Summary
Although histone-modifying enzymes are generally assumed to function in a manner dependent on their enzymatic activities, this assumption remains untested for many factors. Here we show the Tip60 (Kat5) lysine acetyltransferase (KAT), which is essential for embryonic stem cell (ESC) self-renewal and pre-implantation development, performs these functions independently of its KAT activity. Unlike ESCs depleted of Tip60, KAT–deficient ESCs exhibited minimal alterations in gene expression, chromatin accessibility at Tip60 binding sites, and self-renewal, thus demonstrating a critical KAT–independent role of Tip60 in ESC maintenance. In contrast, KAT–deficient ESCs exhibited impaired differentiation into mesoderm and endoderm, demonstrating a KAT–dependent function in differentiation. Consistent with this phenotype, KAT–deficient mouse embryos exhibited post-implantation developmental defects. These findings establish separable KAT–dependent and KAT–independent functions of Tip60 in ESCs and during differentiation, revealing a complex repertoire of regulatory functions for this essential chromatin remodeling complex.
Keywords: chromatin, embryonic stem cells, Tip60, Kat5, acetyltransferase, self-renewal, differentiation, development
Graphical abstract
Introduction
Embryonic stem cells (ESCs)—cells derived from the inner cell mass of the early blastocyst—have been utilized as an in vitro model of differentiation due to their pluripotency and unlimited capacity for self-renewal in culture (Keller, 2005). A complex array of signaling pathways and transcription factors control ESC fate, promoting self-renewal in the presence of either leukemia inhibitory factor (LIF) or inhibitors of differentiation-promoting kinases MEK1/2 and Gsk3β (Ying et al., 2008). In addition to transcription factors, a number of chromatin regulatory proteins help control the expression of pro-self-renewal and pro-differentiation genes (T. Chen and Dent, 2014). Although dozens of chromatin regulators necessary for ESC self-renewal or differentiation have been identified, the specific contributions of many chromatin regulatory proteins to ESC self-renewal and differentiation are poorly understood, due to the redundant and context-dependent contributions of most chromatin modifications to gene expression (Rando and Chang, 2009).
Previously we showed that RNAi-mediated knockdown (KD) of components of the well-conserved Tip60-p400 (also called NuA4) chromatin regulatory complex resulted in multiple defects in ESC pluripotency (Fazzio et al., 2008a). ESCs depleted of Tip60-p400 subunits exhibit cell and colony morphologies indicative of differentiation and reduced expression of some pluripotency markers. However, Tip60-p400-depleted cells are also defective in normal ESC differentiation, forming small, abnormal embryoid bodies under differentiation conditions that fail to upregulate some markers of differentiated cells (P. B. Chen et al., 2013; Fazzio et al., 2008a). Consistent with this self-renewal defect, homozygous knockout of the Tip60 gene in mouse results in embryonic lethality at approximately the blastocyst stage (the stage at which ESCs are derived) (Hu et al., 2009). Tip60-/- blastocysts are morphologically abnormal and fail to hatch from the zona pellucida when cultured in vitro. No post-implantation Tip60-/- embryos were observed, demonstrating an absolute requirement for Tip60 at or before this stage.
Tip60-p400 has two biochemical activities that contribute to its functions within the nucleus. The Tip60 subunit is a lysine acetyltransferase (KAT) that targets histones H4, H2A, H2A variants, and non-histone proteins (Ikura et al., 2000; Keogh et al., 2006; Squatrito et al., 2006; Xu and Price, 2011). Histone acetylation near gene promoters or enhancers is strongly associated with gene expression, consistent with Tip60's known function as a co-activator that collaborates with numerous transcription factors (Squatrito et al., 2006). In addition to its role as a co-activator, Tip60 also directly regulates the activities of numerous transcription factors through acetylation of lysine residues (Farria et al., 2015). Finally, besides regulation of transcription, Tip60 plays important roles in DNA damage repair, senescence, and apoptosis (Doyon et al., 2004; Ikura et al., 2000; Kusch et al., 2004; Xu and Price, 2011) (Jiang et al., 2011; Sykes et al., 2006; Tang et al., 2006; Van Den Broeck et al., 2012). Importantly, the KAT activity of Tip60 has been shown to be essential for its role in each of these processes.
The second chromatin remodeling activity found within Tip60-p400 complex is catalyzed by the p400 subunit (gene name: Ep400). The p400 protein, like its homologs in other eukaryotes, catalyzes ATP-dependent incorporation of histone H2A variant H2A.Z into chromatin via exchange of H2A-H2B dimers within nucleosomes for free H2A.Z-H2B dimers (Gévry et al., 2007; Mizuguchi et al., 2004). Interestingly, p400 was recently shown to incorporate histone H3 variant H3.3 into chromatin (Pradhan et al., 2016). H2A.Z and H3.3 are often enriched near gene regulatory regions, consistent with a role for p400 (like Tip60) as a co-activator of transcription (Melters et al., 2015). However, p400 also appears to repress transcription in some contexts, as well as promote DNA repair in concert with Tip60 (Gévry et al., 2007; Papamichos-Chronakis et al., 2011; Xu et al., 2012).
How does Tip60-p400 promote ESC self-renewal and pre-implantation development? Tip60-p400 binds near the promoters of both active genes and lowly expressed developmental genes in ESCs, and acetylates the promoter-proximal histones of both groups (P. B. Chen et al., 2013; 2015; Fazzio et al., 2008a; Ravens et al., 2015). Given the well-established activating roles of histone acetylation, these data imply that Tip60-p400 may drive expression of highly expressed housekeeping and pluripotency genes, but that its developmental targets are resistant to this activation, possibly due to the repressive activities of Polycomb complexes or other factors (Aloia et al., 2013; Simon and Kingston, 2013). However, this model is unlikely to be correct, since Tip60-p400 is largely dispensable for transcriptional activation in ESCs, and instead functions mainly to repress its developmental targets (P. B. Chen et al., 2013; Fazzio et al., 2008a; 2008b). Therefore, the Tip60 KAT activity must either inhibit transcription of developmental genes in ESCs, or repression of these genes by Tip60-p400 is KAT–independent.
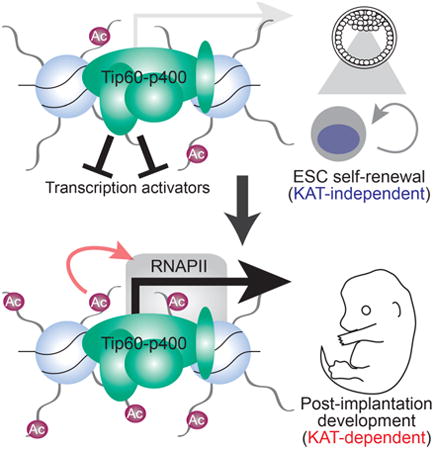
Here we show that Tip60 functions independently of its KAT activity to repress differentiation genes in ESCs and promote ESC self-renewal. Consistent with this repressive function, Tip60 limits promoter-proximal chromatin accessibility at many Tip60 target genes, and this function is similarly KAT–independent. By contrast, KAT–deficient ESCs are impaired for differentiation, revealing a critical role for the Tip60 KAT activity in pluripotency. Upon induction of differentiation, KAT mutant ESCs exhibit defects in production of mesoderm and endoderm cell types, due to reduced induction of numerous key drivers of differentiation. Unlike Tip60 null mice (Hu et al., 2009), KAT–deficient mutant mice proceed past the blastocyst stage, consistent with the ability of KAT mutant ESCs to self-renew. However, KAT mutant mice exhibit post-implantation developmental defects beginning around the start of gastrulation, analogous to the ESC differentiation defect observed in vitro. Together, these findings establish separable KAT–independent and KAT–dependent roles of Tip60 in pluripotency and embryonic development that are both essential, but which act at different stages.
Results
Tip60 KAT activity is dispensable for gene regulation and self-renewal in ESCs
Tip60 is one of several HATs that acetylate the N-terminal tails of histones H4 and H2A, whereas p400 is one of two SWI/SNF family ATPases that mediate H2A.Z deposition (Altaf et al., 2009; Lalonde et al., 2014). To test the importance of these activities in ESCs, we generated independent ESC lines with homozygous mutations encoding amino acid substitutions in the acetyl CoA binding site of Tip60 (Tip60ci/ci) or the ATP-binding pocket of p400 (Ep400ci/ci; Figure S1A-B), both of which were previously shown to block enzymatic activity (Ikura et al., 2000; Xu et al., 2010). We confirmed that these mutations broadly reduced H4 acetylation and H2A.Z deposition, respectively, in ESCs (Figure S1C-D). Since Tip60 or Ep400 depletion in ESCs causes loss of self-renewal (Fazzio et al., 2008a), we utilized previously validated shRNAs (P. B. Chen et al., 2013) to perform acute KD of Tip60 or Ep400, along with an Ep400 hypomorphic mutant (Ep400hypo) that exhibits reduced levels of p400 protein (Figure S1E), for comparison. Surprisingly, Tip60ci/ci and Ep400ci/ci lines had normal ESC morphology and maintained expression of pluripotency markers such as alkaline phosphatase (AP; Figure 1A), and SSEA-1 (Figure S1F), whereas Tip60 KD or Ep400hypo cells exhibited reduced AP and SSEA-1 staining and flattened colony morphologies, as observed previously (P. B. Chen et al., 2015; Fazzio et al., 2008a). Tip60ci/ci and Ep400ci/ci cells proliferated more rapidly than Tip60 KD and Ep400hypo cells (Figure 1B), although Tip60ci/ci cells proliferated slightly less rapidly than wild type controls. Finally, to test for functional redundancy, we constructed double homozygous mutant Tip60ci/ci Ep400ci/ci lines. As with the single mutants, these lines expressed markers of pluripotent stem cells and normal ESC colony morphology, similar to that of Tip60ci/ci single mutants (Figure S1F-G). These data suggest loss of Tip60 KAT activity and p400 ATP-dependent nucleosome remodeling activity have minimal effects on ESC maintenance.
Figure 1. Tip60 KAT and p400 ATPase activities are dispensable for ESC self-renewal and gene regulation.
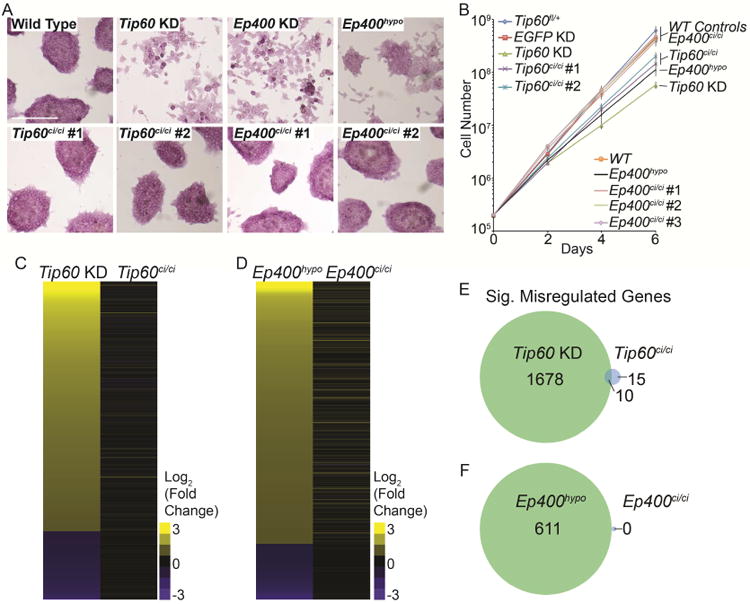
(A) Alkaline phosphatase staining (AP) of Tip60ci/ci and Ep400ci/ci mutants and controls (Tip60fl/+, Tip60 KD, Ep400 KD, and Ep400hypo). Scale bars equal 200 μm. (B) Growth curve, measuring the proliferation rates of the indicated mutant and control ESCs. (C, D) Heatmaps of differentially expressed genes in Tip60ci/ci and Tip60 KD ESCs relative to Tip60fl/+control cells (C), or Ep400ci/ci and Ep400hypo ESCs relative to wild type (E14) control ESCs (D). Genes in the heatmaps are sorted from the most upregulated to the most down regulated genes in the Tip60 KD and Ep400hypo controls, respectively. (E, F) Venn diagrams showing number of genes commonly misregulated in Tip60ci/ci and Tip60 KD ESCs (E), or Ep400ci/ci and Ep400hypo ESCs (F). Genes were considered significantly misregulated in each KD or mutant if their |log2 (fold change)| > 1 and their multiple testing-adjusted p value < 0.05.
To test whether gene expression is altered in Tip60ci/ci and Ep400ci/ci mutant ESCs, in spite of their normal self-renewal, we performed RNA-seq on biological replicates of Tip60ci/ci and Ep400ci/ci mutants, along with positive and negative controls. Consistent with previous findings (P. B. Chen et al., 2013; Fazzio et al., 2008a), Tip60 KD and Ep400hypo cells each exhibited up-regulation of numerous genes enriched for developmental factors, and down-regulation of a smaller group of genes (Figure 1C-F, Figure S2A-B). In contrast, few genes were significantly altered in Tip60ci/ci, Ep400ci/ci, or Tip60ci/ci Ep400ci/ci double mutants (Figure 1C-F, Figure S2C-F). These data demonstrate that while Tip60 and p400 are necessary for gene regulation and self-renewal in ESCs, their catalytic activities are dispensable for these processes.
KAT–independent regulation of promoter-proximal chromatin accessibility by Tip60
Since KATs function mainly as co-activators of gene expression, we next focused on how Tip60 functions independently of its KAT activity to repress transcription in ESCs. We confirmed normal expression of Tip60 and p400 in Tip60ci/ci Ep400ci/ci ESCs (Figure S3A), and found that Tip60ci/ci and Ep400ci/ci ESCs assemble intact Tip60-p400 complexes with compositions similar to that of wild type cells, in contrast to p400hypo mutant ESCs (Figure S3B).
Given its size (∼1.5 MDa; 17 core subunits), we considered the possibility binding of Tip60-p400 complex reduces the accessibility of underlying chromatin, regardless of its enzymatic functions. To test this possibility, we performed ATAC-seq (Buenrostro et al., 2013) to quantify changes in chromatin accessibility at Tip60 binding sites. In Tip60fl/+ control ESCs (expressing wild type Tip60), chromatin accessibility is higher at Tip60 binding sites than flanking regions (Figure 2A-B), consistent with the observed enrichment of Tip60 near gene regulatory elements such as promoters and enhancers (P. B. Chen et al., 2013; Fazzio et al., 2008a; Ravens et al., 2015). Interestingly, we observed significantly increased chromatin accessibility upon Tip60 KD, but minimal changes in accessibility in KAT–deficient ESCs (Figure 2A-B). Clustering of these data identified two prominent patterns of chromatin accessibility, segregated primarily by whether the Tip60–binding sites were promoter-proximal or -distal (Figure 2C). Examination of promoter-proximal regions of Tip60 target genes revealed that Tip60 KD increased chromatin accessibility within a several hundred base pair window extending from the promoter into the gene body, corresponding to Tip60-p400 binding sites on chromatin (Figure 2D) (P. B. Chen et al., 2015; 2013; Ravens et al., 2015). In contrast, KAT-deficient ESCs were minimally affected. Unlike promoter-proximal regions, chromatin accessibility at gene-distal Tip60–binding sites was relatively unaltered by Tip60 KD or KAT mutation (Figure 2E). Consistent with these findings, KAT–deficient Tip60 bound to Tip60-p400–target genes at levels similar to wild type (Figure S3C). These data demonstrate Tip60 functions independently of its KAT activity to regulate promoter-proximal chromatin accessibility in ESCs.
Figure 2. KAT-independent regulation of chromatin accessibility at Tip60 target loci.
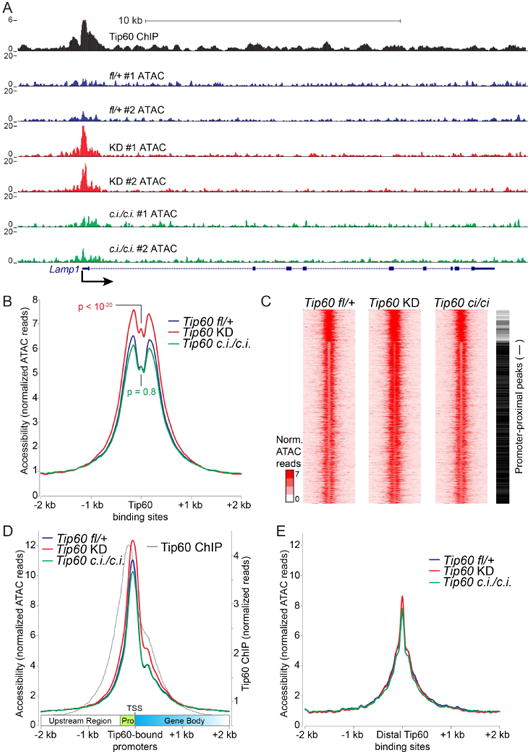
(A) Example Tip60 target gene showing increased promoter-proximal chromatin accessibility in Tip60 KD but not Tip60ci/ci relative to Tip60fl/+ control cells. Shown are normalized ATAC-seq reads ≤ 100bp for each biological replicate, and Tip60 ChIP-seq data from (Ravens et al., 2015). (B) Aggregation plot showing average ATAC-seq signal for two biological replicates of each mutant or KD aggregated over high-quality Tip60 binding sites. A Kolmogorov–Smirnov test of differences in ATAC profiles was used to calculate p values. (C) K-means clustering (K=3) for ATAC-seq data over Tip60 binding sites. Promoter-proximal peaks are marked with a black bar to the right, promoter-distal peaks with a white bar. (D) Aggregation plot of ATAC-seq data (as in B) over Tip60–bound promoter regions aligned such that all gene bodies are to the right. Promoter-proximal regions (pro) and transcription start sites (TSS) are indicated. Tip60 ChIP-seq data (Ravens et al., 2015) are shown for reference. (E) Aggregation plot over Tip60–bound gene-distal regions.
Tip60 KAT activity is necessary for differentiation and post-implantation development
Consistent with the self-renewal defect of Tip60 KD ESCs (Fazzio et al., 2008a), Tip60 homozygous null (Tip60-/-) mice die at the peri-implantation stage: Tip60-/- blastocysts fail to hatch and survive in culture, and no post-implantation Tip60-/- embryos can be recovered (Hu et al., 2009). Since Tip60ci/ci ESCs self-renew normally, we next tested whether the Tip60 KAT activity is also dispensable for mouse development. To this end, we generated and intercrossed Tip60ci/+ heterozygous mice to produce Tip60ci/ci homozygotes (see Experimental Procedures for details). However, we recovered no Tip60ci/ci pups at birth (χ2= 40.45; P < 0.001), suggesting the Tip60 KAT activity is essential for development (Figure 3A). To elucidate the developmental defect of Tip60ci/ci animals, we examined the morphology of embryos at multiple stages. Tip60ci/ci embryos were recovered as late as 10.5 days post fertilization (E10.5; Figure 3A), but were much smaller than Tip60+/+ or Tip60ci/+ littermates (Figure 3B), and exhibited morphological abnormalities as early as E6.5 (Figure S4A-C). The contrasting phenotypes between Tip60-/- and Tip60ci/ci mice reveal an essential KAT–independent role for Tip60 in pre-implantation development, as well as an essential KAT–dependent role in early post-implantation development.
Figure 3. The Tip60 catalytic activity is required for differentiation and post-implantation development.
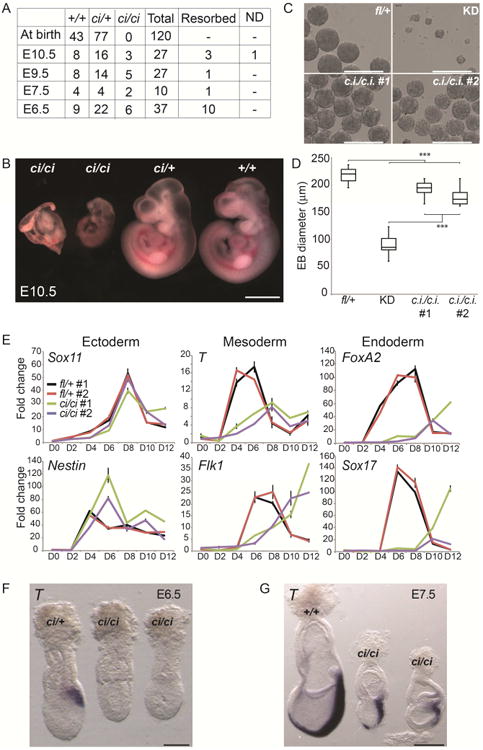
(A) Genotypes of embryos from Tip60ci/+ intercrosses at different developmental stages. (B) Images of E10.5 embryos of the indicated genotypes. Scale bar equals 1 mm. (C) Embryoid body (EB) formation assay comparing EB morphology in Tip60ci/ci mutant ESCs to Tip60fl/+ and Tip60 KD controls. Scale bars equal 400 μm. (D) Quantification of EB size in indicated mutants and controls (n = 49 per genotype). Boxes range from the 25th to the 75th percentile, the dark lines indicate the median, and the whiskers indicate the lesser of either the extreme (max or min) value or 1.5 times the interquartile range (***p < 0.001, calculated using a two-sided t-test). (E) RT-qPCR analysis of indicated germ layer markers during a time course of EB differentiation. (F, G) Whole mount in situ hybridization in E6.5 and E7.5 mouse embryos staining for T transcript. Scale bars equal 100 μm (F) or 250 μm (G).
The phenotypes of Tip60ci/ci embryos are evident at or just before gastrulation, where the three primary germ layers are established, suggesting that although Tip60ci/ci ESCs self-renew normally, they may not differentiate properly. We tested this possibility using embryoid body differentiations of control, Tip60 KD, and Tip60ci/ci ESCs. Previously, we showed that KD of Tip60, Ep400, or (Tip60-p400 subunit) Dmap1 resulted in defects in EB formation (P. B. Chen et al., 2013; Fazzio et al., 2008a), suggesting Tip60-p400 is required for this initial step of differentiation. In contrast, Tip60ci/ci ESCs efficiently formed EBs, which expanded in culture at near wild type levels, although modest differences in EB morphology were observed relative to Tip60fl/+cells (Figure 3C-D). However, induction of mesodermal and endodermal markers was delayed and/or reduced in Tip60ci/ci EBs (Figure 3E) compared to Tip60fl/+ controls. These data suggest that the Tip60 KAT activity is important for specification of mesodermal and endodermal cell types in vitro.
To test whether the ESC differentiation defects were recapitulated in vivo, we stained post-implantation Tip60ci/ci embryos for T (also known as Brachyury), a marker of cells migrating through the primitive streak to become mesodermal or endodermal cell types (Herrmann, 1991; Rivera-Pérez and Magnuson, 2005). Although T staining of Tip60+/+ and Tip60ci/+ embryos was evident at E6.5 and strong at E7.5, Tip60ci/ci embryos exhibited reduced staining at both stages (Figure 3F-G). These data show that gastrulation is delayed or impaired in Tip60ci/ci embryos. This phenotype could result from impaired lineage commitment, poor migration of cells through the primitive streak, or other factors. Regardless, this developmental defect is consistent with the impaired induction of early mesodermal and endodermal markers observed for KAT-defective ESCs in vitro.
Impaired expression of multiple drivers of differentiation in KAT–deficient ESCs
What is the molecular basis for the in vivo and in vitro developmental defects of Tip60ci/ci mutants? These phenotypes could result from failure to upregulate key lineage-specific transcription factors and/or a disruption in signaling pathways that promote lineage commitment. To address these possibilities, we compared the changes in gene expression during a time course of differentiation of control (Tip60fl/+) and Tip60ci/ci ESCs using RNA-seq on biological replicate samples. We observed differences in both the timing and levels of markers of mesoderm and endoderm (Figure 4A; e.g. FoxA2, Gata4, Sox17, T, Hand1, Flk1), expanding on our preliminary analyses (Figure 3E). Next we used k-means clustering to identify groups of genes induced early or late during differentiation in Tip60fl/+ control cells and characterized the effects of the KAT mutation on their induction. We observed 1,338 genes of this type that mainly fall into three clusters based on the timing of their expression peak (Figure 4B). In Tip60ci/ci cells, we observed reduced or delayed induction of numerous genes with key roles in differentiation, including developmental transcription factors and mediators of growth factor signaling, within each of the three clusters (Figure 4B).
Figure 4. Delayed/impaired expression of developmental regulators in differentiating Tip60ci/ci ESCs.
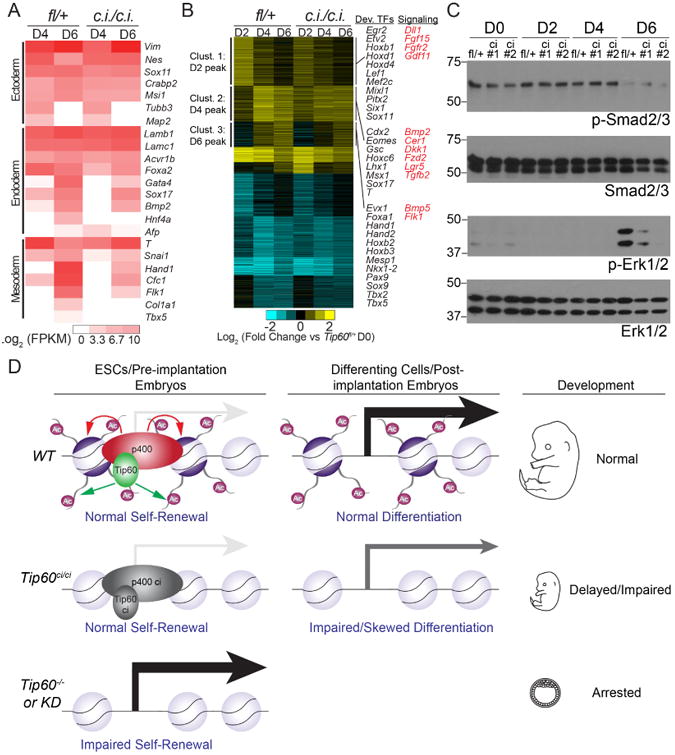
(A) Heatmap indicating induction kinetics of each germ layer markers during differentiation of Tip60fl/+ controls or Tip60ci/ci mutant ESCs. (B) K-means clustering (K = 9) of differentially expressed genes [|log2 (fold change)| > 0.7; multiple testing-adjusted p value < 0.05] in Tip60fl/+ controls or Tip60ci/ci mutant ESCs during the differentiation time course. Large up-regulated clusters are noted. Key regulatory proteins with impaired induction in Tip60ci/ci mutant ESCs are highlighted. (C) Western blots (one of two independent experiments with similar results) of phosphorylated and total Smad2/3 and Erk1/2 during differentiation in Tip60fl/+ or Tip60ci/ci ESCs. (D) Model indicating the KAT-independent role of Tip60 in ESC self-renewal and gene regulation, as well as pre-implantation development, and the KAT-dependent role of Tip60 in differentiation and post-implantation development. See text for additional details.
To test whether impaired induction of key signaling proteins hindered activation of their downstream targets, we examined activation of the FGF/MEK/ERK and TGF-β pathways using antibodies recognizing the phosphorylated (and activated) forms of ERK1/2 and Smad2/3, respectively (Tsang and Dawid, 2004; Whitman and Mercola, 2001). These factors act downstream of FGF and BMP signaling in differentiating ESCs and embryos, and are critical for differentiation (Sui et al., 2013). Although Smad2/3 phosphorylation was unaltered in differentiating Tip60ci/ci ESCs, we observed impaired ERK phosphorylation in these mutants after six days of differentiation (Figure 4C). Together, these data suggest that the differentiation defect observed in Tip60ci/ci ESCs is due to at least two overlapping defects: delayed or reduced activation of ERK, and impaired induction of key developmental transcription factors.
Discussion
Here we showed that Tip60 functions in ESC gene regulation and self-renewal, as well as pre-implantation development, independently of its KAT activity. This finding was unexpected because Tip60 depletion or knockout leads to a self-renewal defect in ESCs and pre-implantation lethality in mice (Fazzio et al., 2008a; Hu et al., 2009). Furthermore, KAT–impaired mutants of esa1, the yeast homolog of Tip60, are severely growth impaired (Selleck et al., 2005), suggesting the critical cellular functions of this KAT are dependent on its acetylation activity.
The fact that Tip60 is largely a repressor of transcription in ESCs (Fazzio et al., 2008a), and this repressive function is independent of its KAT activity, suggests that Tip60 regulates ESC gene expression in a manner that is distinct from other well-studied KATs, at least in part. Consistent with its role as a broadly acting repressor of transcription in ESCs, we found Tip60 functions by a KAT–independent mechanism to limit chromatin accessibility directly over its promoter-proximal binding sites at many target genes. Additional studies will be necessary to determine whether Tip60 also performs this function in somatic cell types.
In contrast, the Tip60 KAT activity is essential during ESC differentiation and post-implantation development. Consequently, these findings demonstrate separable, essential functions of Tip60: its KAT-independent function is sufficient for Tip60's essential role in ESC self-renewal and pre-implantation development, and its KAT–dependent function is required for post-implantation development and ESC differentiation. Interestingly, we found that the ATP-dependent histone exchange activity of p400 was also dispensable for gene regulation and self-renewal in ESCs, revealing that Tip60-p400 complex represses differentiation genes in ESCs independently of its known chromatin remodeling activities (Figure 4D). These findings necessitate a re-evaluation of current models of gene regulation by this essential chromatin regulatory complex.
What is the role of the Tip60 KAT activity during development? Given the defect of KAT-deficient ESCs and embryos in lineage specification, one possibility is that histone acetylation at differentiation genes in ESCs (as observed previously (Fazzio et al., 2008a)) facilitates their up-regulation when differentiation is induced. This provides a potential explanation for the counterintuitive role of Tip60 in repression of differentiation genes in ESCs–occupancy of Tip60-p400 at differentiation gene promoters helps repress these genes by reducing chromatin accessibility, while acetylation at these loci may allow more rapid induction after binding of differentiation-specific transcription factors. Together, these data show that not all functions of Tip60 are reliant on its KAT activity, and raise the possibility that KAT–independent gene regulation by Tip60 plays important roles in additional cell types.
Experimental Procedures
Antibodies
Antibodies used in this study: p400 (A300-541A; Bethyl), StainAlive™ SSEA-1 (09-0067; Stemgent); Smad2/3 (8685; Cell Signaling Technologies); Phospho-Smad2/3 (8828; Cell Signaling Technologies); Erk1/2 (9102; Cell Signaling Technologies); Phospho-Erk1/2 (9101; Cell Signaling Technologies); H2AZ (ab4174, Abcam); Acetyl-H4 (06-598; Millipore); FLAG-M2 (F1804; Sigma); IgG (ab37415; Abcam); β-actin (A5316; Sigma).
Cell Lines
Mouse ESC lines were derived from E14 (129/Ola) (Hooper et al., 1987) and grown as described (P. B. Chen et al., 2013). Tip60ci/ci ESCs were derived from floxed Tip60-H3F cells (P. B. Chen et al., 2013), by introduction of Cre recombinase (Addgene, 20781) to loop out wild type Tip60 regions upstream of exon 11 that harbors two substitution mutations (Q377E and G380E) that eliminate acetyl CoA binding (Ikura et al., 2000) (figure S1A).
Catalytically inactive mutants of p400 (Ep400ci/ci) were generated using homologous recombination stimulated by CRISPR/Cas9-mediated cleavage (Cong et al., 2013; Mali et al., 2013). A repair template (Table S3) was synthesized (Integrated DNA Technologies), cloned into pCR2.1, and introduced together with the CRISPR/Cas9 vector (a variant of plasmid pX330 that expresses puromycin resistance). The Ep400hypo mutant line, described previously (P. B. Chen et al., 2015), was generated using the same CRISPR/Cas9 construct, but without the repair template, resulting in a homozygous 135bp in-frame deletion that disrupts the ATPase domain and results in lower expression of p400 protein (Figure S1E).
ESC differentiation
Embryoid bodies (EBs) for growth/morphology assays were formed using hanging drops containing 100 cells in 10 ul of differentiation medium. Morphology was examined after 48 hours. For gene expression assays, 106 ESCs were plated on non-adherent plates for 48 hours to form EBs, and then transferred into gelatinized 6-well plates at a low density. Cells were harvested using TRIzol reagent (Invitrogen) at indicated time points. RNA was prepared and RT-qPCR was performed as described (P. B. Chen et al., 2013), using primers listed in Table S1.
Cell Staining
105 ESCs were grown on gelatin-coated 6-well plates for 48 hours. Alkaline phosphatase (AP) staining was performed using a kit (EMD Millipore, SCR004), following the manufacturers' guidelines. SSEA-1 staining of live ESCs was also performed per the manufacturers' instructions (Stemgent, 09-0067).
Tip60-p400 Purification
Tip60-p400 complex was purified from nuclear extracts of WT, Tip60ci/ci, p400ci/ci, and p400hypo cells with endogenous 6Xhis/3XFLAG tags at the Tip60 locus, as described previously (P. B. Chen et al., 2013). Purified proteins were separated on SDS-PAGE gels, and Silver Staining was performed using a Silver Staining Kit (ThermoFisher, LC6100).
Western Blotting
30ug of nuclear extract per lane (prepared using the NE-PER kit; ThermoFisher, 78833) were used for Western blotting.
Generation of Tip60ci/ci mice
Tip60ci/+ heterozygous mice were generated by crossing Tip60 floxed mice (P. B. Chen et al., 2013) with the allele described above with Tg(EIIa-cre) mice, which broadly express Cre recombinase (Dooley et al., 1989; Lakso et al., 1996). Mice were genotyped by PCR with primers listed in Table S2. Tip60ci/+ mice were maintained as heterozygotes on an inbred FVB/N background and intercrossed to generate Tip60+/+, Tip60ci/+, and Tip60ci/ci embryos. Animal studies were performed in accordance with guidelines of the Institutional Animal Care and Use Committee at the University of Massachusetts Medical School (A-2165) and NIH.
RNA in situ hybridization
Whole mount in situ hybridization was performed as previously described (Rivera-Pérez and Magnuson, 2005), using a full-length cDNA probe of T (Herrmann, 1991). Embryos were genotyped after staining by PCR, using primers listed in Table S2.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation and deep sequencing were performed as described previously (P. B. Chen et al., 2013; Hainer et al., 2015). ChIP-qPCR was performed using SYBR FAST (KAPA Biosystems) with primers described previously (Fazzio et al., 2008a).
RNA-seq
Strand specific library construction and RNA-seq were performed by Applied Biological Materials, Inc. and the UCLA Clinical Microarray Core for ESCs and differentiating ESCs, respectively. Data analysis is described in Supplemental Experimental Procedures.
ATAC-seq
ATAC-seq was performed essentially as described (Buenrostro et al., 2013; 2015). Two independent ATAC reactions per biological replicate were performed, using 35,000 and 70,000 ESCs each. After library preparation, the two reactions were found to have indistinguishable distributions of fragment sizes, and were therefore combined for sequencing. (Therefore, each biological replicate consisted of two ATAC reactions.) Data analysis is described in Supplemental Experimental Procedures.
Statistical Methods
For non-genomic in vitro experiments, two tailed t-tests were used to calculate statistical significance. A chi-square test was used to evaluate genotypes of offspring from Tip60ci/+ intercrosses. Adjusted p-values were calculated for RNA-seq data using DEseq2. Significance of differences in ATAC-seq read enrichment were calculated by a hypergeometric test using the dhyper package in R.
Supplementary Material
Acknowledgments
We thank J. Benanti and T. Tsukiyama for critical reading of the manuscript. This work was supported by US National Institutes of Health (NIH) grants R01HD072122 (T.G.F.), R01HD083311 (J.A.R.-P.), and R01CA131158 (I.B.). T.G.F. is supported as a Leukemia and Lymphoma Society Scholar. S.J.H. is a Special Fellow of the Leukemia and Lymphoma Society.
Footnotes
Accession numbers: Deep sequencing data are available at Gene Expression Omnibus (accession: GSE85505).
Author Contributions: D.A., J.A.R.-P, and T.G.F. designed experiments. D.A. performed most experiments with help from Y.Y. and J.A.R.-P (early embryo dissection and staining), F.W. and I.B. (late embryo dissection), and T.F. (ATAC-seq). S.J.H. and D.A. analyzed the deep sequencing data.
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aloia L, Di Stefano B, Di Croce L. Polycomb complexes in stem cells and embryonic development. Development. 2013;140:2525–2534. doi: 10.1242/dev.091553. [DOI] [PubMed] [Google Scholar]
- Altaf M, Auger A, Covic M, Côté J. Connection between histone H2A variants and chromatin remodeling complexes. Biochem Cell Biol. 2009;87:35–50. doi: 10.1139/o08-140. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Meth. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol. 2015;109:21.29.1–9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PB, Chen HV, Acharya D, Rando OJ, Fazzio TG. R loops regulate promoter-proximal chromatin architecture and cellular differentiation. Nature Structural & Molecular Biology. 2015;22:999–1007. doi: 10.1038/nsmb.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PB, Hung JH, Hickman TL, Coles AH, Carey JF, Weng Z, Chu F, Fazzio TG. Hdac6 regulates Tip60-p400 function in stem cells. Elife. 2013;2:e01557. doi: 10.7554/eLife.01557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Dent SYR. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet. 2014;15:93–106. doi: 10.1038/nrg3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley TP, Miranda M, Jones NC, DePamphilis ML. Transactivation of the adenovirus EIIa promoter in the absence of adenovirus E1A protein is restricted to mouse oocytes and preimplantation embryos. Development. 1989;107:945–956. doi: 10.1242/dev.107.4.945. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Selleck W, Lane WS, Tan S, Côté J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farria A, Li W, Dent SYR. KATs in cancer: functions and therapies. Oncogene. 2015;34:4901–4913. doi: 10.1038/onc.2014.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008a;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. Chromatin regulation Tip(60)s the balance in embryonic stem cell self-renewal. Cell Cycle. 2008b;7:3302–3306. doi: 10.4161/cc.7.21.6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gévry N, Chan HM, Laflamme L, Livingston DM, Gaudreau L. p21 transcription is regulated by differential localization of histone H2A.Z. Genes and Development. 2007;21:1869–1881. doi: 10.1101/gad.1545707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainer SJ, Gu W, Carone BR, Landry BD, Rando OJ, Mello CC, Fazzio TG. Suppression of pervasive noncoding transcription in embryonic stem cells by esBAF. Genes & Development. 2015;29:362–378. doi: 10.1101/gad.253534.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann BG. Expression pattern of the Brachyury gene in whole-mount TWis/TWis mutant embryos. Development. 1991;113:913–917. doi: 10.1242/dev.113.3.913. [DOI] [PubMed] [Google Scholar]
- Hooper M, Hardy K, Handyside A, Hunter S, Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- Hu Y, Fisher JB, Koprowski S, McAllister D, Kim MS, Lough J. Homozygous disruption of the Tip60 gene causes early embryonic lethality. Dev Dyn. 2009;238:2912–2921. doi: 10.1002/dvdy.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Kamath R, Jin S, Balasubramani M, Pandita TK, Rajasekaran B. Tip60-mediated acetylation activates transcription independent apoptotic activity of Abl. Mol Cancer. 2011;10:88. doi: 10.1186/1476-4598-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes & Development. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, Buratowski S. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes and Development. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde ME, Cheng X, Côté J. Histone target selection within chromatin: an exemplary case of teamwork. Genes and Development. 2014;28:1029–1041. doi: 10.1101/gad.236331.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melters D, Nye J, Zhao H, Dalal Y. Chromatin Dynamics in Vivo: A Game of Musical Chairs. Genes (Basel) 2015;6:751–776. doi: 10.3390/genes6030751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011;144:200–213. doi: 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan SK, Su T, Yen L, Jacquet K, Huang C, Côté J, Kurdistani SK, Carey MF. EP400 Deposits H3.3 into Promoters and Enhancers during Gene Activation. Mol Cell. 2016;61:27–38. doi: 10.1016/j.molcel.2015.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ, Chang HY. Genome-wide views of chromatin structure. Annu Rev Biochem. 2009;78:245–271. doi: 10.1146/annurev.biochem.78.071107.134639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravens S, Yu C, Ye T, Stierle M, Tora L. Tip60 complex binds to active Pol II promoters and a subset of enhancers and co-regulates the c-Myc network in mouse embryonic stem cells. Epigenetics & Chromatin. 2015;8:45. doi: 10.1186/s13072-015-0039-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Pérez JA, Magnuson T. Primitive streak formation in mice is preceded by localized activation of Brachyury and Wnt3. Dev Biol. 2005;288:363–371. doi: 10.1016/j.ydbio.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Selleck W, Fortin I, Sermwittayawong D, Côté J, Tan S. The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the Enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes. Mol Cell Biol. 2005;25:5535–5542. doi: 10.1128/MCB.25.13.5535-5542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell. 2013;49:808–824. doi: 10.1016/j.molcel.2013.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends in Cell Biology. 2006;16:433–442. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Sui L, Bouwens L, Mfopou JK. Signaling pathways during maintenance and definitive endoderm differentiation of embryonic stem cells. Int J Dev Biol. 2013;57:1–12. doi: 10.1387/ijdb.120115ls. [DOI] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Tsang M, Dawid IB. Promotion and attenuation of FGF signaling through the Ras-MAPK pathway. Sci STKE. 2004;2004:pe17–pe17. doi: 10.1126/stke.2282004pe17. [DOI] [PubMed] [Google Scholar]
- Van Den Broeck A, Nissou D, Brambilla E, Eymin B, Gazzeri S. Activation of a Tip60/E2F1/ERCC1 network in human lung adenocarcinoma cells exposed to cisplatin. Carcinogenesis. 2012;33:320–325. doi: 10.1093/carcin/bgr292. [DOI] [PubMed] [Google Scholar]
- Whitman M, Mercola M. TGF-beta superfamily signaling and left-right asymmetry. Sci STKE. 2001;2001:re1–re1. doi: 10.1126/stke.2001.64.re1. [DOI] [PubMed] [Google Scholar]
- Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol Cell. 2012;48:723–733. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Price BD. Chromatin dynamics and the repair of DNA double strand breaks. Cell Cycle. 2011;10:261–267. doi: 10.4161/cc.10.2.14543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Sun Y, Jiang X, Ayrapetov MK, Moskwa P, Yang S, Weinstock DM, Price BD. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J Cell Biol. 2010;191:31–43. doi: 10.1083/jcb.201001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


