Abstract
Alterations to chromatin structure induced by environmental insults have become an attractive explanation for the persistence of exposure effects into subsequent life stages. However, a growing body of work examining the epigenetic impact alcohol and other drugs of abuse exert consistently note a disconnect between induced changes in chromatin structure and patterns of gene transcription. Thus, an important question is whether perturbations in the ‘histone code’ induced by prenatal exposures to alcohol implicitly subvert gene expression, or if the hierarchy of cellular signaling networks driving development is such that they retain control over the transcriptional program. To address this question, we examined the impact of ethanol exposure in mouse embryonic stem cells cultured under 2i conditions, where the transcriptional program is rigidly enforced through the use of small molecule inhibitors. We find that ethanol-induced changes in post-translational histone modifications are dose-dependent, unique to the chromatin modification under investigation, and that the extent and direction of the change differ between the period of exposure and the recovery phase. Similar to in vivo models, we find post-translational modifications affecting histone 3 lysine 9 are the most profoundly impacted, with the signature of exposure persisting long after alcohol has been removed. These changes in chromatin structure associate with dose-dependent alterations in the levels of transcripts encoding Dnmt1, Uhrf1, Tet1, Tet2, Tet3, and Polycomb complex members Eed and Ezh2. However, in this model, ethanol-induced changes to the chromatin template do not consistently associate with changes in gene transcription, impede the process of differentiation or impact the acquisition of monoallelic patterns of expression for the imprinted gene Igf2R. These findings question the inferred universal relevance of epigenetic changes induced by drugs of abuse and suggest changes in chromatin structure cannot unequivocally explain dysgenesis in isolation.
Keywords: Developmental Programming, Epigenetics, Fetal Alcohol Syndrome, Environmental Epigenetics, Chromatin, Teratogen, Neurodevelopmental programming, Embryonic Stem Cells, Epigenetic Inheritance
Introduction
Despite numerous preventative and educational strategies, gestational exposure to alcohol remains the most common cause of environmentally induced birth defects impacting humans. The world-wide prevalence of Fetal Alcohol Spectrum Disorders (FASDs) range from 6 to 55 children per 1000 live births, depending on the region examined (Roozen et al., 2016). While progress towards understanding the toxicological and neurological actions of ethanol have been made, the teratogenic actions of this agent remain very poorly understood (Memo, Gnoato, Caminiti, Pichini & Tarani, 2013). In particular, it has been incredibly challenging to identify relevant molecular alterations arising between the period of initial exposure and the acquisition of developmental defects.
The separation between the period of exposure and the development of structural defects suggest that alcohol imparts a memory of exposure that influences the developmental trajectory of select cellular populations. In recent years, alterations to chromatin structure induced by environmental and/or nutritional insults have become an attractive explanation for the persistence of exposure effects into subsequent life stages (Feil & Fraga, 2011). Indeed, numerous studies have demonstrated that ethanol (EtOH) has the capacity to alter chromatin structure, suggesting that epigenetic mechanisms are relevant to the dysgenesis associated with FASDs (Basavarajappa & Subbanna, 2016; Mead & Sarkar, 2014; Resendiz, Mason, Lo & Zhou, 2014; Ungerer, Knezovich & Ramsay, 2013). For example, work from a number of laboratories have demonstrated both acute and long-term EtOH exposures influence patterns of DNA methylation (Ungerer et al., 2013). Further, our group and others have reported persistent alterations in post-translational histone modifications both in vitro and in vivo (Bekdash, Zhang & Sarkar, 2013; Finegersh et al., 2015; Govorko, Bekdash, Zhang & Sarkar, 2012; Moonat, Sakharkar, Zhang, Tang & Pandey, 2013; Pal-Bhadra et al., 2007; Pan et al., 2014; Subbanna & Basavarajappa, 2014; Veazey, Carnahan, Muller, Miranda & Golding, 2013; Veazey, Parnell, Miranda & Golding, 2015; Zhang, Ho, Vega, Burne & Chong, 2015). Collectively, these published studies suggest alcohol has the capacity to induce alterations in chromatin structure and that in select instances, these changes are heritable through cell division, persist well beyond the initial encounter, and have the potential to contribute to the genesis of FASD birth defects.
Work by our group using an in vitro neurosphere model has previously identified significant EtOH-induced changes in chromatin structure within the regulatory regions of multiple genes controlling both neural precursor cell identity and the processes of cellular differentiation (Veazey et al., 2013). However, despite significant changes in chromatin structure, only a small subset of genes displayed altered patterns of transcription. Further, when EtOH was withdrawn and exposed neurospheres allowed to recover, a distinct profile of chromatin alterations emerged, but yet no correlative alterations in transcription were observed (Veazey et al., 2015). These observations are significant as they suggest that the induced changes in chromatin structure do not impact the transcriptional programs governing cellular identity and would therefore have a limited capacity to impact subsequent developmental stages. Similar to our work, exposures to cocaine and other drugs of abuse are associated with wide-spread alterations in chromatin structure, yet in these studies, a significant number of genes show changes in expression opposite to those that would be predicted based upon the chromatin profiles, while most genes show no change at all (Nestler, 2014).
Recent studies in chromatin biology suggest that rather than being causal in controlling gene transcription, alterations to the chromatin template serve as a mechanism for the integration of cell signaling events with the biochemical machinery controlling transcription (Badeaux & Shi, 2013). In this view, multiple signaling pathways converge on the chromatin fiber, which serves as a regulable platform to facilitate the integration of transcription factor regulatory networks with cell-specific cohorts of genes. The key distinction here being that while chromatin structure facilitates gene expression, protein transcription factors drive it. Thus, an important question that remains to be resolved is whether perturbations in the ‘histone code’ induced by prenatal exposures to drugs of abuse actually have the potential to alter gene transcription, or if the extracellular and intracellular signaling programs regulating cellular identity maintain rigid control over cellular patterns of gene expression. As neurospheres are complex populations of cells, we elected to examine this question using mouse embryonic stem cells, which can be cultured with high uniformity in terms of cellular phenotype, chromatin profiles and transcriptional output (Galonska, Ziller, Karnik & Meissner, 2015; Marks et al., 2012).
Embryonic Stem Cells (ESCs) have an innate cell signaling program maintaining pluripotency, which is balanced by differentiation-promoting signals from the mitogen-activated protein kinase (MAPK) and glycogen synthase kinase 3 (GSK3) signaling pathways (Hirai, Karian & Kikyo, 2011). When input from these differentiation inducing pathways are blocked, the networks governing cellular identity become locked into a transcriptional program promoting perpetual self-renewal and a ground state of pluripotency (Ying et al., 2008). Given the static state in which the transcriptional program of these cells are maintained, we sought to employ this model to determine the extent to which EtOH-induced modifications to chromatin structure could heritably alter transcription. To this end, we employed the 2i embryonic stem cell culture system, which strictly enforces the undifferentiated state through the use of small molecule inhibitors 6-bromoindirubin-3′-oxime, a potent and reversible GSK-3α/β inhibitor and PD0325901, which inhibits MAPK. Although not strictly necessary, Leukemia Inhibitory Factor (LIF) was also used for maximal pluripotency (Miyanari & Torres-Padilla, 2012). Cultures were monitored for changes in the enrichment of key post-translational histone modifications within the regulatory regions of genes controlling pluripotency and core aspects of neural differentiation, during both the period of exposure as well as during the recovery period.
Similar to our previous studies in cultured neural stem cells, we find that EtOH-induced changes in post-translational histone modifications are dose-dependent, unique to the chromatin modification under investigation, and finally that both the extent and direction of the change differ between the period of exposure and the recovery phase. However, these EtOH-induced changes to the chromatin template do not directly associate with changes in gene transcription. To assess the impact of EtOH induced changes on cellular differentiation and a measure of developmental programming, we examined the transcription of Insulin Growth Factor 2 Receptor (Igf2R), which is an imprinted gene that acquires monoallelic patterns of gene expression during embryonic stem cell differentiation (Nagano et al., 2008; Santoro et al., 2013). Genomic imprinting is an epigenetic mechanism of transcriptional regulation that restricts expression to either the maternally- or paternally-inherited copy of the gene; the opposite parental copy is silent (Bartolomei & Tilghman, 1997). During the production of eggs and sperm, a subset of genes are marked or “imprinted” with differential patterns of DNA methylation to remember whether they are transmitted from the mother or father, respectively. In developing embryos, expression of imprinted genes become restricted based on these parental epigenetic marks (Mann et al., 2004). Disruptions in imprinting can have severe consequences for growth and development of the mammalian embryo, as proper acquisition and maintenance of genomic imprints are crucial for both fetal and placental development (McGrath & Solter, 1984). Importantly, alterations in Igf2R were recently identified in studies examining the impact of prenatal EtOH exposures on the developing brain using an ex vivo model of early neurulation (Liu, Balaraman, Wang, Nephew & Zhou, 2009). In our model, EtOH exposure did not appreciably impede the process of differentiation nor impact the acquisition Igf2R imprinted gene expression. Analysis of fully differentiated cells identifies similar trends, suggesting that while EtOH can alter chromatin structure, the induced changes do not necessarily impact the transcriptional program.
Materials and Methods
Cell Culture
Primary embryonic stem cells (ESCs) derived from B6XCAST F1 embryos were employed to examine allele specific patterns of gene expression in a parent of origin specific fashion (Golding et al., 2011). ESCs were maintained in ESC medium under 2i conditions as previously described (Miyanari & Torres-Padilla, 2012). Cells were grown in DMEM culture medium (Catalogue # D5671; Sigma) supplemented with 50μg/ml penicillin/streptomycin (Catalogue # P4333; Sigma), 100 μM B-mercaptoethanol (Catalogue # M3148; Sigma), 1000 units/ml LIF (Catalogue # L5158;Sigma), 3uM 6-bromoindirubin-3′-oxime (BIO) (Catalogue # B1686; Sigma), 1uM PD0325901 (Catalogue # PZ0162 Sigma), 2 mM L-Glutamine (Catalogue # G3126; Sigma), and 1x MEM nonessential amino acids (Catalogue # M7145; Sigma). When passaging, cells were washed twice with 1x PBS, then disassociated with 0.5x Accutase (Catalogue # SF006; Millipore) and split 1:20. Mouse Embryonic Fibroblasts (MEFs) were cultured in DMEM (Catalogue # D5671; Sigma) supplemented with 50μg/ml penicillin/streptomycin (Catalogue # P4333; Sigma) and 10% fetal bovine serum (Atlanta Biologicals Lawrenceville, GA; Cat# S11550).
Ethanol Exposure
The concentrations of alcohol utilized in this study were meant to mimic those obtained from a casual and a binge drinker as described previously (Veazey et al., 2015). The treatment groups were cultured in appropriate medium containing either no ethanol, 80 mg/dL, 160 mg/dL or 240 mg/dL ethanol and the lid sealed with parafilm to prevent evaporation. The medium and treatment were replaced every 48 hours.
Neural Differentiation of Ethanol-Exposed ESCs
ESCs were exposed to ethanol and allowed to recover in 2i medium maintaining a ground level of stemness as described above. Following the 10 day recovery, ESCs were split into flasks containing 50% ESC medium (described above), and 50% Neural Stem Cell (NSC) medium containing a 50%/50% mixture of Neurobasal medium (Catalogue # 21103-049; Invitrogen) and DMEM F-12 (Catalogue # 11320- 033; Invitrogen) supplemented with the N2-supplement (Catalogue # 17502-048; Invitrogen), B27 supplement (Catalogue # 17504-044; Invitrogen), 0.05% TC grade BSA in PBS (Catalogue # A1933 Sigma), 2mM L-glutamine (Catalogue # 25030-081; Invitrogen), 1x Penicillin/Streptomycin (Catalogue # 15140-122; Invitrogen), 20 μg/ml FGF basic (Catalogue # PMG0034; Invitrogen), 20 μg/ml EGF (Catalogue # PHG0311; Invitrogen), and 0.85 units/ml heparin (Catalogue # H3149; Sigma). After 2 days, cells were moved into laminin-coated flasks containing a mixture of 25% ESC medium and 75% NSC medium for final differentiation.
Chromatin Immunoprecipitation Analysis
Cells were grown to 80% confluence, washed twice in warm PBS, dissociated with 0.5x Accutase (Catalogue # SCR005; Millipore), and re-suspended in warm medium (DMEM F-12 Catalogue # 11320-033; Invitrogen) containing 0.1 volume crosslinking solution (Kondo, Shen, Yan, Huang & Issa, 2004). ChIP reactions were performed as described previously (Veazey et al., 2015) followed by DNA purification with a Qiaquick PCR Cleanup kit (Catalogue # 28106; QIAGEN). Antibodies used include: anti-H3K4me3 (Catalogue # 04-745; Millipore), anti- H3K27me3 (Catalogue # 39155; Active Motif), anti-H3K9ac (Catalogue # 07-352; Millipore), and anti-H3K9me2 (Catalogue # 39239; Active Motif). Antibodies for modified histones were used at 1 μg/ChIP reaction. The concentration of IgG (Catalogue # SC-2027; Santa Cruz) was also used at 1 ug/ChIP reaction. For analysis of candidate loci, real-time PCR was performed with the Dynamo Flash supermix (Catalogue # F-415XL; Thermo Scientific) according to the recommended protocol. Reactions were performed on a Bio-Rad CFX384 Touch PCR system. Data was analyzed using the formula previously described (Mukhopadhyay, Deplancke, Walhout & Tissenbaum, 2008). Primer sequences are listed in Table S1- Primer Sequences.
RNA analysis
Cultured cells were grown to 80% confluence, washed twice in warm PBS, and dissociated with 0.5x Accutase (Catalogue # SCR005; Millipore). Cells were spun down, washed once in cold PBS, and RNA isolated using Trizol (Catalogue # 15596026; Invitrogen) according to the manufacturer’s protocol. One microgram of purified total RNA was treated with amplification grade DNase I (Catalogue # AMPD1; Sigma) according to the manufacturer’s recommendations, and 250 ng RNA seeded into a reverse transcription reaction using the SuperScriptII system (Catalogue # 18064-071; Invitrogen) by combining 1 μL random hexamer oligonucleotides (Catalogue # 48190011; Invitrogen), 1 μL 10 mM dNTP (Catalogue # 18427- 013; Invitrogen), and 11 μL RNA plus water. This mixture was brought to 70°C for 5 minutes and then cooled to room temperature. SuperScriptII reaction buffer, DTT, and SuperScriptII were then added according to manufacturer’s protocol, and the mixture brought to 25°C for 5 minutes, 42°C for 50 minutes, 45°C for 20 minutes, 50°C for 15 mi nutes, and then 70°C for 5 minutes. Relative levels of candidate gene transcripts were analyzed using the Dynamo Flash mastermix (Catalogue # F-415XL; Thermo Scientific) according to the recommended protocol. Reactions were performed on a Bio-Rad CFX38. Primers are listed in Table S1 - Primer Sequences.
Analysis of Igf2R
Isolated RNAs were converted to cDNA and a 388 base pair region of the Igf2R transcript amplified using primers Igf2R Forward 5′ATCTGTGACCTCCTCTTGAGCAGG and Igf2R Reverse 5′GGGTTGTTTAGAGCCAATCAA. The amplified product was digested with the restriction enzyme TaqI (Thermo-Fisher Scientific Catalogue # ER0671) which cuts the paternal castaneus allele into fragments 210 and 178 base pairs in length. The fragments were visualized on an agarose gel, and densitometry performed using ImageJ (NIH.gov).
Flow Cytometry
Flow cytometry was performed using conjugated antibodies recognizing CD90.2 (Catalogue #; BD Biosciences) and CD24a (Catalogue #; BD Biosciences). Approximately 5 million cells were washed in 1mL PBS + 0.1% BSA and incubated with 0.3 uL of each antibody in 100ul PBS +BSA for 20 minutes at 4 C. Cells were then washed again in 1mL PBS + BSA, then resuspended in 300 uL PBS+BSA and analyzed using a Beckman Coulter Gallios Flow Cytometer. Populations were visualized using the Kaluza Flow Analysis Software.
Statistical Analysis
For all experiments, statistical significance was set at alpha = 0.05. For analysis of gene expression, the replicate cycle threshold (Ct) values for each transcript were compiled and normalized to the geometric mean of the reference genes peptidylprolyl isomerase A (Ppia—NM_008907), glyceraldehyde 3-phosphate dehydrogenase (Gapdh—NM_008084), and hypoxanthine-phosphoribosyl transferase (Hprt—NM_013556). From our previous studies of 14 candidate reference genes in EtOH-exposed cultures, Ppia, Gapdh, and Hprt have been validated as stable across the range of alcohol treatments utilized in this study (Carnahan et al., 2013). Normalized expression levels were calculated using the ddCt method described previously (Schmittgen & Livak, 2008). Relative fold change values from each biological replicate were transferred into the statistical analysis program GraphPad (GraphPad Software, Inc., La Jolla, CA), verified for normality using the Brown-Forsythe test, and an analysis of variance (ANOVA) utilized to assay differences between experimental treatments. For comparisons with p-values < 0.05, we applied Tukey’s HSD analysis for multiple comparisons, and have marked statistically significant differences with an asterisk.
For quantitative analysis of candidate gene regulatory region enrichment in ESCs, ChIP samples were first normalized to 1% input, using the formula previously described (Mukhopadhyay et al., 2008). To independently examine alterations in each post-translational modification, the means from each independent sample were normalized to the control. The results of 3 independent experiments were then tabulated, cumulative means calculated and standard error of the mean derived. Values from each biological replicate were transferred into the statistical analysis program GraphPad (GraphPad Software, Inc., La Jolla, CA), verified for normality using the Brown-Forsythe test, and an analysis of variance (ANOVA) run to assay differences between experimental treatments. For samples with p-values < 0.05, we applied Tukey’s HSD analysis for multiple comparisons, and have marked statistically significant differences with an asterisk.
Results
The effects of ethanol are dose-dependent and exhibit distinct profiles between the period of exposure and the recovery phase
In order to analyze the impact EtOH exposures have on the profile of chromatin structure and gene transcription in a rigidly enforced culture system (Marks et al., 2012; Ying et al., 2008), mouse ESCs grown under 2i conditions were treated for four days with physiologically relevant concentrations of EtOH (80 mg/dL, 160 mg/dL, and 240 mg/dL). After the four-day exposure period, EtOH was removed from the medium and ESCs were allowed to recover for a ten-day period. Samples were taken at days 4, 8, and 14, which represent the period of exposure, post 4-day recovery, and post 10-day recovery respectively. These samples were analyzed for the enrichment of key post-translational histone modifications associated with transcriptionally permissive chromatin structure (histone 3 lysine 4 trimethylation and histone 3 lysine 9 acetylation) and transcriptionally repressive chromatin structure (histone 3 lysine 27 trimethylation and histone 3 lysine 9 dimethylation) using chromatin immunoprecipitation (ChIP). The selected post-translational modifications are commonly examined as proxies to infer the state of gene expression, and in embryonic stem cells, have been linked to the control of transcriptional programs governing development and differentiation (Bernstein et al., 2006; Moris, Pina & Arias, 2016). qPCR was employed to probe precipitated cellular extracts for the enrichment or loss of these modifications within the regulatory regions of a panel of 25 candidate genes. These genes were selected given their roles in stem cell pluripotency, neural development and their identification in transcriptional studies of FASD developmental defects (Ogony, Malahias, Vadigepalli & Anni, 2013; Rugg-Gunn, Cox, Ralston & Rossant, 2010; Zhou et al., 2011).
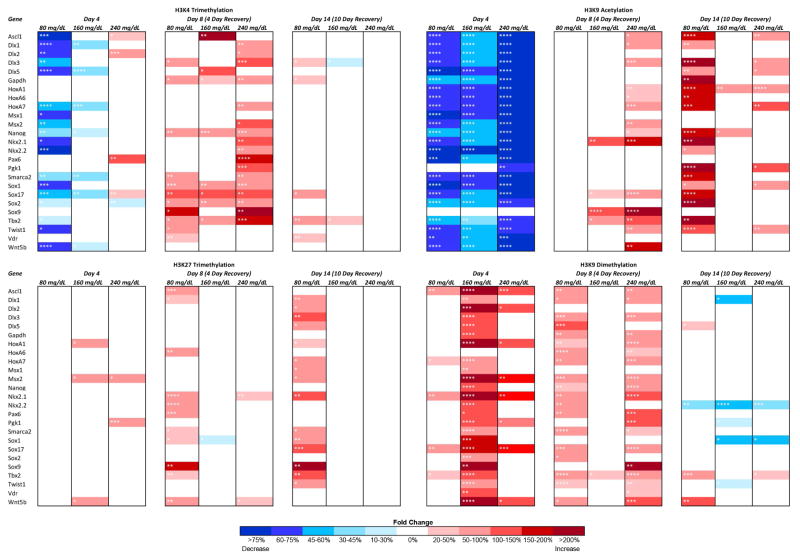
A comprehensive map of the effects differing concentrations of EtOH induced over the experimental time-course is presented in Fig. 1, with the statistical significance demarcated in each cell. After four days of EtOH exposure, especially in low dose groups (80 mg/dL), post-translational modifications associated with a relaxed chromatin configuration are uniformly decreased, while at higher doses, each modification behaves differently. For example, histone 3 lysine 4 trimethylation (H3K4me3) is lost during the initial period of exposure in the 80 mg/dL treatment group while the higher concentrations tested exhibited minor fluctuations in fewer candidate genes. In contrast, during the initial recovery phase (Day-8) the highest concentration group (240mg/dL) exhibited the greatest change, with multiple candidate genes exhibiting states of hypermethylaiton. Interestingly, histone 3 lysine 9 acetylation (H3K9ac) is uniformly reduced upon EtOH exposure in all concentrations tested, and while this reduction is reversed after EtOH is removed from the system, we observe dose-dependent patterns of hyperactylation that persist for at least ten days. With the exception of select candidate genes, only the lower dose of EtOH appeared to effect histone 3 lysine 27 trimethylation (H3K27me3), with a state of hypermethylation persisting out to Day-14 in the 80mg/dL treatment group. Similar to our previous in vitro and in vivo studies in neural stem cells (Veazey et al., 2015), enrichment of histone 3 lysine 9 dimethylation (H3K9me2) is dramatically increased within the regulatory regions of our candidate genes. However, the patterns of enrichment are dependent on dose and exposure period. For instance, we observed a dramatic increase in methylation during the exposure period for the 160mg/dL treatment group, which largely returns to normal after EtOH has been removed. While several candidate genes exhibit hypermethylation during the period of exposure in the 80mg/dL and 240mg/dL treatments, the large majority of changes do not appear until the Day-8 time point, four days after the EtOH was removed. Interestingly, enrichment of H3K9ac and H3K9me2 appear to be inversely affected by EtOH at Day-4, but do not maintain this relationship during the recovery phase. In summary, the cumulative profiles of change observed during the exposure period indicate EtOH induces a loss of H3K4me3, and H3K9ac, while levels of H3K9me2 increase. This would suggest a decrease in transcriptionally permissive chromatin structure and an increase in transcriptionally repressive state during the phase of exposure. The profiles of change observed during the recovery phase did not exhibit a cohesive pattern of change.
Fig. 1.
Dose-dependent and histone modification-specific changes in chromatin structure. Murine embryonic stem cells were cultured in varying concentrations of ethanol (80 mg/dL, 160 mg/dL, or 240 mg/dL) for four days, followed by a no-ethanol recovery period for ten days. Samples were taken at Day-4, Day-8, and Day-14 and analyze for enrichment of H3K4me3, H3K9ac, H3K27me3, and H3K9me2 using chromatin immunoprecipitation. The enrichment of the indicated post-translational histone modifications was then analyzed within the promoter regions of candidate genes using qPCR. The heat map represents fold change compared to the control group. Within the three separate biological replicates (N = 3), three ChIPs were performed, and two qPCR replicates performed on each independent ChIP. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
Changes in chromatin structure at candidate gene promoters do not correlate with transcriptional activity during or after the period of EtOH exposure
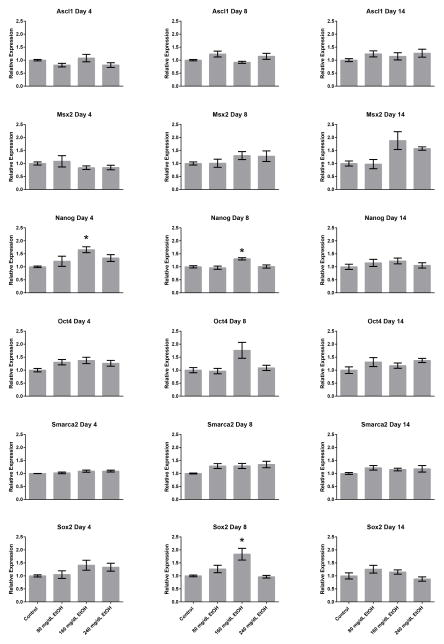
The current widely-accepted belief in the field environmental epigenetics is that changes in post-translational histone modifications induced by toxic exposures directly impact patterns of gene expression within exposed cells. We and others have previously challenged this notion using various models of EtOH exposure (Ponomarev, 2013; Veazey et al., 2015). To determine whether the shifts in chromatin profiles induced by EtOH exposure could influence patterns of gene transcription within our model, RNA was isolated from all treatment groups utilized in our ChIP experiments and gene expression analyzed using reverse transcriptase polymerase chain reaction (RT-qPCR). Of the 25 candidate genes examined, we were able to detect transcripts encoding Ascl1, Msx2, Nanog, Oct4, Smarca2, and Sox2 in embryonic stem cells (Fig. 2). In these analyses, only Nanog and Sox2 displayed significant increases in transcript levels (p < 0.05), and only in the 160mg/dL treatment groups. The observed alterations in the expression of these candidate genes is consistent with previous work in ESCs cultured on feeders (Ogony et al., 2013). Importantly, no consistent correlation could be identified between the observed changes in chromatin structure and the modest changes in gene transcription for these two genes. From these analyses, we conclude that in this culture system, EtOH-induced changes in chromatin structure do not measurably impact patterns of gene transcription.
Fig. 2.
Alterations in gene transcription arising during and after the window of EtOH exposure. Murine embryonic stem cells examined in Figure 1 were cultured in varying concentrations of ethanol (80 mg/dL, 160 mg/dL, or 240 mg/dL) for four days, followed by a no-ethanol recovery period for ten days. Samples were taken at Day-4, Day-8, and Day-14, RNA isolated and transcript levels encoding the candidate genes examined in Figure 1 assayed using quantitative RT-PCR. Ct values were normalized to the geometric mean of Gapdh, Hprt, and Ppia and graphed relative to the control treatment. Graphs represent three independent replicates (N = 3), with two independent RT reactions and three qPCR measurements for each RT. Differences were measured using a one-way ANOVA, error bars represent SEM. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
Alterations in the genetic pathways linked to alcohol metabolism, oxidative stress response and the control of DNA methylation
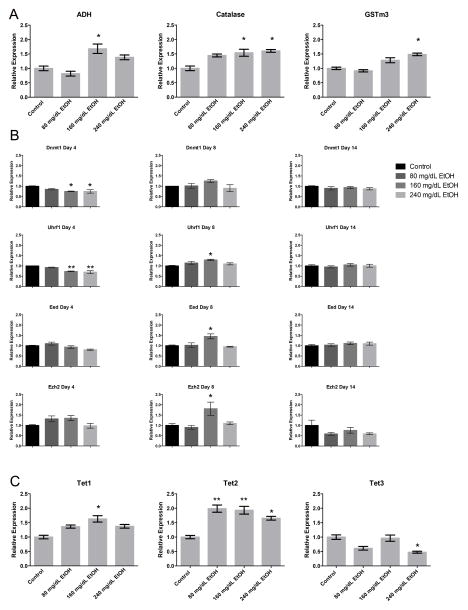
Alcohol metabolism is known to increase reactive oxygen species (ROS), which has been suggested to contribute to the phenotypic effects of EtOH exposure during development (Brocardo, Gil-Mohapel & Christie, 2011; Dong, Sulik & Chen, 2008). As previous studies have established a link between components of the oxidative stress pathway and enzymes regulating chromatin structure (Bosch-Presegué et al., 2011; Chia et al., 2011), we therefore examined the expression of seventeen candidate genes involved in the genetic pathways regulating ethanol metabolism and oxidative stress (Fig. 3A). Of the genes regulating alcohol metabolism (Table 1), only Adh and Catalase were expressed in our ESCs. Both displayed dose-dependent increases in transcription; Adh at 160 mg/dL, and Catalase in both the 160 and 240 mg/dL treatment groups (p < 0.05). Of the genes involved in the oxidative stress response (Table 1), only Gstm3, a glutathione S-transferase that functions in the detoxification of electrophilic compounds, displayed a modest (> 1.5 fold, p < 0.05) increase in the 240 mg/dL- exposed group.
Fig. 3.
Alterations in the genetic pathways controlling alcohol metabolism, oxidative stress response and the control of DNA methylation. Murine embryonic stem cells were cultured in varying concentrations of ethanol (80 mg/dL, 160 mg/dL, or 240 mg/dL) for four days, followed by a no-ethanol recovery period for ten days. Samples were taken at Day-4, Day-8, and Day-14, and transcripts encoding genes involved in A) alcohol metabolism and the oxidative stress response, B) H3K27, H3K9 and DNA methyltransferase enzymes, C) TET family of Fe(II) and a-KG- dependent dioxygenases, and D) histone demethylases examined using RT-qPCR. Ct values were normalized to the geometric mean of Gapdh, Hprt, and Ppia and graphed relative to the control treatment. Graphs represent three independent replicates (N = 3), with two independent RT reactions and three qPCR measurements for each RT. Differences were measured using a one-way ANOVA, error bars represent SEM. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
Table 1.
Candidate genes involved in ethanol metabolism and cellular oxidative stress response.
| CANDIDATE GENE | BIOCHEMICAL FUNCTION |
|---|---|
| Catalase | Antioxidant Enzyme; Reactive Oxygen Species to H2O2 |
| Adh1 | Alcohol Breakdown; Alcohol to acetaldehyde |
| Cyp2e1 | Antioxidant Enzyme; Breakdown of xenobiotics |
| Gpx1 | Antioxidant Enzyme; Breakdown of H2O2 |
| Gpx3 | Antioxidant Enzyme; Breakdown of H2O2 |
| Gsr | Antioxidant Enzyme; Glutathione disulfide to Glutathione (important antioxidant) |
| Gstm2 | Detoxification of electrophilic compounds; Conjugation w/glutathione. |
| Gstm3 | Detoxification of electrophilic compounds; Conjugation w/glutathione. |
| Gstp1 | Role in detoxification; Links Reactive Oxygen Species elements to glutathione for detoxification. |
| Hif1a | Transcription factor; Master hypoxia regulator |
| Nrf2 | Transcription factor; Master antioxidant gene regulator |
| Nqo1 | Prevents production of Reactive Oxygen Species; Reduces quinones to hydroquinones. |
| Prx1 | Antioxidant Enzyme; Control cytokine-induced peroxide levels. |
| Sod1 | Antioxidant Enzyme; Free-radical scavenger. |
| Sod2 | Antioxidant Enzyme; Superoxide to H2O2 & O2 |
| Sod3 | Antioxidant Enzyme; Superoxide to H2O2 & O2 |
| Txn | Antioxidant Enzyme; Reduction of proteins. |
Using primary neural stem cells, we have previously shown that transcripts encoding the H3K27, H3K9 and DNA methyltransferase enzymes, as well as H3K9 demethylases are altered upon exposure to EtOH (Veazey et al., 2015). We therefore sought to examine whether alterations in the expression of these chromatin modifiers occur in pluripotent stem cells, or if this phenomenon is cell type-specific. We examined transcript levels of the DNA methyltransferases and their associated interacting proteins including Dnmt1, Dnmt3b, Uhrf1, Polycomb Repressive Complex 2 (PRC2) members (Eed, Ezh2), and the H3K9 methyltransferases (Ehmt2, Setdb1) (Fig. 3B). After four days of EtOH exposure, only Dnmt1 and Uhrf1 displayed dose-dependent decreases in expression relative to the control. During the initial recovery phase, transcripts encoding Uhrf1 were modestly increased, where as none of the other DNA methyltransferases were significantly different. By Day-14, no significant differences were observed for any of the transcripts examined. We next examined the expression of the TET family of Fe(II) and a-KG- dependent dioxygenases, which utilize oxygen to convert 5-methyl-cytosine (5mC) to 5-hydroxymethyl-cytosine (5hmC), an intermediate in the DNA demethylation cycle (Tahiliani et al., 2009). Both Tet1 and Tet2 displayed increases in expression (Tet1 only in 160 mg/dL, and Tet2 in all EtOH-exposed groups), while Tet3 displayed a reduction in expression in the 240 mg/dL group (Fig. 3C). These results suggest that EtOH exposure modifies the transcriptional control of both the DNA methylating and DNA demethylating machinery within our ESC model.
Both members of PRC2 displayed increased expression in the 160 mg/dL treatment group, but only after the four day recovery period (Day-8). We could not identify any correlations between the expression of these candidates and the enrichment of H3K27me3 at this time point. With the exception of these two candidates during the initial recovery phase, we could not detect alterations in transcript levels for any of the other histone methyltransferases examined, at any other time point. Interestingly, despite a widespread increase in H3K9me2 at Day-4 and Day-8, no changes in the expression of Ehmt2 were detected. One possible explanation to these observations would be that EtOH may possibly interfere with the transcription of histone demethylases; and our observations are the result of a reduced rate of turnover. We therefore measured the transcript levels of two H3K9 demethylases, Kdm3a and Kdm4c (Fig. 3D). No alterations in the transcript levels of either enzyme were detected. These observations suggest that lasting changes to the profile of chromatin structure are not appreciably linked to the transcriptional regulation of enzymes responsible for imparting the examined post-translational histone modifications.
EtOH exposure does not disrupt the ability of exposed ESCs to differentiate into neural stem cells nor interfere with the acquisition of Igf2 imprinted gene expression
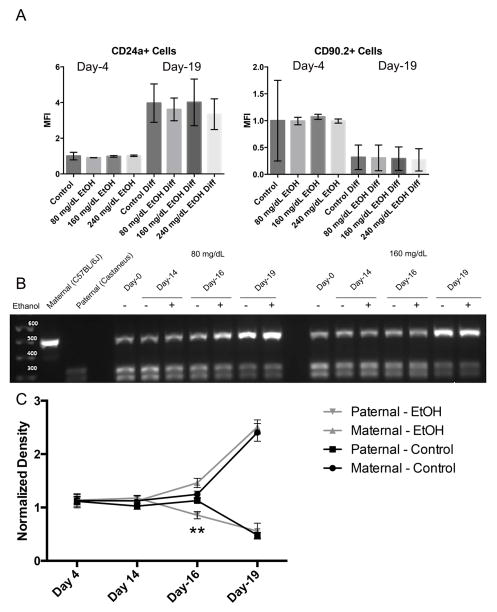
We next sought to determine the extent to which the observed changes in chromatin structure impact the capacity of exposed ESCs to properly establish patterns of imprinted gene expression during differentiation. Using ESCs derived from F1 hybrid crosses between Mus musculus castaneus (paternal) and C57BL/6J-Mus musculus musculus (maternal) strains of mice, we examined the impact of EtOH exposure on the acquisition of imprinted gene expression within the Igf2R locus, located on mouse chromosome 17. When using stem cells derived from the described F1 hybrid crosses, we can distinguish maternal and paternal Igf2R mRNAs using a restriction digest-based assay (i.e. polymorphisms between strains make the paternal allele susceptible to digestion while the maternal allele is not) (Golding et al., 2011). Igf2R acquires imprinted gene expression during ESC differentiation along the neural lineage and is regulated through epigenetic mechanisms involving both DNA and H3K9 methylation (Nagano et al., 2008; Santoro et al., 2013). To determine the impact EtOH-induced changes in chromatin structure have upon the acquisition of imprinted patterns of gene transcription, we exposed ESCs to EtOH for four days, allowed a ten day period of recovery, at which point ESCs were differentiated into neural stem cells (NSCs) using a five day protocol. Samples were taken at Day-0, Day-4 (EtOH-exposed), Day-16 (early differentiation) and Day-19 (after the five day differentiation protocol). To monitor the process of differentiation, cells were assayed for the expression of cell surface markers CD90.2 (embryonic stem cells (Zhao, Ji, Zhang, Li & Ma, 2012)) and CD24a (neuroblasts and neurons (Pruszak, Ludwig, Blak, Alavian & Isacson, 2009)) at Day-4 and Day-19 using flow cytometry (Fig. 4A). As anticipated, levels of CD24a increased over the differentiation protocol while expression of CD90.2 decreased. Importantly, EtOH exposure did not measurably impact the ability of cells to up-regulate CD24a or the suppress CD90.2 during differentiation. As can be seen in Fig 4B and 4C, patterns of Igf2R transcription are biallelic at both Day-4 and Day-14, move towards the maternal (C57BL/6J) allele at Day-16, and are predominantly derived from the maternal allele at Day-19. Only the Day-16 time point displayed a significant difference in allelic expression (p<0.01) where EtOH exposed samples were quicker to restrict expression to the maternal allele. These results indicate that within our model, EtOH-induced disruptions in chromatin structure do not impede the process of differentiation or the acquisition of imprinted gene expression within the Igf2R locus.
Fig. 4.
EtOH exposure does not disrupt the ability of ESCs to acquire imprinted patterns of transcription during differentiation. A) ESCs were seeded into flasks and exposed to varying concentrations of ethanol (0 mg/dL, 80 mg/dL, 160 mg/dL, or 240 mg/dL) for four days, followed by a no-ethanol recovery period for ten days. Cells were then differentiated into neuroblasts using an established 5-day protocol. At Day-4 and Day -19, cells were monitored for changes in the expression of the surface markers CD90.2 and CD24a using flow cytometry. Graphs represent three separate biological replicates (N = 3) examining half a million cells in each replicate. B) Representative graphs examining allelic patterns of Igf2R transcripts through ESC differentiation using a polymophism-based restriction digest assay. In this assay, transcripts derived from the maternal C57BL/6J strain (upper band) are refractory to Taq1 digestion, while transcripts derived from the paternal Mus musculus castaneus strain (lower doublet) are susceptible. C) Densitometry analysis of Igf2R allelic expression. Graph represents three separate biological replicates (N = 3).
EtOH-induced changes in chromatin structure do not impact transcription in a fully differentiated cell type
The results of our experiments suggest that in mouse ESCs cultured under 2i conditions, EtOH induced changes in chromatin structure do not correlatively influence the expression of our candidate genes; at least at the concentrations tested in this study. We therefore sought to contrast these observations with those in a fully differentiated cell type. To this end, mouse embryonic fibroblasts (MEFs) were cultured under standard conditions and exposed to 160 mg/dL EtOH. This concentration was selected as it consistently displayed significant changes in all of the post-translational modifications examined and was the only treatment associated with measurable alterations in the gene transcription (Nanog and Sox2). Cellular extracts were examined for alterations in chromatin structure and gene transcription at Day-4 and Day-8, which represent the period of exposure and after a four-day recovery respectively. As can be seen in Fig. 5A, only H3K9me2 displayed measurable alterations in enrichment within the regulatory regions of multiple genes; becoming hypermethylated after the recovery phase. Importantly, when transcripts encoding the candidate genes were assayed by qRT-PCR, no significant differences in gene expression could be detected during any of the examined time points (Fig. 5B, p > 0.05). Similar to our observations in ESCs, we did observe modest EtOH-induced changes in the expression of Dnmt1, Ehmt2 and Ezh2 (Fig. 5C, p < 0.01) during the exposure phase but not after the recovery phase. These results suggest that within fully differentiated MEFs, EtOH has a more limited capacity to impact chromatin structure, and again the observed changes do not significantly impact the transcription of our candidate genes.
Fig. 5.
Analysis of EtOH-induced changes in chromatin structure in a fully differentiated cell type. A) Alcohol-induced changes in H3K9 acetylation, H3K9 dimethylation H3K4 trimethylation, and H3K27 trimethylation in mouse embryonic fibroblasts. Fibroblast cells were treated with 160 mg/dl EtOH for 4 days followed by a 4-day recovery period in medium without EtOH. Samples were subjected to chromatin immunoprecipitation and enrichment assayed within the regulatory regions of the indicated genes. Fold changes for H3K9 acetylation, H3K9 dimethylation and H3K4 trimethylation relative to the control are displayed; no alterations in H3K27 trimethylation were observed. Within the three separate biological replicates (N = 3), three ChIPs were performed, and two qPCR replicates performed on each independent ChIP. Differences were determined using a two-way ANOVA. B) Transcript levels of candidate genes are not impacted by EtOH-induced changes in chromatin structure. Transcripts encoding the indicated candidate genes were quantified using RT-qPCR. C) EtOH-induced up-regulation of transcripts encoding Dnmt1, Ehmt2 and Ezh2 do not persist beyond window of exposure. Transcripts encoding Dnmt1, Eed, Ehmt1, Ehmt2, Ezh2 and Setdb1 were measured using qPCR both during the period of exposure and after the four-day recovery phase. In analyses using RT-qPCR, measured Ct values were normalized to the geometric mean of Gapdh, Hprt, and Ppia and graphed relative to the control treatment. Graphs represent three independent replicates (N = 3), with two independent RT reactions and three qPCR measurements for each RT. All data are reported as Mean ± SEM. Differences were assessed using an unpaired, two-tailed parametric Student’s t-test. * p<0.05, ** p<0.01 versus untreated control; n=4.* p < 0.05; ** p < 0.01; *** p < 0.001, **** p < 0.0001.
Discussion
In rodent models, work correlating acute EtOH exposures with progressive periods of neural tube patterning indicates that distinct regions of the face and brain are susceptible to alcohol-induced teratogenesis during very specific developmental windows. Interestingly, exposures separated by as little as a day can result in varying patterns of dysgenesis (Godin et al., 2010; O’Leary-Moore et al., 2010; Parnell et al., 2013;, 2009). Despite the fact that many of the impacted structures derive from a common pool of progenitor cells, the developmental stage-dependency of these observed defects suggest that key cellular populations are susceptible to insult while most others are not. This stage specificity is likely a contributing factor to the enormous variation observed in FASD clinical phenotypes, and is an aspect of this disorder that may be tied to core elements of developmental plasticity and differentiation.
In embryonic stem cell models of differentiation, much attention has been focused on the co-localization of H3K4me3 and H3K27me3, two chromatin modifications correlated with transcriptionally active and repressed chromatin states respectively (Bernstein et al., 2006). These bivalent domains are hypothesized to be poised for either rapid up- or down-regulation during the onset of differentiation, and along with several other histone post-translational modifications, have been proposed to provide a chromatin-based blueprint for transcriptional regulation through lineage determination. However, recent work examining the transition from culture conditions employing LIF/feeders towards the ground state imparted by 2i conditions, reveal that epigenetic marks provide limited insights into the transcriptional dynamics of stem cells, and suggest that in the undifferentiated state, chromatin modifications are more likely secondary effects associated with transcription rather then causal (Galonska et al., 2015). These observations are significant as they suggest that in stem cells, environmentally induced changes in chromatin structure may not actually disrupt the transcriptional programs controlling cellular identity, and therefore have a limited capacity to impact subsequent developmental stages.
Consistent with these observations, several recent studies examining in vivo models of EtOH exposure, along with other drugs of abuse, have noted inconsistencies between induced changes in chromatin structure and patterns of gene transcription. For example, Ponomarev and colleagues recently employed a transcriptome profiling approach to examine EtOH-exposed human brains. In this report, the authors noted that while half of the identified up-regulated genes exhibited increased enrichment of transcriptionally permissive chromatin modifications, the other half did not (Ponomarev, Wang, Zhang, Harris & Mayfield, 2012). Similarly, studies employing a rodent model of morphine addiction identified dramatic enrichment of transcriptionally repressive chromatin modifications with very little impact on mRNA expression (Sun et al., 2012). These studies are similar to a growing body of work examining other environmental exposures, which note a disconnect between induced alterations in chromatin structure and consistent impacts on gene transcription (Dhimolea et al., 2014; Lima, Ding, Goetz, Yang & Baulch, 2014).
Similar to our previous observations, we do not observe a linear dose response between alcohol exposure and alterations in chromatin structure (Veazey et al., 2015). The basis for this non-linearity is unclear but may be tied to the duration of exposure and response thresholds. However, such data emphasize that the impacts of ethanol are not diminished with lower doses. One common element emerging from these studies has been the consistent impact of EtOH on post-translational modifications on H3K9. However, these patterns of change are dependent on dose, timing of exposure and cell type. For example, work examining the consequences of EtOH exposure in adult hepatocytes reported significant increases in H3K9me2 during the exposure phase while similar studies in cortical neuronal cultures observed an increase in H3K9ac and a loss of H3K9me2 (Pal-Bhadra et al., 2007; Qiang, Denny, Lieu, Carreon & Li, 2011). Similarly, in a chronic intermittent in vivo model of exposure, EtOH increased H3K9ac within the nucleus accumbens and prefrontal cortex regions of the mouse brain (Finegersh et al., 2015). In contrast, we observe loss of H3K9ac during the exposure period and H3K9 hypermethylation during both the period of exposure, as well as the recovery phase. Alterations in H3K9 post-translational modifications are emerging as a recurring element in multiple studies examining the impact of EtOH, as well as cocaine and other drugs of abuse (Maze et al., 2010; Subbanna & Basavarajappa, 2014; Sun et al., 2012). However the functional relevance of these changes are not immediately clear. In our in vitro stem cell models, we have failed to identify consistent reductions in gene transcription correlating with the increased H3K9me2. In this study, only two candidates genes exhibited significant changes in gene transcription (Nanog and Sox2) and levels of these genes increased despite acquisition of H3K9 hypermethylation. However, work in vivo has identified correlative decreases in gene expression in proopiomelanocortin neurons (Govorko et al., 2012) and in studies of the developing brain, pretreatment of mice with an Ehmt2 (which catalyzes the dimethylation of H3K9) inhibitor prevents postnatal neurobehavioral deficits in a gestational model of exposure (Subbanna & Basavarajappa, 2014). Thus alterations in H3K9me2 induced by EtOH are clearly relevant to patterns of dysgenesis associated with FASDs, however not all cell types appear equally susceptible.
During differentiation, the chromatin landscape of stem cells transition from a plastic phase into one in which the transcriptional programs become locked in. During this transition, the accessibility of the chromatin fiber becomes more restrictive, adopting cell-specific patterns of accessible and inaccessible chromatin (Weintraub & Groudine, 1976). Recent studies suggest that this three-dimensional partitioning of the nucleus organizes the genome into active and silent compartments. Importantly, although histone post-translational modifications seem to be reflective of this organization, they do not appear to be casual in its establishment (Wijchers et al., 2016). Therefore, it is perhaps unsurprising that the data presented in this study indicate that in stem cell types in which the transcriptional programs are rigidly controlled, and in fully differentiated fibroblasts, EtOH-induced changes in chromatin structure have a limited capacity to directly impact patterns of gene transcription. However, the long-term effects EtOH-induced changes in chromatin structure have on genome stability, cellular aging and developmental plasticity remain unknown and of serious concern; even if the transcriptome is not affected in the short term.
During development, epigenetic changes like DNA methylation and a minority of post-translational histone modifications are thought to help stabilize nucleosome structure and lock in the transcriptional profile of the differentiated cell type. In particular, changes in DNA methylation are lasting and have longer-term impacts on gene transcription (Bintu et al., 2016). Our experiments identified alterations in transcripts encoding several core members of the DNA methylation machinery, including Dnmt1, Uhrf1, and Tet2. Collectively, these changes could have major impacts on development that manifest outside our experimental window, or that are only apparent in vivo. Genome-wide epigenetic and transcriptomic studies are required to obtain a comprehensive view of the effect of alcohol exposure in this model. However, it is interesting to note that exposed ESCs retain the ability to correctly establish patterns of imprinted gene expression during differentiation. Thus, by the limited criteria examined in this study, EtOH-induced alterations in chromatin structure do not permanently alter developmental potential or the processes of epigenetic programming. However, we speculate that cells encountering EtOH during specific phases of active differentiation, as in the case of the developing proopiomelanocortin neurons for example, may be more susceptible to the influence of the induced alterations. Thus, while undifferentiated stem cells maintained in a static state retain the ability to correct induced alterations to chromatin structure, the maturing transcriptional regulatory networks of differentiating cells are more susceptible, where the induced changes lock-in, permanently altering the cohorts of genes available/unavailable for transcription and negatively impacting patterns of gene expression.
How a common exposure can induce errors in the developmental trajectory of specific sub-populations of cells remains a key question broadly relevant to the field developmental toxicology. We believe that EtOH-induced changes in chromatin structure are relevant to dysgenesis associated with FASDs, but not universally so. Additional studies are needed to better resolve the links between differentiation and alcohol-induced teratogenesis to clearly distinguish EtOH-induced changes in chromatin structure that are symptoms versus those that are truly epigenetic; heritably influencing gene expression through development and into the next generation.
Supplementary Material
Primer sequences used in ChIP and qRT-PCR analysis.
Alterations to chromatin induced by alcohol do not universally impact transcription.
Exposure to ethanol dramatically alters histone 3 lysine 9 in multiple cell types.
Induced changes to chromatin do not impede the acquisition of genomic imprints.
Acknowledgments
This work was supported by the NIH–NIAA Grant 1R21AA022484. KJV was supported through the Texas A&M University College of Veterinary Medicine and Biomedical Sciences Graduate Student Research Trainee Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Badeaux AI, Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nature Reviews. Molecular Cell Biology. 2013;14(4):211–24. doi: 10.1038/nrm3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei MS, Tilghman SM. Genomic imprinting in mammals. Annual Review of Genetics. 1997;31(1):493–525. doi: 10.1146/annurev.genet.31.1.493. [DOI] [PubMed] [Google Scholar]
- Basavarajappa BS, Subbanna S. Epigenetic mechanisms in developmental alcohol-induced neurobehavioral deficits. Brain Sci. 2016;6(2) doi: 10.3390/brainsci6020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekdash RA, Zhang C, Sarkar DK. Gestational choline supplementation normalized fetal alcohol-induced alterations in histone modifications, DNA methylation, and proopiomelanocortin (POMC) gene expression in β-endorphin-producing POMC neurons of the hypothalamus. Alcoholism, Clinical and Experimental Research. 2013;37(7):1133–42. doi: 10.1111/acer.12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bintu L, Yong J, Antebi YE, McCue K, Kazuki Y, Uno N, et al. Dynamics of epigenetic regulation at the single-cell level. Science (New York, NY) 2016;351(6274):720–4. doi: 10.1126/science.aab2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch-Presegué L, Raurell-Vila H, Marazuela-Duque A, Kane-Goldsmith N, Valle A, Oliver J, et al. Stabilization of suv39h1 by sirt1 is part of oxidative stress response and ensures genome protection. Molecular Cell. 2011;42(2):210–23. doi: 10.1016/j.molcel.2011.02.034. [DOI] [PubMed] [Google Scholar]
- Brocardo PS, Gil-Mohapel J, Christie BR. The role of oxidative stress in fetal alcohol spectrum disorders. Brain Research Reviews. 2011;67(1–2):209–25. doi: 10.1016/j.brainresrev.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Carnahan MN, Veazey KJ, Muller D, Tingling JD, Miranda RC, Golding MC. Identification of cell-specific patterns of reference gene stability in quantitative reverse-transcriptase polymerase chain reaction studies of embryonic, placental and neural stem models of prenatal ethanol exposure. Alcohol (Fayetteville, NY) 2013;47(2):109–120. doi: 10.1016/j.alcohol.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia N, Wang L, Lu X, Senut MC, Brenner C, Ruden DM. Hypothesis: Environmental regulation of 5-hydroxymethylcytosine by oxidative stress. Epigenetics : Official Journal of the DNA Methylation Society. 2011;6(7):853–6. doi: 10.4161/epi.6.7.16461. [DOI] [PubMed] [Google Scholar]
- Dhimolea E, Wadia PR, Murray TJ, Settles ML, Treitman JD, Sonnenschein C, et al. Prenatal exposure to BPA alters the epigenome of the rat mammary gland and increases the propensity to neoplastic development. PloS One. 2014;9(7):e99800. doi: 10.1371/journal.pone.0099800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Sulik KK, Chen SY. Nrf2-mediated transcriptional induction of antioxidant response in mouse embryos exposed to ethanol in vivo: Implications for the prevention of fetal alcohol spectrum disorders. Antioxidants & Redox Signaling. 2008;10(12):2023–33. doi: 10.1089/ars.2007.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Fraga MF. Epigenetics and the environment: Emerging patterns and implications. Nature Reviews Genetics. 2011;13(2):97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- Finegersh A, Ferguson C, Maxwell S, Mazariegos D, Farrell D, Homanics GE. Repeated vapor ethanol exposure induces transient histone modifications in the brain that are modified by genotype and brain region. Front Mol Neurosci. 2015;8:39. doi: 10.3389/fnmol.2015.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galonska C, Ziller MJ, Karnik R, Meissner A. Ground state conditions induce rapid reorganization of core pluripotency factor binding before global epigenetic reprogramming. Cell Stem Cell. 2015 doi: 10.1016/j.stem.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin EA, O’Leary-Moore SK, Khan AA, Parnell SE, Ament JJ, Dehart DB, et al. Magnetic resonance microscopy defines ethanol-induced brain abnormalities in prenatal mice: Effects of acute insult on gestational day 7. Alcoholism, Clinical and Experimental Research. 2010;34(1):98–111. doi: 10.1111/j.1530-0277.2009.01071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding MC, Magri LS, Zhang L, Lalone SA, Higgins MJ, Mann MR. Depletion of kcnq1ot1 non-coding RNA does not affect imprinting maintenance in stem cells. Development (Cambridge, England) 2011 doi: 10.1242/dev.057778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govorko D, Bekdash RA, Zhang C, Sarkar DK. Male germline transmits fetal alcohol adverse effect on hypothalamic proopiomelanocortin gene across generations. Biological Psychiatry. 2012;72(5):378–88. doi: 10.1016/j.biopsych.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H, Karian P, Kikyo N. Regulation of embryonic stem cell self-renewal and pluripotency by leukaemia inhibitory factor. The Biochemical Journal. 2011;438(1):11–23. doi: 10.1042/BJ20102152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Yan PS, Huang TH, Issa JP. Chromatin immunoprecipitation microarrays for identification of genes silenced by histone H3 lysine 9 methylation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(19):7398–403. doi: 10.1073/pnas.0306641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima F, Ding D, Goetz W, Yang AJ, Baulch JE. High LET (56)fe ion irradiation induces tissue-specific changes in DNA methylation in the mouse. Environmental and Molecular Mutagenesis. 2014;55(3):266–77. doi: 10.1002/em.21832. [DOI] [PubMed] [Google Scholar]
- Liu Y, Balaraman Y, Wang G, Nephew KP, Zhou FC. Alcohol exposure alters DNA methylation profiles in mouse embryos at early neurulation. Epigenetics : Official Journal of the DNA Methylation Society. 2009;4(7):500–11. doi: 10.4161/epi.4.7.9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann MR, Lee SS, Doherty AS, Verona RI, Nolen LD, Schultz RM, et al. Selective loss of imprinting in the placenta following preimplantation development in culture. Development (Cambridge, England) 2004;131(15):3727–35. doi: 10.1242/dev.01241. [DOI] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149(3):590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Covington HE, Dietz DM, LaPlant Q, Renthal W, Russo SJ, et al. Essential role of the histone methyltransferase g9a in cocaine-induced plasticity. Science (New York, NY) 2010;327(5962):213–6. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37(1):179–83. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- Mead EA, Sarkar DK. Fetal alcohol spectrum disorders and their transmission through genetic and epigenetic mechanisms. Frontiers in Genetics. 2014;5:154. doi: 10.3389/fgene.2014.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memo L, Gnoato E, Caminiti S, Pichini S, Tarani L. Fetal alcohol spectrum disorders and fetal alcohol syndrome: The state of the art and new diagnostic tools. Early Human Development. 2013;89(Suppl 1):S40–3. doi: 10.1016/S0378-3782(13)70013-6. [DOI] [PubMed] [Google Scholar]
- Miyanari Y, Torres-Padilla ME. Control of ground-state pluripotency by allelic regulation of nanog. Nature. 2012;483(7390):470–3. doi: 10.1038/nature10807. [DOI] [PubMed] [Google Scholar]
- Moonat S, Sakharkar AJ, Zhang H, Tang L, Pandey SC. Aberrant histone deacetylase2-mediated histone modifications and synaptic plasticity in the amygdala predisposes to anxiety and alcoholism. Biological Psychiatry. 2013;73(8):763–73. doi: 10.1016/j.biopsych.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moris N, Pina C, Arias AM. Transition states and cell fate decisions in epigenetic landscapes. Nature Reviews Genetics. 2016;17(11):693–703. doi: 10.1038/nrg.2016.98. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A, Deplancke B, Walhout AJ, Tissenbaum HA. Chromatin immunoprecipitation (chip) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in caenorhabditis elegans. Nature Protocols. 2008;3(4):698–709. doi: 10.1038/nprot.2008.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, et al. The air noncoding RNA epigenetically silences transcription by targeting g9a to chromatin. Science (New York, NY) 2008;322(5908):1717–20. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. Epigenetic mechanisms of drug addiction. Neuropharmacology. 2014;76(Pt B):259–68. doi: 10.1016/j.neuropharm.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogony JW, Malahias E, Vadigepalli R, Anni H. Ethanol alters the balance of sox2, oct4, and nanog expression in distinct subpopulations during differentiation of embryonic stem cells. Stem Cells and Development. 2013 doi: 10.1089/scd.2012.0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary-Moore SK, Parnell SE, Godin EA, Dehart DB, Ament JJ, Khan AA, et al. Magnetic resonance microscopy-based analyses of the brains of normal and ethanol-exposed fetal mice. Birth Defects Research Part A, Clinical and Molecular Teratology. 2010;88(11):953–64. doi: 10.1002/bdra.20719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Jackson DE, Mamatha L, Park PH, Shukla SD. Distinct methylation patterns in histone H3 at lys-4 and lys-9 correlate with up- & down-regulation of genes by ethanol in hepatocytes. Life Sciences. 2007;81(12):979–87. doi: 10.1016/j.lfs.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B, Zhu J, Lv T, Sun H, Huang X, Tian J. Alcohol consumption during gestation causes histone3 lysine9 hyperacetylation and an alternation of expression of heart development-related genes in mice. Alcoholism, Clinical and Experimental Research. 2014;38(9):2396–402. doi: 10.1111/acer.12518. [DOI] [PubMed] [Google Scholar]
- Parnell SE, Holloway HT, O’Leary-Moore SK, Dehart DB, Paniaqua B, Oguz I, et al. Magnetic resonance microscopy-based analyses of the neuroanatomical effects of gestational day 9 ethanol exposure in mice. Neurotoxicology and Teratology. 2013;39:77–83. doi: 10.1016/j.ntt.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell SE, O’Leary-Moore SK, Godin EA, Dehart DB, Johnson BW, Allan Johnson G, et al. Magnetic resonance microscopy defines ethanol-induced brain abnormalities in prenatal mice: Effects of acute insult on gestational day 8. Alcoholism, Clinical and Experimental Research. 2009;33(6):1001–11. doi: 10.1111/j.1530-0277.2009.00921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I. Epigenetic control of gene expression in the alcoholic brain. Alcohol Research : Current Reviews. 2013;35(1):69–76. doi: 10.35946/arcr.v35.1.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. The Journal of Neuroscience : The Official Journal of the Society for Neuroscience. 2012;32(5):1884–97. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruszak J, Ludwig W, Blak A, Alavian K, Isacson O. CD15, CD24, and CD29 define a surface biomarker code for neural lineage differentiation of stem cells. Stem Cells (Dayton, Ohio) 2009;27(12):2928–40. doi: 10.1002/stem.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang M, Denny A, Lieu M, Carreon S, Li J. Histone H3K9 modifications are a local chromatin event involved in ethanol-induced neuroadaptation of the NR2B gene. Epigenetics : Official Journal of the DNA Methylation Society. 2011;6(9):1095–104. doi: 10.4161/epi.6.9.16924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resendiz M, Mason S, Lo CL, Zhou FC. Epigenetic regulation of the neural transcriptome and alcohol interference during development. Frontiers in Genetics. 2014;5:285. doi: 10.3389/fgene.2014.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roozen S, Peters GJ, Kok G, Townend D, Nijhuis J, Curfs L. Worldwide prevalence of fetal alcohol spectrum disorders: A systematic literature review including meta-analysis. Alcoholism, Clinical and Experimental Research. 2016;40(1):18–32. doi: 10.1111/acer.12939. [DOI] [PubMed] [Google Scholar]
- Rugg-Gunn PJ, Cox BJ, Ralston A, Rossant J. Inaugural article: Distinct histone modifications in stem cell lines and tissue lineages from the early mouse embryo. Proceedings of the National Academy of Sciences of the United States of America. 2010 doi: 10.1073/pnas.0914507107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro F, Mayer D, Klement RM, Warczok KE, Stukalov A, Barlow DP, et al. Imprinted igf2r silencing depends on continuous airn lncrna expression and is not restricted to a developmental window. Development (Cambridge, England) 2013;140(6):1184–95. doi: 10.1242/dev.088849. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature Protocols. 2008;3(6):1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Subbanna S, Basavarajappa BS. Pre-administration of g9a/GLP inhibitor during synaptogenesis prevents postnatal ethanol-induced LTP deficits and neurobehavioral abnormalities in adult mice. Experimental Neurology. 2014;261:34–43. doi: 10.1016/j.expneurol.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Maze I, Dietz DM, Scobie KN, Kennedy PJ, Damez-Werno D, et al. Morphine epigenomically regulates behavior through alterations in histone H3 lysine 9 dimethylation in the nucleus accumbens. The Journal of Neuroscience : The Official Journal of the Society for Neuroscience. 2012;32(48):17454–64. doi: 10.1523/JNEUROSCI.1357-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science (New York, NY) 2009;324(5929):930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer M, Knezovich J, Ramsay M. In utero alcohol exposure, epigenetic changes, and their consequences. Alcohol Research : Current Reviews. 2013;35(1):37–46. doi: 10.35946/arcr.v35.1.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veazey KJ, Carnahan MN, Muller D, Miranda RC, Golding MC. Alcohol-induced epigenetic alterations to developmentally crucial genes regulating neural stemness and differentiation. Alcoholism, Clinical and Experimental Research. 2013;37(7):1111–22. doi: 10.1111/acer.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veazey KJ, Parnell SE, Miranda RC, Golding MC. Dose-dependent alcohol-induced alterations in chromatin structure persist beyond the window of exposure and correlate with fetal alcohol syndrome birth defects. Epigenetics & Chromatin. 2015;8(39):1–19. doi: 10.1186/s13072-015-0031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976;193(4256):848–56. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Wijchers PJ, Krijger PH, Geeven G, Zhu Y, Denker A, Verstegen MJ, et al. Cause and consequence of tethering a subtad to different nuclear compartments. Molecular Cell. 2016;61(3):461–73. doi: 10.1016/j.molcel.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453(7194):519–23. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CR, Ho MF, Vega MC, Burne TH, Chong S. Prenatal ethanol exposure alters adult hippocampal VGLUT2 expression with concomitant changes in promoter DNA methylation, H3K4 trimethylation and mir-467b-5p levels. Epigenetics & Chromatin. 2015;8:40. doi: 10.1186/s13072-015-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Ji X, Zhang F, Li L, Ma L. Embryonic stem cell markers. Molecules (Basel, Switzerland) 2012;17(6):6196–236. doi: 10.3390/molecules17066196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FC, Zhao Q, Liu Y, Goodlett CR, Liang T, McClintick JN, et al. Alteration of gene expression by alcohol exposure at early neurulation. BMC Genomics. 2011;12(1):124. doi: 10.1186/1471-2164-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences used in ChIP and qRT-PCR analysis.