Abstract
Metastasis accounts for most cancer-related deaths. The majority of solid cancers, including those of the breast, colorectum, prostate and skin, metastasize at significant levels to the liver due to its hemodynamic as well as tumor permissive microenvironmental properties. As this occurs prior to detection and treatment of the primary tumor, we need to target liver metastases to improve patients’ outcomes. Animal models, while proven to be useful in mechanistic studies, do not represent the human population heterogeneity, drug metabolism or cell-cell interactions, and this gap between animals and humans results in costly and inefficient drug discovery. This underscores the need to accurately model the human liver for disease studies and drug development. Further, the occurrence of liver metastases is influenced by the primary tumor type, sex and race; thus, modeling these specific settings will facilitate the development of personalized/targeted medicine for each specific group. We have adapted such all-human 3D ex vivo hepatic microphysiological system (MPS) (a.k.a. liver-on-a-chip) to investigate human micrometastases. This review focuses on the sources of liver resident cells, especially the iPS cell-derived hepatocytes, and examines some of the advantages and disadvantages of these sources. In addition, this review also examines other potential challenges and limitations in modeling human liver.
Keywords: 3D ex vivo hepatic microphysiological system (MPS), Liver metastasis, Hepatocytes, Non-parenchymal cells (NPC), Liver sinusoidal endothelial cells (LSE cells), Induced pluripotent stem cells (iPS cells), Embryonic stem cells (ES cells), Personalized/precision medicine
INTRODUCTION
Metastasis, the dreadful disease state that causes about 90% of cancer-related mortality [1], is a the hallmark of solid cancers wherein tumor cells from the primary site acquire invasive traits, such as loss of E-cadherin during epithelial-to-mesenchymal transition (EMT) to allow the tumor cells to escape and seed into secondary organs [2]. Other than EMT, there are several other hypotheses that have been proposed to be important in driving metastasis such as tumor angiogenesis [3], tumor-initiating or cancer stem cells [4,5], and recently proposed EMT-independent metastases [6]. Still, regardless of the initial change that enables dissemination, the intricate processes of establishing secondary distant tumors largely depend on the cancer cells and the new tumor microenvironment; the cancer cells undergo genetic and phenotypic alterations that allow them to integrate, survive and proliferate within the modified tumor permissive foreign microenvironments. In most cases, patients succumb to this disease due to increased tumor burden that alters the microenvironment of the secondary organs resulting in acute organ failure [7]. Although some primary and metastatic pairs share mutations in commonly affected genes such as TP53 and KRAS, metastatic lesions also show independent and distinct genomic profiles when compared to their respective primary pairs [8]. Furthermore, the microenvironments of secondary organs such as the liver can nurture the metastatic nodules to develop resistance against chemotherapeutic agents [9–12]. Consequently, tumor heterogeneity and distinct genomic and phenotypic profiles often thwart the treatment and management of patients with metastatic diseases and these highlight the need to independently evaluate and study metastatic tumors for the development of more efficacious targeted therapies.
In this perspective, we will describe a situation for which human-derived stem cells can be co-opted to provide a solution to the problems of studying tumor metastasis and developing effective therapies. To understand this lethal stage of cancers, a complex tissue-level system is needed. No animal models truly recapitulate the early and dormant stages that vex our current approaches. We, and a few others, have turned to organotypic models (‘tissue-on-a-chip’) to study this progression. However, the availability of cells hampers the widespread adoption of such powerful tools. Stem cells hold promise of overcoming the shortages and can be derived from the patients themselves enabling a personalized approach. Herein, we will discuss the clinical situation and the most currently developed lab-on-a-chip solution. This will also highlight how stem cells would contribute to these technical developments.
Liver metastasis
The majority of solid tumors including those of the breast, prostate, colon and skin preferentially metastasize to the liver, lung and bone [13–15]. There are several key factors that allow for the cancer cells to extravasate into the liver. First of all, the liver contains complex network of blood circulation that delivers oxygenated blood via the hepatic artery and nutrient-enriched blood via the portal vein [16]. The extensive network of blood vessels, coupled with slow hepatic microcirculation in the sinusoidal capillaries potentiate cancer cell retention in the liver [16]. Secondly, the liver endothelium lacks basement membrane, thus allowing tumor cells to access the extracellular matrix proteins for motility cues and efficient attachment to establish micrometastases [17,18]. Lastly, liver resident cells express high levels of surface molecules to enable them to uptake nutrients and clear gut-derived toxins and microbes. However, these surface molecules, such as cell adhesion molecules, endocytic receptors, toll-like receptors and oligosaccharides moieties [16], also play some roles in aiding cancer cell diapedesis from the lumen into the space of Disse [18].
The liver as a metastatic site poses significant challenges in treatment as it is the main site for drug metabolism. Most current and past studies on metastasis rely on animal models, specifically mouse models that can be genetically altered to mimic human diseases [19]. Although these models offer a lot of advantages and have led to remarkable scientific breakthroughs, these models pose some serious limitations with the difference in therapy response and drug metabolism between human and animal being the prime disadvantages [20] . Almost 95% of tested compounds fail clinical trials [21,22] and this astonishingly high attrition rate, primarily due to drug toxicity often only in subsets of patients, results in cumulative expenditures of over two billion dollars to achieve a successful drug approval by the US Food and Drug Administration (FDA) [23]. Therefore, there is a pressing need for a more reliable and robust model to study liver metastases and drug discovery as they pertain to the human condition.
Liver metastases occur at different rates in different primary tumor types, sex and race. Modeling these specific settings is needed to understand the biological factors contributing to these incidences and this will help in the development of personalized/targeted medicine for each setting. The following section discusses the rate and some of the causes and consequences of liver metastases in these different settings.
Liver metastases incidence in different primary tumor types
The incidence of liver metastases is different among the primary tumor types. Most studies on liver metastases focus on colorectal and pancreatic metastases since these cells originate from the gastrointestinal (GI) tract that drains directly into the liver [24]. Accordingly, liver metastasis affects more than 50% of colorectal cancer patients during their lifespan [25]. However, it is important to note that liver metastases also occur at high frequencies in other solid tumors but are usually detected in the late stage of tumor progression. About 8% of prostate cancer patients develop clinically evident liver metastases during their treatment period [26] and a much higher percentage (25–45%) of prostate cancer patients show liver involvement at autopsy [15,27]. These metastases are typically hormone-refractory and aggressive prostate cancer cells [26]. A similar pattern is also observed in breast cancer patients where less than 5% of the patients have clinically evident liver metastases at the time of diagnosis [14,28]. However, liver involvement in breast cancer patients dramatically increases to affect approximately 60–70% of the patients at autopsy [14,29]. Breast cancer patients with liver metastases show the worst prognosis when compared to lung and bone metastases with a 5-year survival percentage of merely 5.5–8.5% and a median of 14–16 months of survival times [14]. The vast majority of patients with these primary carcinomas are ‘cured’ upon resection (or radiologic extirpation) of the primary mass, and therefore the fraction of liver involvement in persons with advanced disease is much greater.
In addition to different rates of liver metastases, these primary tumors also differently affect the liver. For example, the right hemi-liver, due to blood circulation and its larger size, is more frequently affected in patients with body-tail primary pancreatic adenocarcinoma [30] and primary colorectal carcinoma originating from the right hemi-colon [31]. In addition to lobar distribution, liver metastases from these primary tumors often show distinct pathological features such as size, shape, growth pattern and vascularity [13]. Neuroendocrine liver metastases show increased frequency of fluid-fluid levels, which can be used as independent predictor to distinguish neuroendocrine and non-neuroendocrine (colon, rectum, breast, pancreatic, melanoma and several other primary tumor types) liver metastases [32]. These findings can be useful in disease management including screening, diagnosis, targeted therapies and surveillance post primary tumor treatment.
It should be noted that isolated or limited liver metastatic nodules have long been recognized as candidates for removal. Colorectal carcinomas initially disseminate mainly to the liver (due to both vascular drainage and ontologic derivation considerations) and have been removed for decades with great improvement in survival and even ‘cures’ [33]. More recently, liver metastases of other solid tumors have been targeted surgically. For instance, the removal of breast cancer metastases has doubled the one-year survival rate to 80% in advanced breast cancers [34]. Thus, liver metastases are targetable tumors even in the likely presence of second metastatic seeding.
In short, most solid primary malignancies progress to the liver, at least in the later stages, and may differently affect the liver and cause distinct pathological conditions. This has implications for survival and even treatment approaches, as the liver is the main organ for drug metabolism and limiting toxicities. Thus, reflective models are necessary to unravel the underlying causes and consequences of liver metastases originating not only from the gastrointestinal tract and pancreas, but from other primary sites as well in order to develop more precise therapies.
Liver metastases incidence by sex
The second factor that affects the incidence of liver metastases is sex. Studies show that women are more susceptible to cirrhosis, benign liver lesions, alcohol liver disease and toxin-induced acute liver injury than men [35]. Biological factors that can contribute to these differences include different profiles of metabolizing enzymes such as cytochrome P540 (CYP) [36,37] and nuclear receptors [38] leading to varying sensitivity to drugs. Men, on the other hand, have a higher rate of primary liver malignancy [35]. The ratio of hepatocellular carcinoma (HCC), which is one of the most common forms of primary liver malignancy, between men and women is 3–4:1 [39]. Different hormonal activities and signaling are among the factors that contribute for sexual dimorphism in liver malignancy. In female mice, estrogen prevents diethylnitrosamine (DEN)-induced HCC by inhibiting MyD88-mediated NF-κB transcriptional activity to reduce tumor-promoting interleukin-6 (IL-6) mRNA and serum level [39]. In male, the activation of androgen receptor promotes hepatocarcinogenesis by promoting cell growth, increasing cellular oxidative stress to elevate cellular DNA damage and inhibiting tumor suppressor P53 and its downstream targets [40]. These sexual dimorphism effects are dependent on FOXA1/2 transcription factors that play differential roles in recruiting estrogen receptor α and androgen receptor to co-regulate the expression of their targets [41].
In addition to primary liver cancer, several lines of evidence suggest that men are also more prone to liver metastases [24,42–44] and, in some cases, show a poorer prognosis [45]. Separate studies on two European cohorts revealed a higher rate of liver metastases in men regardless of the origin of the primary tumor [24,42]. Furthermore, the frequency of both synchronous and metachronous colorectal cancer liver metastases are higher in men [43]. This suggests that there is a cancer-related predilection rather than metastases seeding into a diseased liver. It is also reported that both men and women can benefit from liver resection for improved survival [43]. In addition to colorectal carcinoma, a univariate analysis on patients with neuroendocrine liver metastases showed significant reduced survival in men when compared to women and multivariate analysis revealed that male sex was significantly associated with three times greater mortality risk [45].
In all, men have a higher tendency of developing liver metastases than women. The underlying biological factors are still unclear but could be due to differences in hormonal signaling and distinct CYP and nuclear receptor profiles. However, these studies, mainly performed in rodents, warrant for further validation and we are, at the moment, actively pursuing this subject using our all human 3D ex vivo hepatic MPS.
Racial disparities in liver metastases
Lastly, the incidence of liver metastases also differs among different races and ethnicities. Most clinical studies on racial disparity in tumor incidence and treatment have always been confounded by other external factors such as environmental and socioeconomic status that may increase the risks of exposure to liver toxins and limit access to medical care. However, biological differences may still play some roles in the discrepancies of the distribution of liver metastases among different races and ethnicities. In general, African Americans show high incidence and mortality rate for colorectal, cervical, prostate and liver tumors when compared to other races and are more frequently diagnosed with advanced stage cancer [46]. When adjusted for census tract poverty level, African Americans and American Indians/Alaska Natives still show a lower 5-year survival rate when compared to non-Hispanic whites, indicating that socioeconomic status is not the only factor for racial disparity in cancer incidence and mortality [46]. Other than higher incidence rate, African American HCC patients are more likely to present with regional and distant diseases at diagnosis [47]. A recent study reported that a single nucleotide polymorphism on TP53 (P47S) gene, found to be specifically restricted in African-descent populations, can cause inefficient apoptosis induction when challenged with cisplatin and increase the susceptibility of spontaneous tumorigenesis and increase the risk of metastasis [48] thus cementing the contribution of genetic variations in promoting tumorigenesis and metastasis in these races/ethnicities.
For liver metastases, a Surveillance, Epidemiology and End Results (SEER)-based study on colorectal cancer reveals a higher percentage of metastases to the liver and lung in African American patients when compared to Caucasians, Asians and other ethnicities [49]. Interestingly, for gastric cancer patients, non-Hispanic whites showed higher incidences of liver metastases when compared to other ethnicities [44]. Additionally, basal type or triple negative breast cancer, which is more prevalent in pre-menopausal African American women [50,51], shows greater propensity to metastasize to the liver and brain when compared to other breast cancer types [52]. In all, biological differences in these ethnic groups, together with environmental factors and socioeconomic status, may contribute to racial disparity of liver metastases.
MODELING LIVER METASTASIS
All of the above indicate that liver metastasis is not simply ‘one disease’ and each specific setting should be separately addressed for better and efficacious targeted therapies. Further, the differences noted by sex and ethnicity/race of individuals argue for using a diversity of primary cells to capture this heterogeneity ex vivo. Current studies on spontaneous liver metastases mainly utilize animal models that poorly replicate clinical conditions [53], while standard in vitro culture limits primary cell viability and functionality [54].
Three-dimensional (3D) cell culture re-establishes the structural and signaling relationships of the tissues that are important for organ function. Several conditions in constructing the liver and modeling liver metastases ex-vivo need to be properly addressed in order to overcome the limitations associated with mouse models and standard 2D in vitro cultures. First, the liver itself must be reasonably recreated to allow for homeostatic functions such as drug metabolism. The ex vivo liver must also be responsive to external factors such as lipopolysaccharide (LPS) that can promote metastasis and has been shown to be elevated in patients with liver metastases [55]. Second, the tissue structure must be challenged with appropriate tumor cells that can recreate the metastatic phenotypes noted in persons, including both dormant and outgrowing nodules. This means a relatively comprehensive regeneration of the pathologically involved organ that will require reliable sources of human parenchymal and non-parenchymal cells (NPC) to recreate 3D ex vivo liver tissues on perfusable scaffolds that can maintain tissue integrity and function while minimizing the effects of the construct. Other considerations that should be included are common medium to accommodate heterogeneous cell types in the system and precise mechatronics to continuously supply nutrients at physiologic concentration and remove cellular wastes. Comprehensive requirements to ‘engineer’ the liver are extensively discussed elsewhere [56]. There are several platforms that have been constructed according to the aspects mentioned above such as LiverChip microphysiologic system (MPS), PEARL perfusion liver system and sequentially layered, self-assembly liver (SQL-SAL) platform. These platforms have been validated to improve and maintain liver functions for extended period of times. The advantages and disadvantages of these platforms are reviewed extensively elsewhere by us and others [57,58].
In addition to the platforms discussed previously, there are several liver platforms that have been recently developed including a novel microfluidic device that can simultaneously monitor oxygen consumption, glucose uptake and lactate production in real time [59]. The precise measurements of these parameters allow for accurate detection of early mitochondrial dysfunction that is critical in liver toxicity. This device, similar to other platforms, provides continuous perfusion and maintains hepatic functions for an extended period of time [59]. Another platform, metastasis-on-a-chip (MOC), connects gut and liver organoids through an elastic tube and allows for real-time monitoring of cancer migration from the gut to the liver [60]. The system also permits the investigators to manipulate the stiffness of the tumor foci separately from the stiffness of the tissue microenvironment (i.e. soft tumor foci and stiff gut/liver microenvironment and vice versa) that may be crucial in studying the effects of physical microenvironment on tumor cell migration. Recently, bio-printing, a state-of-the-art technology to construct 3D structure using live cells into viable and functional tissues, has been utilized to construct 3D tissues including the liver for both static culture and perfused bioreactor [61–64]. Table 1 summarizes some of the properties of these recent platforms.
Table 1.
Select 3D ex vivo hepatic MPS
| Platform | Cell types | Time | Features |
|---|---|---|---|
| LiverChip MPS | Fresh and cryopreserved primary human hepatocytes Fresh primary human NPCs |
15–29 days | - Continuous/controlled perfusion - Spontaneously induces tumor dormancy [69] - Utilizes soft (PEG hydrogel) and stiff (polystyrene) scaffolds to more accurately model normal and diseased liver [73] - Establishes oxygen gradients that mimics liver physiology [70] - Successfully predicts drug clearance and metabolism that correlates with clinical observations [71,78] |
| Bavli et al [59] | HepG2/C3A derivative | 28 days | - Measures oxygen consumption using tissue embedded two- frequency phase modulation phosphorescent microprobes - Simultaneously measures glucose uptake and lactate production using computer controlled microfluidic switchboard - Continuous/controlled perfusion |
| 3D bio-printed liver | HepG2 and HUVEC [61] Cryopreserved primaryhuman hepatocytes [63,64] iPS/ES cells [62,64] |
6–35 days | - Bio-mimetic perfusion [61] - Allows for the fabrication of complex structure in only one-step 3D cell printing [61] - Improves liver functions/differentiation |
| MOC [Metastasis on a Chip) [60] | HepG2 Human intestine epithelial cells (INT-407) |
24 days | - Combines two organoids for early cancer metastasis study - Micro-peristaltic pump for controlled medium flow - Real-time microscopy to monitor cells migration - Tunable hydrogel system to modulate the stiffness of tumor foci and tumor microenvironment |
This review, fitting with the theme of stem cells, will focus on liver resident cells and their derivation in modeling the liver. The liver is composed of several different cell types. The liver parenchyma, hepatocytes, makes up about 80% of the liver weight and about 60% of liver cell number [65]. The basal side of the hepatocytes is connected to the liver sinusoid and the apical side forms bile canaliculi [16]. The liver sinusoids are lined with liver sinusoidal endothelial (LSE) cells and their scavenger function together with the fenestrated feature allow for particles to migrate through the endothelial barrier [16]. The liver also has a population of resident macrophages, the Kupffer cells, to protect the liver and clear foreign bodies. Other liver resident cells include hepatic stellate cells, oval cell and cholangiocytes [16]. The parenchymal cells mediate most of liver functions with the support from the NPC. These cells also play differential roles in liver pathology. Thus, suitable and reliable sources of human hepatocytes and NPC are instrumental in order to properly recapitulate functional liver microenvironment for disease modeling. The advantages and disadvantages of several sources of human liver resident cells are discuss in the next section and summarized in Table 2 and currently available sources of liver parenchymal and NPC are listed in Table 3.
Table 2.
Advantages and disadvantages of hepatocytes sources
| Cell source | Advantages | Disadvantages |
|---|---|---|
| Fresh primary hepatocytes | • Fully functional • Abundant yield |
• Limited and haphazard availability • Non-replicative • Limited viability in-vitro/ex- vivo |
| Cryopreserved hepatocytes | • Fully functional • Less variability • Multiple different sources |
• Non-replicative • Limited viability in-vitro/ex- vivo and following cell thawing • Costly • Limited cell number |
| Immortalized hepatocytes | • Cost-effective • Unlimited supply |
• Poor hepatocyte morphology and hepatic functions |
| iPS/ES-derived hepatocytes | • Unlimited supply • Suitable for personalized medicine • Rapidly improving field |
• Moderate/poor hepatic induction • Could be labor intensive and costly • Ethical issues with ES cells |
Table 3.
Available sources of liver resident cells
| Cells | Source |
|---|---|
| Hepatocytes | Fresh or cryopreserved primary cells Cell lines iPS cell-derived hepatocytes |
| Liver sinusoidal endothelial cells | Fresh or cryopreserved primary cells Cell lines: TMNK-1, TRP3 |
| Kupffer cells | Fresh or cryopreserved primary cells Cell lines: KUP5 (mouse), RKC1 and RKC2 (rat) |
| Hepatic stellate cells | Fresh or cryopreserved primary cells Cell lines: LX1, LX2, TWNT-1, LI90 |
| Cholangiocytes | Fresh or cryopreserved primary cells Cell lines: H69 iPS cell-derived hepatocytes |
| Oval cells | Primary cells Cell lines: LE/2, LE/6,p53−/−, OC/CDE (all are rat cells) |
Fresh human primary hepatocytes
One of the main and arguably the best source of hepatocytes are fresh hepatocytes that are isolated from resected liver sections. The status and functionality of these cells are self-evident thus making them the gold standard in liver research. Unfortunately, the cells have limited survival in standard 2D cultures (up to 7 days) and in sandwich cultures (up to 15 days) [66,67]. Moreover, hepatocytes under these conditions tend to lose the functionality of differentiated cells in terms of CYP activities and albumin (ALB) secretion [68]. Under physiological flow in perfused 3D ex vivo hepatic MPS, this functionality can be maintained for 30 days [56,57,69–73]. Another main advantage of this source is that millions of cells can be isolated from each gram of liver tissues [74,75]. Although fresh hepatocytes are the gold standard in liver research, there are several limitations of these fresh cells beyond cell stability. The main disadvantage of these cells is their erratic availability and variable health of the cells, both of which are dependent on the patient’s condition and the isolation processes [74] since these cells are excess pathological specimens from therapeutic procedures and thus reflect the underlying clinical conditions. Liver cells isolated from patients with relatively benign conditions (e.g. removal of hemangiomas) are typically very healthy and robust but underlying pathological conditions such as alcoholic liver disease and other end-stage cirrhotic liver could further complicate the isolation process and severely compromise the cells’ viability and functions (e.g. ALB secretion) [74]. Other than viability and functionality, the presence of liver malignancy and metastases can also negatively affect the isolation yield [75]. Another disadvantage of this source is restricted accessibility. There are several regional medical centers in the US that distribute patient-derived fresh primary human hepatocytes to various laboratories through NIH-funded Liver Tissue Cell Distribution System (LTCDS) service. Laboratories outside these centers will not have immediate access to the cells and shipment could further affect the cells’ viability. Therefore, investigators should also take logistical issues into consideration when planning for experiments utilizing fresh human hepatocytes.
Cryopreserved human hepatocytes
In order to overcome the drawbacks with fresh human hepatocytes, efforts have been made to cryopreserve the hepatocytes. Since these cells are derived from human patients, they are functionally, in term of drugs metabolism, comparable to the fresh primary hepatocytes [76,77]. Similarly, these cells can also survive in vitro but 3D culture and continuous perfusion in the 3D ex vivo hepatic MPS could prolong cell survival and functionality ex vivo [71,78]. An added advantage of using cryopreserved hepatocytes is that the experiments can be performed using the same batch of the primary cells, thus reducing the cells variability for better experimental plan and data analysis. However, this will neglect population diversity but could be simply addressed by utilizing different sources of cryopreserved hepatocytes. One disadvantage of this source is that the cryopreservation and thawing processes can significantly compromise cell viability following cell thawing [54]. One study found that pre-incubation of the freshly isolated hepatocytes with glucose, fructose or lipoic acid at low temperature significantly ameliorated the cells and resulted in improved viability and attachment efficiency following thawing [79]. This pre-incubation step helps the cells to recover from the enzymatic isolation procedure, maintains cell membrane integrity, boosts intracellular adenosine triphosphate (ATP) and reduces osmotic stress on the hepatocytes which collectively bolster the cells to withstand the preservation process [79]. Another study has shown that cryopreservation using a specially formulated hypothermic preservation solution could enhance the viability as well as maintaining hepatocyte functions in secreting albumin, synthesizing urea and maintaining CYP enzymatic activity after 14 days of in vitro culture following thawing [54]. Alternatively, cryopreserved hepatocytes can be purchased from several vendors. These cells are well-characterized and well-maintained under stringent laboratory practices but they are prohibitively expensive and severely limited in the number of cells.
Immortalized human hepatocyte cell lines
Several strategies have been employed to immortalize human hepatocytes such as transduction with viral oncogenes simian virus 40 large T antigen (SV40 Tag) [80], transfection with human telomerase reverse transcription retrovirus [81], conditional immortalization using Cre-loxP recombinase and tet-on and tet-off system [80]. Some of the commonly used hepatocytes or hepatoma cells in toxicology studies include HepaRG, Huh7, Hepato cells, HepG2, cBal111 and others (summarized in Table 4). While these cell lines offer unlimited supply of hepatocytes possibly at minimal cost, extra precautions and controls should be taken to verify the status of the cells. For instance, HepaRG cells, which were serially selected from hepatitis C infected liver tumor [82], can be differentiated into biliary-like or hepatocyte-like cells and have been shown to retain some hepatic functions upon differentiation [83]. However, these cells show distinct morphology and significant alteration in CYP activity when compared to both fresh and cryopreserved primary human hepatocytes [84]. Similarly, other cell lines also display distinct morphology and inferior hepatic functions especially the expression and activity of CYP enzymes [81,85–87].
Table 4.
Commonly used immortalized human hepatocytes/hepatoma lines
| Cells | Source | Immortalization | Reference(s) |
|---|---|---|---|
| HepaRG | Hepatocellular carcinoma | Serially selected | [82] |
| Huh7 | Hepatocellular carcinoma | Spontaneous replication | [158] |
| HepatoCells | Human primary hepatocytes | Proprietary method | |
| HepG2 | Hepatocellular carcinoma | Serially selected | [159] |
| cBal111 | Human fetal hepatocytes | hTERT | [87] |
| Hc-3716-hTERT | Human fetal hepatocytes | hTERT | [81] |
| TPH-1 | Human adult hepatocytes | HCV core | [160] |
Induced-pluripotent stem cell-derived hepatocytes
An emerging source of hepatocytes is induced-pluripotent stem cell (iPS) cell-derived hepatocytes. Since their discovery in 2006 [88], extensive efforts have been made to produce functional human tissues and organs in vitro. One main advantage of iPS cells is they can be derived from individuals with diverse genetic backgrounds using nearly any type of cell such as fibroblasts and blood cells and can be differentiated back into almost all cell types. The plasticity of iPS cells is valuable and suitable for personalized medicine. Tremendous breakthroughs using iPS cells have been reported including successful liver bud development and implantation to treat liver failure in mice [89] and whole mice generation from iPS cells [90]. However, translation from bench to bedside proves to be quite challenging, and regretfully, most iPS cell-derived human hepatocytes are still functionally inferior to adult human primary cells and more closely resemble immature, fetal hepatocytes instead [91–93]. There are several factors that can significantly affect the differentiation of the iPS cells. First and foremost, differentiation efficiency highly depends on the quality of the iPS cells. There are several reprogramming methods with varying levels of pluripotency induction [94]. Somatic cell nuclear transfer (SCNT) into enucleated egg and somatic cell fusion with pluripotent stem cell methods create iPS cells more rapidly and efficiently than the revolutionary method of transducing cells with OCT3/4, SOX2, KLF4 and c-MYC (OSKM or Yamanaka factors) but are more technically challenging and pose ethical and safety issues [94]. The latter method can be performed with several different vectors [94] and requires appropriate stoichiometry of the four reprogramming factors to increase reprogramming efficacy where higher pluripotency induction rate can be achieved by increasing KLF4 [95] and OCT4 [95,96] or by decreasing SOX2 level [97]. Although most iPS cell lines have been validated to express stem cell markers, form teratomas in mice and mimic the genetic and epigenetic status of embryonic stem cells, only a small subset of the iPS cell lines have been shown to achieve complete pluripotency as validated with tetraploid blastocyst complementation assay to produce live pups [90,98]. Other than the quality of the iPS cells, successful hepatic differentiation is also heavily dependent on the epigenetic status of the transformed cells [99]. It is important to note that the epigenetic marks between iPS cells and embryonic stem (ES) cells are distinct from one another [100] although this issue is still debatable as there are also some conflicting reports suggesting these two cell types show similar somatic epigenetic marks (reviewed in [99]). Some iPS cells tend to retain residual transcriptional memory of their original somatic cells due to incomplete gene repression during reprogramming which leads to distinct genetic and DNA methylation profiles when compared to ES cells [100]. Accordingly, generation of hepatocytes with ES cells is more efficient and convenient than iPS cells [101–105]. A recent report also demonstrated that even proficient ALB-secreting ES-derived hepatocytes could not recapitulate the CYP profile of primary human hepatocytes due to DNA hypermethylation on CYP enzymes promoters and this could be reversed by inhibiting of DNA methyltranferase and/or histone deacethylases [106]. Thus, in brief, iPS cells differentiation into hepatocyte-like cells depends on 1) the quality of the iPS cells that should closely mimic the genetic and epigenetic status of ES cells or in vivo pluripotent stem cells and 2) complete replication of primary human hepatocyte genetics including the reversion of DNA methylation specifically on CYP enzymes promoters for functional mature hepatocytes differentiation.
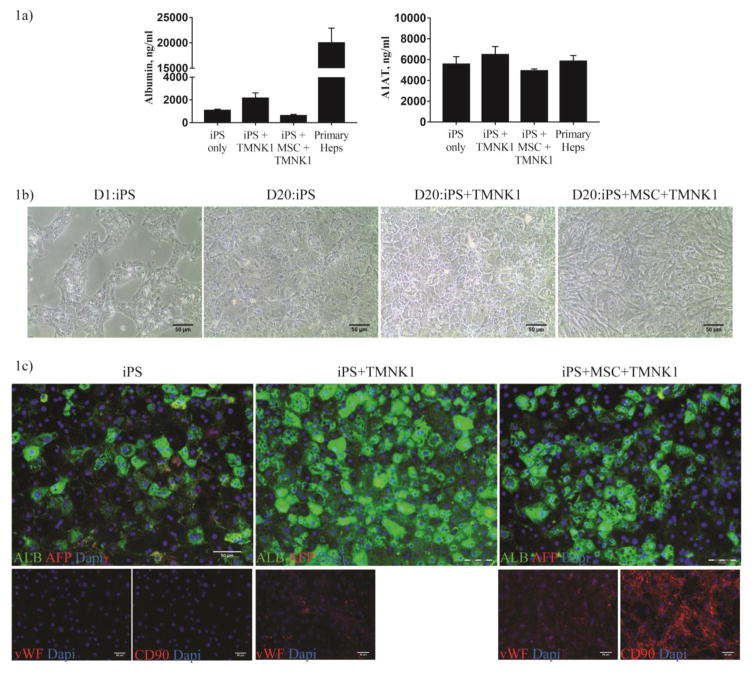
Successful induction of functional mature hepatocytes from iPS cells also relies on the differentiation protocols. Most of these protocols mainly utilize a cocktail of growth factors to induce the cells into different stages of hepatocyte development [93,107]. Hepatic induction is mainly validated by increased expression of hepatocyte markers especially albumin, reduced α-fetoprotein (AFP), elevated secretion of fibrinogen, albumin and α-1 anti-trypsin (A1AT), glycogen storage, incorporation and storage of low density lipoprotein and glycogen, increased CYP enzymes activity and liver failure rescue in mice. A brief summary of commonly used markers for iPS hepatocytes differentiation is summarized in Table 5. However, different parameters/units were used to measure these validation assays and some of these papers do not include primary hepatocytes as positive control [108,109] , thus making it impossible to compare the efficiency of hepatic induction among these different protocols. Other than growth factors, investigators also screen for small molecules as cheaper and stable alternatives to improve differentiation. Combinations of activin A with glycogen synthase-3 inhibitor (CHIR99021) markedly improved definitive endoderm induction [110] and novel compounds, functional proliferation hit (FPH1 and FPH2) and functional hit (FH1), were able to induce primary hepatocytes proliferation in vitro as well as improved iPS cell-derived hepatocytes differentiation marked by significant increment in albumin secretion and reduction in AFP secretion as well as elevated level of CYP3A4 and CYP2A6 activities [111]. Another strategy to improve hepatic differentiation is to co-culture immature hepatocyte-like cells with stromal cells or other liver resident cells. The basis of this strategy is that during human liver development, stromal cells in the septum transversum mesenchyme and cardiogenic mesoderm provide a plethora of growth factors such as fibroblast growth factor (FGF), Wnt, bone morphogenic factors (BMP) and hepatocyte growth factor (HGF) in a carefully orchestrated manner to activate and deactivate genes in the foregut endoderm [112]. Thus, co-culture with stromal cells will help to provide other various soluble factors at cellular specific concentration. Mouse and human iPS or ES cell co-cultured with endothelial cells [102], fibroblasts [113] or both endothelial and mesenchymal cells [89] showed increased hepatic differentiation. We also validated these observations with our preliminary work with human iPS cells hepatic differentiation in vitro (Figure 1). Co-culture of iPS cells with an immortalized LSE cell line alone, TMNK-1, drastically improved ALB expression and secretion when compared to without co-culture or co-culture with TMNK-1 and mesenchymal stem cells. Co-culture with both TMNK-1 and MSC promoted extensive growth of the supporting cells and resulted in less efficient hepatic differentiation.
Table 5.
Commonly used markers in iPS cells differentiation towards hepatocyte-like cells
| Gene/protein | Value in Hepatocyte Differentiation |
|---|---|
| SRY (sex determining region Y)-box17 (SOX17), chemokine (C-X-C motif) receptor 4 (CXCR4), forkhead box A2 (FOXA2) | Endoderm markers [110,161]) |
| HNF4α | Expressed during hepatic specification stage and persists to mature hepatocytes [162] |
| CYP3A7 | High expression in fetal liver [163] |
| AFP | Abundant in fetal/immature liver [105]. Very low/undetectable in normal adult liver [164] |
| Albumin | High gene expression and protein secretion in adult hepatocytes [162,165] |
| CYP1A2, 2E1, 2D6, 2C9, 2C19, 3A4 | Low or no activity in fetus but high activity in adults [166] |
Figure 1.
Liver sinusoidal endothelial cells augment hepatic differentiation of human iPS cells. iPS cells were differentiated into hepatocytes using protocol by Jun Cai et al [157]. TMNK-1 cells and/or mesenchymal stem cells were added into the culture on day 11 of the differentiation (1:0.7:0.25 ratio of iPSC:TMNK-1:MSC). Figure 1a) Supernatants were collected for albumin (Bethyl laboratories) and A1AT (Genway Biotech Inc) ELISA analyses and normalized to the number of cell. Figure 1b) Brightfield imaging of the cocultures demonstrates the changes in cell morphology of the iPS hepatocytes driven, in part, by the TMNK-1 cells. Figure 1c) Cells were fixed and stained for albumin (Bethyl laboratories) and AFP (Santa Cruz Biotech) (bottom panel). Stromal cells (TMNK-1 and MSC) co-cultured with the NFIF-iPS cells were stained with their respective markers, vWF (Santa Cruz Biotechnology) and CD90 (BD Biosciences). Bar= 50um. N=2
Consistency and robustness are major issues with these protocols and since some of the published protocols utilized only one iPS cell line [91,101,105,114,115], significant optimizations that could be costly and labor-intensive are required. There are some protocols that were validated in multiple cell lines. Hannan et al. were able to differentiate 20 iPS cell lines and 5 ES cell lines [116]. The authors acknowledged the consistency issue and stated that about 20% of the cell lines maintained in their lab were resistant to definitive endoderm induction, the first and important step of hepatic differentiation process [116]. Although the protocol was validated in a broad range of iPS cell lines, proper optimizations and validations are still required to address the variability outcomes. Sullivan et al. induced iPS cells from three different genetic backgrounds into ALB+ and E-cadherin+ hepatic endoderm and were able to detect the secretion of AFP, fibronectin and fibrinogen in all cell lines that also displayed functional CYP1A2 and CYP3A4 activity but with significant variability in iPS cells derived from diabetic North American Indian when compared to male and female Caucasians [108]. Rashid et al. generated iPS cell-derived hepatocytes from patients with A1AT deficiency, glycogen storage disease type 1a (GSD1a) and familial hypercholesterolemia (FH). Successful differentiation was demonstrated with varying degrees in which the iPS cell-derived hepatocytes from A1AT deficient donors showed the highest level of albumin secretion and CYP3A4 activity whereas GSD1a iPS cells gave rise to hepatocytes with the lowest albumin secretion and CYP3A4 activity [109]. One could argue that lower hepatic functions (i.e. reduced ALB secretion) in these patient-derived iPS cells are due to inherent problems caused by the diseases. However, it was reported that ALB serum level in GSD1 patients slightly increased or unchanged when compared to controls, suggesting that albumin production is not negatively affected in GSD1 patients [117]. The inconsistencies of hepatocyte induction in these protocols could also be due to heterogeneous population of immature (ALB+/−/AFP+ cells) and mature cells (ALB+/AFP−) [109,114,118]. Some protocols demonstrated impressive hepatocyte induction but may require prolonged maturation period to completely abolish AFP expression [101,103,115]. Isolation of differentiated cells using cell sorting for ASGPR+ cells improved the analysis for mature hepatocyte markers and improved albumin secretion in Alb-uPA SCID mice and Nagase analbuminemic rat that underwent partial hepatectomy [101].
Efforts have been made to utilize 3D culture methods, mainly by generating cell aggregates or spheroids, to improve hepatic differentiation. Hepatic differentiation was significantly improved when iPS cells and stromal cells were seeded into polydimethylsiloxane (PDMS) microwell platform to uniformly form ~120μm aggregates [119]. Another 3D culture method utilized a commercially available RAFTTM 3D cell culture system to culture iPS aggregates for over 40 days [120]. Both culture systems demonstrated significant reduction in the markers of immature hepatocyte, AFP and CYP3A7, when compared to 2D culture and fetal hepatocytes as well as increased the secretion of albumin, the expression of MDR1 transporter and the activity of CYP3A4 [120] and CYP2C9 [119]. More recently, 3D-bio-printing has been employed to generate human liver using iPS and ES cells [121]. iPS and ES cells, printed into 12-well plate with alginate-derived hydrogel for support, retained their viability and the expression of stem cell markers post-printing. However, following hepatic induction, only the ES cells formed hepatocyte-like cells with cobblestone morphology whereas the iPS cells formed a mixture of cell morphologies and the ES-derived hepatocytes 3D tissues secreted a higher level of albumin when compared to 2D culture [121]. Improved iPS-derived hepatocyte maturation was observed when iPS-derived hepatic progenitor cells and stromal cells (human umbilical vein endothelial cells (HUVEC) and adipose-derived stem cells) were printed into 3D hepatic construct [62]. Interestingly, the stromal cells re-aligned themselves along the edges of the hydrogel scaffold, resembling the liver sinusoid in liver lobule. The 3D-printed tri-culture liver tissue showed higher gene expression of hepatocyte markers (albumin, HNF4α, transthyretin, CYP3A4, CYP2C9 and CYP2C19) and increased albumin and urea production [62]. Schepers et al expanded the 3D culture system into a perfused liver model where iPS-derived hepatocytes and 3T3-J2 fibroblasts were firstly aggregated in pyramidal microwells using low speed centrifugation [122]. The aggregates were then encapsulated with polyethylene glycol (PEG) hydrogel to prevent uncontrollable growth that would limit oxygen and nutrient diffusion. Finally, these encapsulated aggregates were cultured under a range of medium flow in PDMS microfluidic device. This system promoted higher secretion of albumin when compared to the cells in 2D culture and could be maintained for 38 days. The system also induced the activity of CYP1A1 and CYP2C9 but lower than the values observed in primary human hepatocyte aggregates. The aggregates were also positive for both HNF4a and HNF1b, markers for hepatocytes and biliary epithelium. Although the maturity of these cells is still unclear as the CYPs activities were inferior to those of primary human hepatocytes and AFP expression is unknown, this study shows that encapsulation allows the aggregates to withstand the sheer stress produced by medium perfusion and this provides the flexibility for multi-organ systems integration [122]. All of these technological advancements may hopefully overcome the obstacle in producing uniform and fully functional hepatocytes for proper liver modeling.
Human non-parenchymal cells (NPC)
Similar to primary hepatocytes, there are also limited sources of fully functional liver NPC. Primary LSE cells, Kupffer cells and hepatic stellate cells can be isolated from human liver using several methods [65]. However, the sources of human NPC are very scarce. The LTCDS service is one of the few sources for primary human NPC. Alternatively, the individual components of the NPC can be obtained in cryopreserved form from several vendors. Some studies utilize common cell lines such as HUVEC [123] and U937 cells [72] as surrogate to LSE and Kupffer cells. However, it is important to note that the liver NPC have specialized morphology and exhibit functions distinct from general cellular types (e.g. endothelial cells and macrophages, etc.). Subsequently, it is important when recreating the liver ex vivo and/or in vivo to use appropriate phenotypic subset of cells.
Fenestration is a specific feature of primary human LSE cells when compared to most other endothelial cells. Fenestrae, with diameters ranging from 20–300nm, are small windows or pores through the cytoplasm of endothelial cells that provide the cancer cells with direct access to the underlying (limited) basement membrane and hepatocytes; this enables macromolecule trafficking. Many primary, immortalized, iPS cell-derived endothelial cells and human LSE cell lines including TMNK-1 and TRP3 cells express endothelial cell markers such as CD34 and von Willebrand factor (vWF) and exhibit normal endothelial cell functions such as uptake of acetylated-low density lipoprotein or soluble materials, responsiveness to vascular endothelial growth factor (VEGF) and FGF and formation of vascular tube-like structure on basement membrane gel but the ability to establish true fenestration awaits confirmation [124–126]. Primary LSE cells are highly fenestrated but tend to lose this feature within 1–2 days of in vitro culture [17]. Nevertheless, Huebert et al. were able to generate immortalized mouse LSE cells that retained some fenestrations (100–150 nm open holes) organized in sieve plates [127]. The relevance of this feature in modeling human liver, however, depends on the design of the liver systems. For example, PEARL perfusion liver system, specifically designed to mimic the liver sinusoid, is equipped with artificial porous endothelial-like barrier to continuously supply nutrients into 3D tissue aggregates and thus eliminates the need for fenestration in LSE cells [58]. However, this system not only introduces a non-biological foreign body, but also now lacks the active signaling from the LSE cells.
Kupffer cells, resident macrophages of the liver, are genetically different from the infiltrating monocyte-derived macrophages [128]. Additionally, there are different subsets of Kupffer cells, at least in mice, that show different functions. F4/80+/CD68+ Kupffer cells have potent phagocytic activity and reactive oxygen species production following LPS stimulation whereas F4/80+/CD11b+ Kupffer cells displayed cytokines producing phenotype following LPS treatment [129]. However as with general macrophages of the body, the Kupffer cells, can also be polarized into pro-inflammatory M1 and anti-inflammatory M2 macrophages that can differently affect tumor growth [130].
Lastly, there are several hepatic stellate cell lines that sufficiently replicate primary stellate cell functions [131]. LX-1 and LX-2 stellate cells, immortalized with SV40-Tag, display stellate cells characteristics that include the expression of stellate cell markers such as glial fibrillary acidic protein (GFAP), alpha-smooth muscle actin (αSMA), vimentin, the secretion of collagen in response to TGF-β1 stimulation and the ability to store and metabolize retinoic acid [132]. Similarly, TWNT-1 and HSC-T6 cells exhibit hepatic stellate cell functions and express stellate cell markers [133,134]. All cell lines, however, are in the activated state which is marked by increased level of αSMA expression and collagen secretion, a component extracellular matrix that has been shown to promote fibrosis and hepatocellular carcinogenesis [135]. Activation of stellate, caused by the stiff culture condition [136] and the isolation process that simulates liver injury, could be reversed or prevented by culturing the cells on laminin-coated surface [137] or soft culture surface [138]. Thus, proper cultivation and maintenance of these cells such as use of soft scaffold and suitable coating matrix are essential to limit and prevent hepatic stellate activation that can affect the cancer cells in the liver model.
In summary, primary hepatocytes, either fresh or cryopreserved, remain as the ‘gold standard’ in modeling the liver. iPS cell-derived hepatocytes are still a promising alternative to primary hepatocytes but require substantial breakthroughs to overcome major hurdles in this fast-paced but relatively young field. Some of the currently available non-parenchymal cell lines and primary cells may not fully recapitulate in vivo liver resident cells and conventional cell culture techniques could result in artefactual changes in these cells. Appropriate sources of liver resident cells and proper culture techniques and conditions, as developed in some of the 3D liver systems, is crucial to closely replicate liver functions and retain the specialized features of the NPC.
CHALLENGES TOWARDS PERSONALIZED MEDICINE
Personalized or precision medicine utilizes knowledge about an individual’s genes, proteins, and environment to determine the most appropriate and efficacious therapeutic regime. The United States recently announced a national Precision Medicine Initiative that aims, among other things, to improve cancer therapies and generate large-scale biomedical databases that can aid in the assessment of disease risks, mechanisms and optimal therapies [139]. Individual modeling of the liver for metastasis study and therapy might not be a feasible approach at the moment but a similar strategy of stratifying breast cancer patients into subgroups might not be so improbable. The iPS cells, due to their plasticity property, are a perfect tool to model the liver with different genetic backgrounds. However, as discussed in previous section, there are still several improvements required before these liver models can yield meaningful clinical benefits. In addition to the issues with the sources of the liver resident cells, there are also several other challenges in modeling human liver that will be discussed in the next section.
Integration of parenchymal and NPC liver cells into 3D platforms
First of all, as previously mentioned, there are several organotypic liver models currently available. These models, though differently constructed, generally have been shown to improve cellular longevity, basic liver functions and metabolic functions [57,58]. However, one of the challenging tasks in modeling the liver is recapitulating the histologic structure of liver sinusoid, a key functional unit and site of metastatic seeding. For example, a recent publication on liver inflammation demonstrates that the Kupffer and LSE cells, due to their strategic position in the sinusoid, initiate inflammatory reactions upon stimulation [140]. The stellate cells, which are primarily found in the space of Disse, regulate the inflammatory reactions from the sinusoid to the hepatocytes through prostaglandin D2 receptor, DP1, to reduce hepatitis and improve microcirculation [140]. In liver metastases, the LSE cells are the first point of contact with the cancer cells and play important roles in facilitating cancer cells adhesion and recruitment [18]. Within hours of entry into the space of Disse, the growth of these cancer cells is suppressed by the Kupffer cells that can bind to the tumor cells to phagocytose the cells or induce apoptosis [141]. The surviving cancer cells, typically in mesenchymal-like phenotype, are nurtured by the hepatocytes to undergo phenotypic reversion into epithelial phenotype for successful seeding into the liver [142,143]. These cells can also collectively suppress, maintain and promote tumor growth by secreting various factors. These sequential events may be technically difficult to replicate and further works are required to determine the relevance of mimicking the specific cellular arrangement. Nonetheless, sequential addition of cells in the SQL-SAL platform allows for the separation of hepatocytes layer from the LSE-stellate cells layer, replicating the space of Disse and the liver tissue structure in vivo [72]. Importantly, this platform maintains hepatic functions and sufficiently models liver fibrosis after 30 days of culture [72].
Utilizing iPS cell-derived hepatocytes for personalize medicine further complicates ex vivo liver modeling. Our preliminary study using iPS cell-derived hepatocytes in the 3D ex vivo hepatic MPS showed that although the cells could be well-differentiated towards hepatocyte-like cells in vitro, they still could not properly form cell-to-cell contact and maintain 3D-tissue structure in the 3D ex vivo hepatic MPS. This could be due to lack of cadherin junctions and although the cells displayed some hepatic functions, they were still immature and behave like iPS cells that are sensitive to enzymatic digestion into single cells prior to seeding into the scaffold. In a different liver system, fibroblasts could prolong the morphology and functions of commercially available iPS cell-derived hepatocytes for four weeks on micro patterned co-culture (MCPP) platform [113]. Monolayer seeding of hepatocytes, prior to 3D tissue construction using other cell types, also permits the utility of iPS-derived hepatocytes in the SQL-SAL platform [72]. Thus, the applicability of iPS cell-derived hepatocytes in modeling the liver may be dependent on the design of the systems.
Universal medium to support the viability and functionality of liver resident cells
Another important aspect of modeling the liver in 3D ex vivo culture is common medium required to maintain all the different types of cells in liver microenvironment. An ideal universal medium should support long-term survival of the heterogeneous population of cells at physiologic concentration and does not artificially affect the hepatocytes. Our lab has developed phenol red- and bovine serum albumin-free physiologic medium (PHM) supplemented with physiologic concentration of hydrocortisone, glucose, and insulin that can support the viability of fresh and cryopreserved primary hepatocytes and NPC. However, since the PHM lacks some of the factors that are required to induce or maintain the differentiation status of iPS-derived hepatocytes such as Oncostatin M and HGF [144,145], PHM may not be able to sustain the viability and the maturation status of iPS cell-derived hepatocytes. This dependency could be mitigated by co-culturing the iPS cells with other stromal cells that will supply these various factors at cellular physiologic concentration.
Use of immortalized cancer cell lines for drug development
Immortalized cancer cell lines are important tools for research in a variety of fields. These lines are widely available, thus allowing for validation of research findings from various different laboratories. Importantly, these cell lines are relatively homogeneous and have been extensively characterized and verified to replicate numerous of human cancers and associated subtypes [146–148]. However, these cells are susceptible to genetic drift after continuous in vitro culture [149]. In fact, a comparison of breast cancer cell lines that were independently cultured in different laboratories showed aberrant and distinct karyotype profiles [150,151]. In prostate cancer, three subpopulations of DU145 with varying phenotype and E-cadherin expression have been identified [152]. The more epithelial sublines were found to have augmented capacity to form bone metastases in xenograft model whereas the low E-cadherin mesenchymal-like subline could engraft the mice [152]. This might be due to heterogeneity within the cancer cell lines that mimic clinical observation but could affect the results and validation by other groups if this issue is not addressed and analyzed properly. Similarly, our lab also found different phenotypes of DU145 cells. The identities of these cells were validated but showed contradicting E-cadherin expression where DU145 cells with high E-cadherin expression displayed intense heterotypic membranous E-cadherin expression in the liver that protected the cells against chemo drugs-induced apoptosis [11]. Genetic comparison between 47 ovarian cancer cell lines and high grade serous ovarian cancer (HGSOC) revealed that the cell lines overall showed genomic similarity with the HGSOC tissues but closer inspection uncovered five “hypermutated” cell lines including commonly used IGROV1 cells [148]. In addition, cell line models for ovarian cancer, glioblastoma, colorectal cancer and metastatic melanoma failed to mimic the gene expression pattern of multi-drug receptor of primary cultures [153].
Most of the cell lines were derived from metastatic sites and require little or no genetic manipulation for immortalization [147,149]. Careful comparison between cell lines and primary cultures of primary or metastatic tumors should be made for correct conclusions. For example, mammospheres from breast cancer cell lines were tumorigenic but not mammopsheres from primary cultures of fresh invasive ductal carcinoma [154]. This is unexpected because the tumorigenic potential of these two sources are different since the cell lines, MCF-7 and MDA-MB-231, were both derived from metastatic sites whereas the primary cultures were established from primary cancer [154]. Similarly, CpG island methylation was higher in cancer cell lines T47D, MDA-MB-435 and HS578T when compared to 14 breast primary cancer cells [155]. This comparison, performed to determine if the lines represent their primary malignancies, is inappropriate since the first two cell lines are metastatic cells derived from pleural effusion (T47D: breast cancer and MB-435: melanoma). However, the findings with other cancer types namely lung, colon, head and neck, glioblastoma, acute myeloid leukemia, medullablastoma, and testicular germ cell cancer support the conclusion that most cell lines poorly represent their primary malignancies, at the context of CpG island methylation [155].
Thus, true personalization of the liver metastases would require both primary and metastatic tumor specimens. This is challenging since most tumors, available as excess pathologic specimens, are from therapeutic interventions of a primary lesion; metastatic lesions are rarely excised or if sampled, are usually small biopsies or FNA yielding few excess cells (secondary debulking of ovarian carcinoma may represent a specialized and non-generalizable situation). Both genetic tracing and phenotypic analyses demonstrate that tumor cells in the metastatic environment are qualitatively different from those in the primary site, especially in regards to responsiveness to therapy. One potential source of such metastatic cells would be the therapeutic resections of liver metastases [34,156]. These would be limited in source and may not be representative of metastatic cells in other organs. Still, demonstration of theranostics value of such cultures may then allow for sampling of more metastatic lesions as clinically indicated to develop and guide therapies.
CONCLUSIONS
Although significant advances in modeling human liver have been achieved, there are several aspects that require further improvement. Current capabilities allow for all human-derived mini-organs that approach the patient situation for liver metastases. However, as the cells and tumors are derived distinctly, they are not fully personalized as to the involved patient, and thus are better suited for discovery of tumor biology and general drug development. To truly bring this promise to afflicted persons, iPS cell-derived hepatocytes retain promise as viable alternatives to primary human hepatocytes but require major breakthroughs to efficiently and sufficiently generate fully mature hepatocytes in vitro.
Acknowledgments
We thank members of the Wells lab, and Drs. Linda Griffith (MIT), Raman Venkataramanan (UPitt), Donna Stolz (UPitt), Alex Soto-Gutierrez (UPitt) and Ira Fox (UPitt) and their teams for ideas and excellent suggestions. The studies that underlie these perspectives were supported by grants from NIH/NCATS/NCI and the VA Merit Award program.
References
- 1.Chaffer CL, Weinberg RA. A Perspective on Cancer Cell Metastasis. Science. 2011;331:1559–64. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 3.Weidner N, Semple JP, Welch WR, Folkman J. Tumor angiogenesis and metastasis--correlation in invasive breast carcinoma. N Engl J Med. 1991;324:1–8. doi: 10.1056/NEJM199101033240101. [DOI] [PubMed] [Google Scholar]
- 4.Zhou BB, Zhang H, Damelin M, Geles KG, Grindley JC, Dirks PB. Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nat Rev Drug Discov. 2009;8:806–23. doi: 10.1038/nrd2137. [DOI] [PubMed] [Google Scholar]
- 5.Liu HP, Patel MR, Prescher JA, et al. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc Natl Acad Sci U S A. 2010;107:18115–20. doi: 10.1073/pnas.1006732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng X, Carstens JL, Kim J, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–30. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mogrovejo E, Manickam P, Amin M, Cappell MS. Characterization of the syndrome of acute liver failure caused by metastases from breast carcinoma. Dig Dis Sci. 2014;59:724–36. doi: 10.1007/s10620-013-2943-z. [DOI] [PubMed] [Google Scholar]
- 8.Lee SY, Haq F, Kim D, et al. Comparative genomic analysis of primary and synchronous metastatic colorectal cancers. PLoS One. 2014;9:e90459. doi: 10.1371/journal.pone.0090459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gunasinghe NPAD, Wells A, Thompson EW, Hugo HJ. Mesenchymal–epithelial transition (MET) as a mechanism for metastatic colonisation in breast cancer. Cancer Metastasis Rev. 2012;31:469–78. doi: 10.1007/s10555-012-9377-5. [DOI] [PubMed] [Google Scholar]
- 10.Chao Y, Wu Q, Shepard C, Wells A. Hepatocyte induced re-expression of E-cadherin in breast and prostate cancer cells increases chemoresistance. Clin Exp Metastasis. 2011;29:39–50. doi: 10.1007/s10585-011-9427-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma B, Wheeler SE, Clark AM, Whaley DL, Yang M, Wells A. Liver protects metastatic prostate cancer from induced death by activating E-cadherin signaling. Hepatology. 2016;64:1725–42. doi: 10.1002/hep.28755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meads MB, Hazlehurst LA, Dalton WS. The bone marrow microenvironment as a tumor sanctuary and contributor to drug resistance. Clin Cancer Res. 2008;14:2519–26. doi: 10.1158/1078-0432.CCR-07-2223. [DOI] [PubMed] [Google Scholar]
- 13.Minami Y, Kudo M. Hepatic malignancies: Correlation between sonographic findings and pathological features. World J Radiol. 2010;2:249–56. doi: 10.4329/wjr.v2.i7.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tabariès S, Siegel P. Breast Cancer Liver Metastasis. In: Brodt P, editor. Liver Metastasis: Biology and Clinical Management. Springer Science; 2011. pp. 273–306. [Google Scholar]
- 15.Bubendorf L, Schöpfer A, Wagner U, et al. Metastatic patterns of prostate cancer: An autopsy study of 1,589 patients. Hum Pathol. 2000;31:578–83. doi: 10.1053/hp.2000.6698. [DOI] [PubMed] [Google Scholar]
- 16.Vidal-Vanaclocha F. Architectural and Functional Aspects of the Liver with Implications for Cancer Metastasis. In: Brodt P, editor. Liver Metastasis: Biology and Clinical Management. New York: Springer; 2011. pp. 9–42. [Google Scholar]
- 17.Elvevold K, Smedsrod B, Martinez I. The liver sinusoidal endothelial cell: a cell type of controversial and confusing identity. Am J Physiol Gastrointest Liver Physiol. 2008;294:G391–400. doi: 10.1152/ajpgi.00167.2007. [DOI] [PubMed] [Google Scholar]
- 18.Porquet N, Huot J. Signal Transduction Tumor-Endothelial Cell Communication. In: Brodt P, editor. Liver Metastasis: Biology and Clinical Management. New York: Springer; 2011. pp. 187–212. [Google Scholar]
- 19.Vandamme TF. Use of rodents as models of human diseases. J Pharm Bioallied Sci. 2014;6:2–9. doi: 10.4103/0975-7406.124301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheung C, Gonzalez FJ. Humanized mouse lines and their application for prediction of human drug metabolism and toxicological risk assessment. J Pharmacol Exp Ther. 2008;327:288–99. doi: 10.1124/jpet.108.141242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubin LL. Stem Cells and Drug Discovery: The Beginning of a New Era? Cell. 2008;132:549–52. doi: 10.1016/j.cell.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 22.Moreno L, Pearson ADJ. How can attrition rates be reduced in cancer drug discovery? Expert Opinion on Drug Discovery. 2013;8:363–8. doi: 10.1517/17460441.2013.768984. [DOI] [PubMed] [Google Scholar]
- 23.DiMasi JA, Grabowski HG, Hansen RW. Innovation in the pharmaceutical industry: New estimates of R&D costs. J Health Econ. 2016;47:20–33. doi: 10.1016/j.jhealeco.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 24.Turdean S, Gurzu S, Turcu M, Voidazan S, Anca S. Liver Metastases: Incidence and Clinicopathological Data. Acta Medica Marisiensis. 2012;58:5. [Google Scholar]
- 25.Misiakos EP, Karidis NP, Kouraklis G. Current treatment for colorectal liver metastases. World J Gastroenterol. 2011;17:4067–75. doi: 10.3748/wjg.v17.i36.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pouessel D, Gallet B, Bibeau F, et al. Liver metastases in prostate carcinoma: clinical characteristics and outcome. BJU Int. 2007;99:807–11. doi: 10.1111/j.1464-410X.2006.06663.x. [DOI] [PubMed] [Google Scholar]
- 27.de la Monte SM, Moore GW, Hutchins GM. Metastatic behavior of prostate cancer. Cluster analysis of patterns with respect to estrogen treatment. Cancer. 1986;58:985–93. doi: 10.1002/1097-0142(19860815)58:4<985::aid-cncr2820580432>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 28.Lee Y-TM. Patterns of metastasis and natural courses of breast carcinoma. Cancer Metastasis Rev. 1985;4:153–72. doi: 10.1007/BF00050693. [DOI] [PubMed] [Google Scholar]
- 29.Cummings MC, Simpson PT, Reid LE, et al. Metastatic progression of breast cancer: insights from 50 years of autopsies. J Pathol. 2014;232:23–31. doi: 10.1002/path.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ambrosetti MC, Zamboni GA, Mucelli RP. Distribution of liver metastases based on the site of primary pancreatic carcinoma. Eur Radiol. 2016;26:306–10. doi: 10.1007/s00330-015-3843-8. [DOI] [PubMed] [Google Scholar]
- 31.Konopke R, Distler M, Ludwig S, Kersting S. Location of liver metastases reflects the site of the primary colorectal carcinoma. Scand J Gastroenterol. 2008;43:192–5. doi: 10.1080/00365520701677755. [DOI] [PubMed] [Google Scholar]
- 32.Sommer WH, Zech CJ, Bamberg F, et al. Fluid-fluid level in hepatic metastases: a characteristic sign of metastases of neuroendocrine origin. Eur J Radiol. 2012;81:2127–32. doi: 10.1016/j.ejrad.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 33.DeGreef K, Rolfo C, Russo A, et al. Multisciplinary management of patients with liver metastasis from colorectal cancer. World Journal of Gastroenterology. 2016;22:7215–25. doi: 10.3748/wjg.v22.i32.7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weinrich M, Weiss C, Schuld J, Rau BM. Liver resections of isolated liver metastasis in breast cancer: results and possible prognostic factors. HPB Surg. 2014;2014:893829. doi: 10.1155/2014/893829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guy J, Peters MG. Liver disease in women: the influence of gender on epidemiology, natural history, and patient outcomes. Gastroenterol Hepatol (N Y) 2013;9:633–9. [PMC free article] [PubMed] [Google Scholar]
- 36.Wolbold R, Klein K, Burk O, et al. Sex is a major determinant of CYP3A4 expression in human liver. Hepatology. 2003;38:978–88. doi: 10.1053/jhep.2003.50393. [DOI] [PubMed] [Google Scholar]
- 37.Mennecozzi M, Landesmann B, Palosaari T, Harris G, Whelan M. Sex differences in liver toxicity-do female and male human primary hepatocytes react differently to toxicants in vitro? PLoS One. 2015;10:e0122786. doi: 10.1371/journal.pone.0122786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rando G, Wahli W. Sex differences in nuclear receptor-regulated liver metabolic pathways. Biochim Biophys Acta. 2011;1812:964–73. doi: 10.1016/j.bbadis.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 39.Naugler WE, Sakurai T, Kim S, et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–4. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 40.Ma WL, Hsu CL, Wu MH, et al. Androgen receptor is a new potential therapeutic target for the treatment of hepatocellular carcinoma. Gastroenterology. 2008;135:947–55. 55 e1–5. doi: 10.1053/j.gastro.2008.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z, Tuteja G, Schug J, Kaestner KH. Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell. 2012;148:72–83. doi: 10.1016/j.cell.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoyer M, Erichsen R, Gandrup P, Norgaard M, Jacobsen JB. Survival in patients with synchronous liver metastases in central and northern Denmark, 1998 to 2009. Clin Epidemiol. 2011;3(Suppl 1):11–7. doi: 10.2147/CLEP.S20613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, Bouvier AM. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg. 2006;244:254–9. doi: 10.1097/01.sla.0000217629.94941.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao JC, Tseng JF, Worah S, et al. Clinicopathologic behavior of gastric adenocarcinoma in Hispanic patients: analysis of a single institution's experience over 15 years. J Clin Oncol. 2005;23:3094–103. doi: 10.1200/JCO.2005.08.987. [DOI] [PubMed] [Google Scholar]
- 45.Mazzaglia PJ, Berber E, Milas M, Siperstein AE. Laparoscopic radiofrequency ablation of neuroendocrine liver metastases: a 10-year experience evaluating predictors of survival. Surgery. 2007;142:10–9. doi: 10.1016/j.surg.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 46.Ward E, Jemal A, Cokkinides V, et al. Cancer disparities by race/ethnicity and socioeconomic status. CA Cancer J Clin. 2004;54:78–93. doi: 10.3322/canjclin.54.2.78. [DOI] [PubMed] [Google Scholar]
- 47.Sloane D, Chen H, Howell C. Racial disparity in primary hepatocellular carcinoma: tumor stage at presentation, surgical treatment and survival. J Natl Med Assoc. 2006;98:1934–9. [PMC free article] [PubMed] [Google Scholar]
- 48.Jennis M, Kung CP, Basu S, et al. An African-specific polymorphism in the TP53 gene impairs p53 tumor suppressor function in a mouse model. Genes Dev. 2016;30:918–30. doi: 10.1101/gad.275891.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiu M, Hu J, Yang D, Cosgrove DP, Xu R. Pattern of distant metastases in colorectal cancer: a SEER based study. Oncotarget. 2015;6:38658–66. doi: 10.18632/oncotarget.6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 51.Morris GJ, Naidu S, Topham AK, et al. Differences in breast carcinoma characteristics in newly diagnosed African-American and Caucasian patients: a single-institution compilation compared with the National Cancer Institute's Surveillance, Epidemiology, and End Results database. Cancer. 2007;110:876–84. doi: 10.1002/cncr.22836. [DOI] [PubMed] [Google Scholar]
- 52.Yuan N, Meng M, Liu C, et al. Clinical characteristics and prognostic analysis of triple-negative breast cancer patients. Mol Clin Oncol. 2014;2:245–51. doi: 10.3892/mco.2013.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Francia G, Cruz-Munoz W, Man S, Xu P, Kerbel RS. Mouse models of advanced spontaneous metastasis for experimental therapeutics. Nat Rev Cancer. 2011;11:135–41. doi: 10.1038/nrc3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sosef MN, Baust JM, Sugimachi K, Fowler A, Tompkins RG, Toner M. Cryopreservation of isolated primary rat hepatocytes: enhanced survival and long-term hepatospecific function. Ann Surg. 2005;241:125–33. doi: 10.1097/01.sla.0000149303.48692.0f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gul N, Grewal S, Bogels M, et al. Macrophages mediate colon carcinoma cell adhesion in the rat liver after exposure to lipopolysaccharide. Oncoimmunology. 2012;1:1517–26. doi: 10.4161/onci.22303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Griffith LG, Wells A, Stolz DB. Engineering Liver. Hepatology. 2014:1426–34. doi: 10.1002/hep.27150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clark AM, Ma B, Taylor DL, Griffith L, Wells A. Liver metastases: Microenvironments and ex-vivo models. Exp Biol Med. 2016;241:1639–52. doi: 10.1177/1535370216658144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.LeCluyse EL, Witek RP, Andersen ME, Powers MJ. Organotypic liver culture models: meeting current challenges in toxicity testing. Crit Rev Toxicol. 2012;42:501–48. doi: 10.3109/10408444.2012.682115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bavli D, Prill S, Ezra E, et al. Real-time monitoring of metabolic function in liver-on-chip microdevices tracks the dynamics of mitochondrial dysfunction. Proc Natl Acad Sci U S A. 2016;113:E2231–40. doi: 10.1073/pnas.1522556113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Skardal A, Devarasetty M, Forsythe S, Atala A, Soker S. A reductionist metastasis- on-a-chip platform for in vitro tumor progression modeling and drug screening. Biotechnol Bioeng. 2016;113:2020–32. doi: 10.1002/bit.25950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee H, Cho DW. One-step fabrication of an organ-on-a-chip with spatial heterogeneity using a 3D bioprinting technology. Lab Chip. 2016;16:2618–25. doi: 10.1039/c6lc00450d. [DOI] [PubMed] [Google Scholar]
- 62.Ma XY, Qu X, Zhu W, et al. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc Natl Acad Sci U S A. 2016;113:2206–11. doi: 10.1073/pnas.1524510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen DG, Funk J, Robbins JB, et al. Bioprinted 3D Primary Liver Tissues Allow Assessment of Organ-Level Response to Clinical Drug Induced Toxicity In Vitro. Plos One. 2016:11. doi: 10.1371/journal.pone.0158674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robbins JB, Gorgen V, Min P, Shepherd BR, Presnell SC. A novel in vitro three- dimensional bioprinted liver tissue system for drug development. FASEB J. 2013:27. [Google Scholar]
- 65.Damm G, Pfeiffer E, Burkhardt B, Vermehren J, Nussler AK, Weiss TS. Human parenchymal and non-parenchymal liver cell isolation, culture and characterization. Hepatol Int. 2013;7:951–8. doi: 10.1007/s12072-013-9475-7. [DOI] [PubMed] [Google Scholar]
- 66.Xiao W, Perry G, Komori K, Sakai Y. New physiologically-relevant liver tissue model based on hierarchically cocultured primary rat hepatocytes with liver endothelial cells. Integr Biol (Camb) 2015;7:1412–22. doi: 10.1039/c5ib00170f. [DOI] [PubMed] [Google Scholar]
- 67.Genove E, Schmitmeier S, Sala A, et al. Functionalized self-assembling peptide hydrogel enhance maintenance of hepatocyte activity in vitro. J Cell Mol Med. 2009;13:3387–97. doi: 10.1111/j.1582-4934.2009.00970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dash A, Simmers MB, Deering TG, et al. Hemodynamic flow improves rat hepatocyte morphology, function, and metabolic activity in vitro. Am J Physiol Cell Physiol. 2013;304:C1053–63. doi: 10.1152/ajpcell.00331.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wheeler SE, Clark AM, Taylor DP, et al. Spontaneous dormancy of metastatic breast cancer cells in an all human liver microphysiologic system. Br J Cancer. 2014 doi: 10.1038/bjc.2014.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Domansky K, Inman W, Serdy J, Dash A, Lim MH, Griffith LG. Perfused multiwell plate for 3D liver tissue engineering. Lab Chip. 2010;10:51–8. doi: 10.1039/b913221j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tsamandouras N, Kostrzewski T, Stokes CL, Griffith LG, Hughes DJ, Cirit M. Quantitative Assessment of Population Variability in Hepatic Drug Metabolism Using a Perfused Three-Dimensional Human Liver Microphysiological System. J Pharmacol Exp Ther. 2017;360:95–105. doi: 10.1124/jpet.116.237495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vernetti LA, Senutovitch N, Boltz R, et al. A human liver microphysiology platform for investigating physiology, drug safety, and disease models. Exp Biol Med (Maywood) 2016;241:101–14. doi: 10.1177/1535370215592121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Clark AM, Wheeler SE, Young CL, et al. A liver microphysiological system of tumor cell dormancy and inflammatory responsiveness is affected by scaffold properties. Lab Chip. 2016;17:156–68. doi: 10.1039/c6lc01171c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bhogal RH, Hodson J, Bartlett DC, et al. Isolation of primary human hepatocytes from normal and diseased liver tissue: a one hundred liver experience. PLoS One. 2011;6:e18222. doi: 10.1371/journal.pone.0018222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vondran FW, Katenz E, Schwartlander R, et al. Isolation of primary human hepatocytes after partial hepatectomy: criteria for identification of the most promising liver specimen. Artif Organs. 2008;32:205–13. doi: 10.1111/j.1525-1594.2007.00524.x. [DOI] [PubMed] [Google Scholar]
- 76.McGinnity DF, Soars MG, Urbanowicz RA, Riley RJ. Evaluation of fresh and cryopreserved hepatocytes as in vitro drug metabolism tools for the prediction of metabolic clearance. Drug Metab Dispos. 2004;32:1247–53. doi: 10.1124/dmd.104.000026. [DOI] [PubMed] [Google Scholar]
- 77.Li AP, Lu C, Brent JA, et al. Cryopreserved human hepatocytes: characterization of drug-metabolizing enzyme activities and applications in higher throughput screening assays for hepatotoxicity, metabolic stability, and drug-drug interaction potential. Chem Biol Interact. 1999;121:17–35. doi: 10.1016/s0009-2797(99)00088-5. [DOI] [PubMed] [Google Scholar]
- 78.Sarkar U, Rivera-Burgos D, Large EM, et al. Metabolite profiling and pharmacokinetic evaluation of hydrocortisone in a perfused three-dimensional human liver bioreactor. Drug Metab Dispos. 2015;43:1091–9. doi: 10.1124/dmd.115.063495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Terry C, Dhawan A, Mitry RR, Lehec SC, Hughes RD. Preincubation of rat and human hepatocytes with cytoprotectants prior to cryopreservation can improve viability and function upon thawing. Liver Transpl. 2005;11:1533–40. doi: 10.1002/lt.20503. [DOI] [PubMed] [Google Scholar]
- 80.Ramboer E, De Craene B, De Kock J, et al. Strategies for immortalization of primary hepatocytes. J Hepatol. 2014;61:925–43. doi: 10.1016/j.jhep.2014.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Waki K, Anno K, Ono T, Ide T, Chayama K, Tahara H. Establishment of functional telomerase immortalized human hepatocytes and a hepatic stellate cell line for telomere-targeting anticancer drug development. Cancer Sci. 2010;101:1678–85. doi: 10.1111/j.1349-7006.2010.01576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gripon P, Rumin S, Urban S, et al. Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci U S A. 2002;99:15655–60. doi: 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marion M, Hantz O, Durantel D. The HepaRG Cell Line: Biological Properties and Relevance as a Tool for Cell Biology, Drug Metabolism, and Virology Studies. In: Maurel P, editor. Hepatocytes. Springer Science+Business Media; 2010. pp. 261–72. [DOI] [PubMed] [Google Scholar]
- 84.Lubberstedt M, Muller-Vieira U, Mayer M, et al. HepaRG human hepatic cell line utility as a surrogate for primary human hepatocytes in drug metabolism assessment in vitro. J Pharmacol Toxicol Methods. 2011;63:59–68. doi: 10.1016/j.vascn.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 85.Wilkening S, Stahl F, Bader A. Comparison of primary human hepatocytes and hepatoma cell line Hepg2 with regard to their biotransformation properties. Drug Metab Dispos. 2003;31:1035–42. doi: 10.1124/dmd.31.8.1035. [DOI] [PubMed] [Google Scholar]
- 86.Lin J, Schyschka L, Muhl-Benninghaus R, et al. Comparative analysis of phase I and II enzyme activities in 5 hepatic cell lines identifies Huh-7 and HCC-T cells with the highest potential to study drug metabolism. Arch Toxicol. 2012;86:87–95. doi: 10.1007/s00204-011-0733-y. [DOI] [PubMed] [Google Scholar]
- 87.Deurholt T, van Til NP, Chhatta AA, et al. Novel immortalized human fetal liver cell line, cBAL111, has the potential to differentiate into functional hepatocytes. BMC Biotechnol. 2009;9:89. doi: 10.1186/1472-6750-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 89.Takebe T, Sekine K, Enomura M, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–4. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 90.Zhao XY, Li W, Lv Z, et al. iPS cells produce viable mice through tetraploid complementation. Nature. 2009;461:86–90. doi: 10.1038/nature08267. [DOI] [PubMed] [Google Scholar]
- 91.Yu Y, Liu HL, Ikeda Y, et al. Hepatocyte-like cells differentiated from human induced pluripotent stem cells: Relevance to cellular therapies. Stem Cell Research. 2012;9:196–207. doi: 10.1016/j.scr.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schwartz RE, Fleming HE, Khetani SR, Bhatia SN. Pluripotent stem cell-derived hepatocyte-like cells. Biotechnology Advances. 2014;32:504–13. doi: 10.1016/j.biotechadv.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gerbal-Chaloin S, Funakoshi N, Caillaud A, Gondeau C, Champon B, Si-Tayeb K. Human Induced Pluripotent Stem Cells in Hepatology Beyond the Proof of Concept. Am J Pathol. 2014;184:332–47. doi: 10.1016/j.ajpath.2013.09.026. [DOI] [PubMed] [Google Scholar]
- 94.Kim JS, Choi HW, Choi S, Do JT. Reprogrammed pluripotent stem cells from somatic cells. Int J Stem Cells. 2011;4:1–8. doi: 10.15283/ijsc.2011.4.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Carey BW, Markoulaki S, Hanna JH, et al. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell. 2011;9:588–98. doi: 10.1016/j.stem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 96.Tiemann U, Sgodda M, Warlich E, et al. Optimal reprogramming factor stoichiometry increases colony numbers and affects molecular characteristics of murine induced pluripotent stem cells. Cytometry A. 2011;79:426–35. doi: 10.1002/cyto.a.21072. [DOI] [PubMed] [Google Scholar]
- 97.Yamaguchi S, Hirano K, Nagata S, Tada T. Sox2 expression effects on direct reprogramming efficiency as determined by alternative somatic cell fate. Stem Cell Research. 2011;6:177–86. doi: 10.1016/j.scr.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 98.Kang L, Wang J, Zhang Y, Kou Z, Gao S. iPS cells can support full-term development of tetraploid blastocyst-complemented embryos. Cell Stem Cell. 2009;5:135–8. doi: 10.1016/j.stem.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 99.Takahashi K, Yamanaka S. A developmental framework for induced pluripotency. Development. 2015;142:3274–85. doi: 10.1242/dev.114249. [DOI] [PubMed] [Google Scholar]
- 100.Ohi Y, Qin H, Hong C, et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol. 2011;13:541–9. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Basma H, Soto–Gutiérrez A, Yannam GR, et al. Differentiation and Transplantation of Human Embryonic Stem Cell–Derived Hepatocytes. Gastroenterology. 2009;136:990–9.e4. doi: 10.1053/j.gastro.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Soto-Gutierrez A, Navarro-Alvarez N, Zhao D, et al. Differentiation of mouse embryonic stem cells to hepatocyte-like cells by co-culture with human liver nonparenchymal cell lines. Nat Protoc. 2007;2:347–56. doi: 10.1038/nprot.2007.18. [DOI] [PubMed] [Google Scholar]
- 103.Si-Tayeb K, Noto FK, Nagaoka M, et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Espejel S, Roll GR, McLaughlin KJ, et al. Induced pluripotent stem cell-derived hepatocytes have the functional and proliferative capabilities needed for liver regeneration in mice. J Clin Invest. 2010;120:3120–6. doi: 10.1172/JCI43267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Touboul T, Hannan NR, Corbineau S, et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology. 2010;51:1754–65. doi: 10.1002/hep.23506. [DOI] [PubMed] [Google Scholar]
- 106.Park HJ, Choi YJ, Kim JW, et al. Differences in the Epigenetic Regulation of Cytochrome P450 Genes between Human Embryonic Stem Cell-Derived Hepatocytes and Primary Hepatocytes. PLoS One. 2015;10:e0132992. doi: 10.1371/journal.pone.0132992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Han S, Bourdon A, Hamou W, Dziedzic N, Goldman O, Gouon-Evans V. Generation of functional hepatic cells from pluripotent stem cells. J Stem Cell Res Ther. 2012;(Suppl 10):1–7. doi: 10.4172/2157-7633.S10-008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sullivan GJ, Hay DC, Park IH, et al. Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology. 2010;51:329–35. doi: 10.1002/hep.23335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rashid ST, Corbineau S, Hannan N, et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J Clin Invest. 2010;120:3127–36. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Teo AK, Valdez IA, Dirice E, Kulkarni RN. Comparable generation of activin-induced definitive endoderm via additive Wnt or BMP signaling in absence of serum. Stem Cell Reports. 2014;3:5–14. doi: 10.1016/j.stemcr.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shan J, Schwartz RE, Ross NT, et al. Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat Chem Biol. 2013;9:514–20. doi: 10.1038/nchembio.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zorn AM. Liver Development. Community TSCR, editor. StemBook. 2008 doi: 10.3824/stembook.1.52.1. at http://www.stembook.org. [DOI]
- 113.Berger DR, Ware BR, Davidson MD, Allsup SR, Khetani SR. Enhancing the functional maturity of induced pluripotent stem cell-derived human hepatocytes by controlled presentation of cell-cell interactions in vitro. Hepatology. 2015;61:1370–81. doi: 10.1002/hep.27621. [DOI] [PubMed] [Google Scholar]
- 114.Du C, Narayanan K, Leong MF, Wan AC. Induced pluripotent stem cell-derived hepatocytes and endothelial cells in multi-component hydrogel fibers for liver tissue engineering. Biomaterials. 2014;35:6006–14. doi: 10.1016/j.biomaterials.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 115.Chen Y, Li Y, Wang X, et al. Amelioration of Hyperbilirubinemia in Gunn Rats after Transplantation of Human Induced Pluripotent Stem Cell-Derived Hepatocytes. Stem Cell Reports. 2015;5:22–30. doi: 10.1016/j.stemcr.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hannan NR, Segeritz CP, Touboul T, Vallier L. Production of hepatocyte-like cells from human pluripotent stem cells. Nat Protoc. 2013;8:430–7. doi: 10.1038/nprot.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Labrune P, Benattar C, Ammoury N, Chalas J, Lindenbaum A, Odievre M. Serum concentrations of albumin, C-reactive protein, alpha 2-macroglobulin, prealbumin, fibronectin, fibrinogen, transferrin, and retinol binding protein in 55 patients with hepatic glycogen storage diseases. J Pediatr Gastroenterol Nutr. 1994;18:41–4. doi: 10.1097/00005176-199401000-00007. [DOI] [PubMed] [Google Scholar]
- 118.Chen YF, Tseng CY, Wang HW, Kuo HC, Yang VW, Lee OK. Rapid generation of mature hepatocyte-like cells from human induced pluripotent stem cells by an efficient three-step protocol. Hepatology. 2012;55:1193–203. doi: 10.1002/hep.24790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Song W, Lu YC, Frankel AS, An D, Schwartz RE, Ma M. Engraftment of human induced pluripotent stem cell-derived hepatocytes in immunocompetent mice via 3D co-aggregation and encapsulation. Sci Rep. 2015;5:16884. doi: 10.1038/srep16884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gieseck RL, 3rd, Hannan NR, Bort R, et al. Maturation of induced pluripotent stem cell derived hepatocytes by 3D-culture. PLoS One. 2014;9:e86372. doi: 10.1371/journal.pone.0086372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Faulkner-Jones A, Fyfe C, Cornelissen DJ, et al. Bioprinting of human pluripotent stem cells and their directed differentiation into hepatocyte-like cells for the generation of mini-livers in 3D. Biofabrication. 2015;7:044102. doi: 10.1088/1758-5090/7/4/044102. [DOI] [PubMed] [Google Scholar]
- 122.Schepers A, Li C, Chhabra A, Seney BT, Bhatia S. Engineering a perfusable 3D human liver platform from iPS cells. Lab Chip. 2016;16:2644–53. doi: 10.1039/c6lc00598e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Groger M, Rennert K, Giszas B, et al. Monocyte-induced recovery of inflammation-associated hepatocellular dysfunction in a biochip-based human liver model. Sci Rep. 2016;6:21868. doi: 10.1038/srep21868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Matsumura T, Takesue M, Westerman KA, et al. Establishment of an immortalized human-liver endothelial cell line with SV40T and hTERT. Transplantation. 2004;77:1357–65. doi: 10.1097/01.tp.0000124286.82961.7e. [DOI] [PubMed] [Google Scholar]
- 125.Adams WJ, Zhang Y, Cloutier J, et al. Functional vascular endothelium derived from human induced pluripotent stem cells. Stem Cell Reports. 2013;1:105–13. doi: 10.1016/j.stemcr.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Parent R, Durantel D, Lahlali T, et al. An immortalized human liver endothelial sinusoidal cell line for the study of the pathobiology of the liver endothelium. Biochem Biophys Res Commun. 2014;450:7–12. doi: 10.1016/j.bbrc.2014.05.038. [DOI] [PubMed] [Google Scholar]
- 127.Huebert RC, Jagavelu K, Liebl AF, et al. Immortalized liver endothelial cells: a cell culture model for studies of motility and angiogenesis. Lab Invest. 2010;90:1770–81. doi: 10.1038/labinvest.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zigmond E, Samia-Grinberg S, Pasmanik-Chor M, et al. Infiltrating monocyte-derived macrophages and resident kupffer cells display different ontogeny and functions in acute liver injury. J Immunol. 2014;193:344–53. doi: 10.4049/jimmunol.1400574. [DOI] [PubMed] [Google Scholar]
- 129.Kinoshita M, Uchida T, Sato A, et al. Characterization of two F4/80-positive Kupffer cell subsets by their function and phenotype in mice. J Hepatol. 2010;53:903–10. doi: 10.1016/j.jhep.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 130.Yang M, Ma B, Shao H, Clark AM, Wells A. Macrophage phenotypic subtypes diametrically regulate epithelial-mesenchymal plasticity in breast cancer cells. BMC Cancer. 2016:16. doi: 10.1186/s12885-016-2411-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Herrmann J, Gressner AM, Weiskirchen R. Immortal hepatic stellate cell lines: useful tools to study hepatic stellate cell biology and function? Journal of Cellular and Molecular Medicine. 2007;11:704–22. doi: 10.1111/j.1582-4934.2007.00060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xu L, Hui AY, Albanis E, et al. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142–51. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Watanabe T, Shibata N, Westerman KA, et al. Establishment of immortalized human hepatic stellate scavenger cells to develop bioartificial livers. Transplantation. 2003;75:1873–80. doi: 10.1097/01.TP.0000064621.50907.A6. [DOI] [PubMed] [Google Scholar]
- 134.Vogel S, Piantedosi R, Frank J, et al. An immortalized rat liver stellate cell line (HSC-T6): a new cell model for the study of retinoid metabolism in vitro. J Lipid Res. 2000;41:882–93. [PubMed] [Google Scholar]
- 135.Zhao W, Zhang L, Yin Z, et al. Activated hepatic stellate cells promote hepatocellular carcinoma development in immunocompetent mice. Int J Cancer. 2011;129:2651–61. doi: 10.1002/ijc.25920. [DOI] [PubMed] [Google Scholar]
- 136.Dingal PC, Bradshaw AM, Cho S, et al. Fractal heterogeneity in minimal matrix models of scars modulates stiff-niche stem-cell responses via nuclear exit of a mechanorepressor. Nat Mater. 2015;14:951–60. doi: 10.1038/nmat4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Stone LC, Thorne LS, Weston CJ, Graham M, Hodges NJ. Cytoglobin expression in the hepatic stellate cell line HSC-T6 is regulated by extracellular matrix proteins dependent on FAK-signalling. Fibrogenesis Tissue Repair. 2015;8:15. doi: 10.1186/s13069-015-0032-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Caliari SR, Perepelyuk M, Soulas EM, Lee GY, Wells RG, Burdick JA. Gradually softening hydrogels for modeling hepatic stellate cell behavior during fibrosis regression. Integr Biol (Camb) 2016;8:720–8. doi: 10.1039/c6ib00027d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–5. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fujita T, Soontrapa K, Ito Y, et al. Hepatic stellate cells relay inflammation signaling from sinusoids to parenchyma in mouse models of immune-mediated hepatitis. Hepatology. 2016;63:1325–39. doi: 10.1002/hep.28112. [DOI] [PubMed] [Google Scholar]
- 141.Brodt P. Role of the Microenvironment in Liver Metastasis: From Pre- to Prometastatic Niches. Clin Cancer Res. 2016;22:5971–82. doi: 10.1158/1078-0432.CCR-16-0460. [DOI] [PubMed] [Google Scholar]
- 142.Chao YL, Shepard CR, Wells A. Breast carcinoma cells re-express E-cadherin during mesenchymal to epithelial reverting transition. Molecular Cancer. 2010;9:179. doi: 10.1186/1476-4598-9-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yates CC, Shepard CR, Stolz DB, Payne M. Co-culturing human prostate carcinoma cells with hepatocytes leads to increased expression of E-cadherin. Br J Cancer. 2007;96:1246–52. doi: 10.1038/sj.bjc.6603700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kamiya A, Kinoshita T, Ito Y, et al. Fetal liver development requires a paracrine action of oncostatin M through the gp130 signal transducer. EMBO J. 1999;18:2127–36. doi: 10.1093/emboj/18.8.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kamiya A, Kinoshita T, Miyajima A. Oncostatin M and hepatocyte growth factor induce hepatic maturation via distinct signaling pathways. FEBS Lett. 2001;492:90–4. doi: 10.1016/s0014-5793(01)02140-8. [DOI] [PubMed] [Google Scholar]
- 146.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Russell P, Kingsley E. Human Prostate Cancer Cell Lines. In: Russell P, Jackson P, Kingsley E, editors. Prostate Cancer Methods and Protocols. Totowa, NJ: Humana Press; 2003. pp. 21–39. [DOI] [PubMed] [Google Scholar]
- 148.Domcke S, Sinha R, Levine DA, Sander C, Schultz N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun. 2013;4:2126. doi: 10.1038/ncomms3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Burdall SE, Hanby AM, Lansdown MR, Speirs V. Breast cancer cell lines: friend or foe? Breast Cancer Res. 2003;5:89–95. doi: 10.1186/bcr577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bahia H, Ashman JN, Cawkwell L, et al. Karyotypic variation between independently cultured strains of the cell line MCF-7 identified by multicolour fluorescence in situ hybridization. Int J Oncol. 2002;20:489–94. doi: 10.3892/ijo.20.3.489. [DOI] [PubMed] [Google Scholar]
- 151.Watson M, Greenman J, Drew PJ, Lind M, Cawkwell L. Variation between independently cultured strains of the MDA-MB-231 breast cancer cell line identified by multicolour fluorescence in situ hybridisation. Cancer Therapy. 2004;2:167–72. [Google Scholar]
- 152.Putzke AP, Ventura AP, Bailey AM, et al. Metastatic progression of prostate cancer and e-cadherin regulation by zeb1 and SRC family kinases. Am J Pathol. 2011;179:400–10. doi: 10.1016/j.ajpath.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gillet JP, Calcagno AM, Varma S, et al. Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance. Proc Natl Acad Sci U S A. 2011;108:18708–13. doi: 10.1073/pnas.1111840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang R, Lv Q, Meng W, et al. Comparison of mammosphere formation from breast cancer cell lines and primary breast tumors. J Thorac Dis. 2014;6:829–37. doi: 10.3978/j.issn.2072-1439.2014.03.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Smiraglia DJ, Rush LJ, Fruhwald MC, et al. Excessive CpG island hypermethylation in cancer cell lines versus primary human malignancies. Hum Mol Genet. 2001;10:1413–9. doi: 10.1093/hmg/10.13.1413. [DOI] [PubMed] [Google Scholar]
- 156.Nanashima A, Yamaguchi H, Sawai T, et al. Prognostic factors in hepatic metastases of colorectal carcinoma: immunohistochemical analysis of tumor biological factors. Dig Dis Sci. 2001;46:1623–8. doi: 10.1023/a:1010680815954. [DOI] [PubMed] [Google Scholar]
- 157.Cai J, DeLaForest A, Fisher J, et al. Protocol for directed differentiation of human pluripotent stem cells toward a hepatocyte fate. Community TSCR, editor. StemBook. 2012 doi: 10.3824/stembook.1.52.1. at http://www.stembook.org. [DOI] [PubMed]
- 158.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–63. [PubMed] [Google Scholar]
- 159.Knowles BB, Howe CC, Aden DP. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980;209:497–9. doi: 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- 160.Ray RB, Meyer K, Ray R. Hepatitis C virus core protein promotes immortalization of primary human hepatocytes. Virology. 2000;271:197–204. doi: 10.1006/viro.2000.0295. [DOI] [PubMed] [Google Scholar]
- 161.Wong JC, Gao SY, Lees JG, Best MB, Wang R, Tuch BE. Definitive endoderm derived from human embryonic stem cells highly express the integrin receptors alphaV and beta5. Cell Adh Migr. 2010;4:39–45. doi: 10.4161/cam.4.1.10627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.DeLaForest A, Nagaoka M, Si-Tayeb K, et al. HNF4A is essential for specification of hepatic progenitors from human pluripotent stem cells. Development. 2011;138:4143–53. doi: 10.1242/dev.062547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Betts S, Bjorkhem-Bergman L, Rane A, Ekstrom L. Expression of CYP3A4 and CYP3A7 in Human Foetal Tissues and its Correlation with Nuclear Receptors. Basic Clin Pharmacol Toxicol. 2015;117:261–6. doi: 10.1111/bcpt.12392. [DOI] [PubMed] [Google Scholar]
- 164.Ball D, Rose E, Alpert E. Alpha-fetoprotein levels in normal adults. Am J Med Sci. 1992;303:157–9. doi: 10.1097/00000441-199203000-00004. [DOI] [PubMed] [Google Scholar]
- 165.Tilghman SM, Belayew A. Transcriptional control of the murine albumin/alpha-fetoprotein locus during development. Proc Natl Acad Sci U S A. 1982;79:5254–7. doi: 10.1073/pnas.79.17.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lu H, Rosenbaum S. Developmental pharmacokinetics in pediatric populations. J Pediatr Pharmacol Ther. 2014;19:262–76. doi: 10.5863/1551-6776-19.4.262. [DOI] [PMC free article] [PubMed] [Google Scholar]