, Garcia-Garcia L, Leon CI, Bose M, Chaves F, Murray M, Eisenach KD, Sifuentes-Osornio J, Cave MD, Ponce de Leon A, Alland D. 2006. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother 50:2640–2649. doi: 10.1128/AAC.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allix-Béguec C, Wahl C, Hanekom M, Nikolayevskyy V, Drobniewski F, Maeda S, Campos-Herrero I, Mokrousov I, Niemann S, Kontsevaya I, Rastogi N, Samper S, Sng LH, Warren RM, Supply P. 2014. Proposal of a consensus set of hypervariable mycobacterial interspersed repetitive-unit-variable-number tandem-repeat loci for subtyping of Mycobacterium tuberculosis Beijing isolates. J Clin Microbiol 52:164–172. doi: 10.1128/JCM.02519-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo T, Yang C, Gagneux S, Gicquel B, Mei J, Gao Q. 2012. Combination of single nucleotide polymorphism and variable-number tandem repeats for genotyping a homogenous population of Mycobacterium tuberculosis Beijing strains in China. J Clin Microbiol 50:633–639. doi: 10.1128/JCM.05539-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakravorty S, Aladegbami B, Motiwala AS, Dai Y, Safi H, Brimacombe M, Helb D, Alland D. 2008. Rifampin resistance, Beijing-W clade-single nucleotide polymorphism cluster group 2 phylogeny, and the Rv2629 191-C allele in Mycobacterium tuberculosis strains. J Clin Microbiol 46:2555–2560. doi: 10.1128/JCM.00666-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walusimbi S, Bwanga F, De Costa A, Haile M, Joloba M, Hoffner S. 2013. Meta-analysis to compare the accuracy of GeneXpert, MODS and the WHO 2007 algorithm for diagnosis of smear-negative pulmonary tuberculosis. BMC Infect Dis 13:507. doi: 10.1186/1471-2334-13-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin L, Coronel J, Faulx D, Valdez M, Metzler M, Crudder C, Castillo E, Caviedes L, Grandjean L, Rodriguez M, Friedland JS, Gilman RH, Moore DA. 2014. A field evaluation of the Hardy TB MODS kit for the rapid phenotypic diagnosis of tuberculosis and multi-drug resistant tuberculosis. PLoS One 9:e107258. doi: 10.1371/journal.pone.0107258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rigouts L, Coeck N, Gumusboga M, de Rijk WB, Aung KJ, Hossain MA, Fissette K, Rieder HL, Meehan CJ, de Jong BC, Van Deun A. 2016. Specific gyrA gene mutations predict poor treatment outcome in MDR-TB. J Antimicrob Chemother 71:314–323. doi: 10.1093/jac/dkv360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tagliani E, Cabibbe AM, Miotto P, Borroni E, Toro JC, Mansjo M, Hoffner S, Hillemann D, Zalutskaya A, Skrahina A, Cirillo DM. 2015. Diagnostic performance of the new version (v2.0) of GenoType MTBDRsl assay for detection of resistance to fluoroquinolones and second-line injectable drugs: a multicenter study. J Clin Microbiol 53:2961–2969. doi: 10.1128/JCM.01257-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson R, Warren R, Strauss OJ, Jordaan AM, Falmer AA, Beyers N, Schaaf HS, Murray M, Cloete K, van Helden PD, Victor TC. 2006. An outbreak of drug-resistant tuberculosis caused by a Beijing strain in the western Cape, South Africa. Int J Tuberc Lung Dis 10:1412–1414. [PubMed] [Google Scholar]
- 26.Hanekom M, Gey van Pittius NC, McEvoy C, Victor TC, Van Helden PD, Warren RM. 2011. Mycobacterium tuberculosis Beijing genotype: a template for success. Tuberculosis (Edinb) 91:510–523. doi: 10.1016/j.tube.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 27.Yeboah-Manu D, Asante-Poku A, Bodmer T, Stucki D, Koram K, Bonsu F, Pluschke G, Gagneux S. 2011. Genotypic diversity and drug susceptibility patterns among M. tuberculosis complex isolates from South-Western Ghana. PLoS One 6:e21906. doi: 10.1371/journal.pone.0021906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Couvin D, Rastogi N. 2015. Tuberculosis—a global emergency: tools and methods to monitor, understand, and control the epidemic with specific example of the Beijing lineage. Tuberculosis (Edinb) 95(Suppl 1):S177–S189. doi: 10.1016/j.tube.2015.02.023. [DOI] [PubMed] [Google Scholar]
- 29.Ouassa T, Borroni E, Loukou GY, Faye-Kette H, Kouakou J, Menan H, Cirillo DM. 2012. High prevalence of shared international type 53 among Mycobacterium tuberculosis complex strains in retreated patients from Côte d'Ivoire. PLoS One 7:e45363. doi: 10.1371/journal.pone.0045363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomgnimbou MK, Hernandez-Neuta I, Panaiotov S, Bachiyska E, Palomino JC, Martin A, del Portillo P, Refregier G, Sola C. 2013. Tuberculosis-spoligo-rifampin-isoniazid typing: an all-in-one assay technique for surveillance and control of multidrug-resistant tuberculosis on Luminex devices. J Clin Microbiol 51:3527–3534. doi: 10.1128/JCM.01523-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helb D, Jones M, Story E, Boehme C, Wallace E, Ho K, Kop J, Owens MR, Rodgers R, Banada P, Safi H, Blakemore R, Lan NT, Jones-Lopez EC, Levi M, Burday M, Ayakaka I, Mugerwa RD, McMillan B, Winn-Deen E, Christel L, Dailey P, Perkins MD, Persing DH, Alland D. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol 48:229–237. doi: 10.1128/JCM.01463-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coll F, McNerney R, Preston MD, Guerra-Assuncao JA, Warry A, Hill-Cawthorne G, Mallard K, Nair M, Miranda A, Alves A, Perdigao J, Viveiros M, Portugal I, Hasan Z, Hasan R, Glynn JR, Martin N, Pain A, Clark TG. 2015. Rapid determination of anti-tuberculosis drug resistance from whole-genome sequences. Genome Med 7:51. doi: 10.1186/s13073-015-0164-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bradley P, Gordon NC, Walker TM, Dunn L, Heys S, Huang B, Earle S, Pankhurst LJ, Anson L, de Cesare M, Piazza P, Votintseva AA, Golubchik T, Wilson DJ, Wyllie DH, Diel R, Niemann S, Feuerriegel S, Kohl TA, Ismail N, Omar SV, Smith EG, Buck D, McVean G, Walker AS, Peto TE, Crook DW, Iqbal Z. 2015. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat Commun 6:10063. doi: 10.1038/ncomms10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown AC, Bryant JM, Einer-Jensen K, Holdstock J, Houniet DT, Chan JZ, Depledge DP, Nikolayevskyy V, Broda A, Stone MJ, Christiansen MT, Williams R, McAndrew MB, Tutill H, Brown J, Melzer M, Rosmarin C, McHugh TD, Shorten RJ, Drobniewski F, Speight G, Breuer J. 2015. Rapid whole-genome sequencing of Mycobacterium tuberculosis isolates directly from clinical samples. J Clin Microbiol 53:2230–2237. doi: 10.1128/JCM.00486-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karlsson E, Larkeryd A, Sjodin A, Forsman M, Stenberg P. 2015. Scaffolding of a bacterial genome using MinION nanopore sequencing. Sci Rep 5:11996. doi: 10.1038/srep11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.International Union Against Tuberculosis and Lung Disease. 2000. Technical guide, sputum examination for tuberculosis by direct microscopy in low income countries, 5th ed, p 14 International Union Against Tuberculosis and Lung Disease, Paris, France. [Google Scholar]
- 37.Ramirez-Busby SM, Valafar F. 2015. Systematic review of mutations in pyrazinamidase associated with pyrazinamide resistance in Mycobacterium tuberculosis clinical isolates. Antimicrob Agents Chemother 59:5267–5277. doi: 10.1128/AAC.00204-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB. 2009. Tuberculosis drug resistance mutation database. PLoS Med 6:e2. doi: 10.1371/journal.pmed.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alame-Emane AK, Xu P, Pierre-Audigier C, Cadet-Daniel V, Shen X, Sraouia M, Siawaya JF, Takiff H, Gao Q, Gicquel B. 2015. Pyrazinamide resistance in Mycobacterium tuberculosis arises after rifampicin and fluoroquinolone resistance. Int J Tuberc Lung Dis 19:679–684. doi: 10.5588/ijtld.14.0768. [DOI] [PubMed] [Google Scholar]
- 40.Crudu V, Stratan E, Romancenco E, Allerheiligen V, Hillemann A, Moraru N. 2012. First evaluation of an improved assay for molecular genetic detection of tuberculosis as well as rifampin and isoniazid resistances. J Clin Microbiol 50:1264–1269. doi: 10.1128/JCM.05903-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sola C, Abadia E, Le Hello S, Weill FX. 2015. High-throughput CRISPR typing of Mycobacterium tuberculosis complex and Salmonella enterica serotype Typhimurium. Methods Mol Biol 1311:91–109. doi: 10.1007/978-1-4939-2687-9_6. [DOI] [PubMed] [Google Scholar]
- 42.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 35:907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.